Abstract
Background
Schwann cells, which arise from the neural crest, are the myelinating glia of the peripheral nervous system. During development neural crest and their Schwann cell derivatives engage in a sequence of events that comprise delamination from the neuroepithelium, directed migration, axon ensheathment and myelin membrane synthesis. At each step neural crest and Schwann cells are polarized, implying important roles for molecules that create cellular asymmetries. In this work we investigated the possibility that one polarity protein, Pard3, contributes to the polarized features of neural crest and Schwann cells that are associated with directed migration and myelination.
Results
We analyzed mutant zebrafish embryos deficient for maternal and zygotic pard3 function. Time-lapse imaging revealed that neural crest delamination was normal but that migrating cells were disorganized with substantial amounts of overlapping membrane. Nevertheless, neural crest cells migrated to appropriate peripheral targets. Schwann cells wrapped motor axons and, although myelin gene expression was delayed, myelination proceeded to completion.
Conclusions
Pard3 mediates contact inhibition between neural crest cells and promotes timely myelin gene expression but is not essential for neural crest migration or myelination.
Keywords: Pard3, Par proteins, cell polarity, neural crest migration, Schwann cell, myelination, zebrafish
INTRODUCTION
Establishing a functional peripheral nervous system requires the coordinated development of axons and myelin-forming Schwann cells. During embryonic development Schwann cells undergo dynamic changes in both tissue location and cellular behavior to associate along and myelinate peripheral axons. Schwann cell precursors arise from delaminating neural crest cells along the dorsal neural tube. During delamination neural crest cells alter their cellular morphology by losing their apical membrane adhesion and detaching from the dorsal midline, establishing a rounded morphology along the basal side of the neuroepithelium (Ahlstrom and Erickson, 2009; Clay and Halloran, 2013). Following delamination Schwann cell precursors undergo extensive migration to associate along target axons in the peripheral tissue (Bronner-Fraser and LeDouarin, 2012). Association along target axons signals Schwann cell precursors to stop their migration and promotes differentiation into an immature Schwann cell state (Birchmeier, 2009; Woodhoo and Sommer, 2008; Woodhoo et al., 2009). Immature Schwann cells initiate the process of radial sorting, wherein Schwann cells determine whether to wrap multiple small caliber axons as Remak bundles or transition into a mature myelinating Schwann cell, which forms a single compact myelin sheath on large caliber axons (Jessen and Mirsky, 2005). The dynamic changes in cell behavior that occur during Schwann cell development have been extensively studied, but the events that govern the transitions between these behaviors remain poorly understood.
The transitions in neural crest and Schwann cell behavior leading to peripheral nerve myelination are associated with changes in cell polarity (Etienne-Manneville, 2008; Özçelik et al., 2010). Consistent with this, proteins required for regulating cell polarity have been implicated in neural crest and Schwann cell development. One example is Partitioning-Defective 3 (Pard3), a member of the evolutionary conserved partitioning-defective (PAR) multiprotein complex required for many polarized cellular processes (Özçelik et al., 2010; Pegtel et al., 2007) reviewed in (Chen and Zhang, 2013; Goldstein and Macara, 2007; Nance and Zallen, 2011; Thompson, 2013). Prior to neural crest delamination, Pard3 was localized to cell-cell adhesion complexes within the apical domain of neuroepithelial cells (Clay and Halloran, 2013; Takekuni et al., 2003). Subsequently, during neural crest cell migration, Pard3 was concentrated at the membrane leading edge where it mediated contact inhibition between neural crest cells, which might contribute to their directed migration (Moore et al., 2013). In cell culture, Pard3 localized to the Schwann cell-axon interface and Pard3 knockdown inhibited myelination (Chan et al., 2006; Lewallen et al., 2011; Özçelik et al., 2010). However, previous functional tests of Pard3 in neural crest and Schwann cell development have been limited to knock down approaches and no studies have investigated Pard3 function from neural crest delamination to myelination in vivo. Consequently, the degree to which Pard3-mediated cell polarity contributes to the development of neural crest cells and their Schwann cell descendants remains unclear.
We hypothesized that Pard3 provides polarity information for Schwann cells that facilitates their migration, axon interaction and differentiation as myelinating cells. To test this hypothesis we used a loss of gene function approach in combination with time-lapse imaging of transgenic zebrafish reporters that mark the Schwann cell lineage. Here we provide evidence that Pard3 mediates contact inhibition between neighboring Schwann cells, but that this function is not necessary for Schwann cell migration to peripheral targets. Additionally, we show that loss of Pard3 function delays but does not prevent myelin gene expression and myelination. Our data are most compatible with the possibility that Pard3 coordinates other molecular mechanisms that drive Schwann cell development to ensure timely myelination.
RESULTS
Neural Crest Delamination From the Neuroepithelium Proceeds Normally in the Absence of pard3 Function
To investigate the role of Pard3 in regulating Schwann cell behavioral transitions we utilized pard3fh305 mutant zebrafish, which have a chemically induced point mutation that changes a tyrosine at amino acid position 203 to a stop codon. This mutation is predicted to truncate the protein after the conserved oligomerization domain and before the PDZ domains (Fig. 1A), which bind cytoskeletal regulator proteins, adhesion complex proteins, and Protein Kinase C, iota (Prkci) (Wei et al., 2004). Three cDNA variants of the zebrafish pard3 locus have been described and are predicted to encode distinct protein isoforms (Fig. 1A) (Geldmacher-Voss, 2003; Trotha et al., 2006; Wei et al., 2004). The premature stop codon introduced by the pard3fh305 allele truncates all three predicted isoforms. At 5 days post fertilization (dpf) homozygous pard3fh305 mutant larvae produced by matings of heterozygous parents (Z pard3fh305) had a subtle upward body curvature (Fig. 1B) and most mutant animals survived to adulthood. Genotyping confirmed that larvae with curved bodies were homozygous for the pard3fh305 allele (Fig. 1C).
Fig. 1.
Characterization of maternal and zygotic pard3 functions. A: Schematic representation of zebrafish Pard3 isoforms. Each isoform has a conserved oligomerzation domain (CR), three PDZ domains (PDZ1-3) and a Prkci binding domain (PBD). The pard3fh305 lesion, changing a tyrosine to a stop codon at amino acid position 203 (Y203*) occurs after the oligomerization domain and before the PDZ binding domains (red line). B: Images of 5 dpf wild-type, Zpard3fh305, pard3fh305/+, and MZpard3fh305 larvae. MZpard3fh305 larvae fail to form full swim bladders (arrow) and have a more extreme body curvature than Zpard3fh305 mutants. C: Genotyping test for the pard3fh305 allele. Heterozygotes generate bands of 155 base pairs (wild-type allele) and 68 and 87 base pairs, which appear as one band on the gel (mutant allele). Homozygous mutants, selected on the basis of body curvature phenotype, produce only the 68 and 87 base pair fragments. D: Representative images of 5 dpf MZpard3fh305 larvae either without (“negative”, top row) or with (“positive”, bottom row) the Tg(hsp70I:pard3-GFP) transgene. Larvae in left column are control, non-heat shocked and those in right column were heat shocked. Pard3-GFP expression rescued the body curvature (arrowheads) and swim bladder (arrows) phenotypes. E: Graph showing quantification of heat shock rescue experiment. Larvae were produced by crossing MZpard3fh305 females to pard3fh305/+; Tg(hsp70I:pard3-GFP) males. Non-heat shocked control and heat shocked groups therefore consist of approximately 50% MZpard3fh305 and 50% pard3fh305/+ larvae. Larvae were scored at 5 dpf for severity of body deformation and swim bladder formation. Control, n=211; heat shock, n=182.
To investigate development in the absence of maternal contribution of pard3, we raised homozygous mutant animals and crossed them to produce embryos lacking both maternal and zygotic pard3 function (MZpard3fh305). MZpard3fh305 larvae at 5 dpf had shortened bodies and more pronounced body curvature when compared with wild-type and Zpard3fh305 larvae (Fig. 1B). Furthermore, MZpard3fh305 larvae failed to develop full swim bladders, and only approximately 10% survived past 12 dpf. Embryos and larvae produced by MZpard3fh305 females and receiving one wild-type pard3 allele from either wild-type or heterozygous pard3fh305 males (pard3fh305/+) appeared phenotypically normal (Fig. 1B). To confirm that loss of pard3 function is responsible for the morphological defects of mutant larvae, we introduced the transgene Tg(hsp70I:pard3-GFP) (Hudish et al., 2013), which expresses Pard3 fused to GFP (Trotha et al., 2006) under control of heat-responsive regulatory elements (Shoji et al., 1998). Repeated induction of Pard3-GFP expression using elevated temperature during the first three days of development suppressed the body curvature defects and partially rescued swim bladder formation (Fig. 1D,E). Together these observations indicate that Pard3 is required for viability but that embryonic development can proceed with only maternally contributed Pard3.
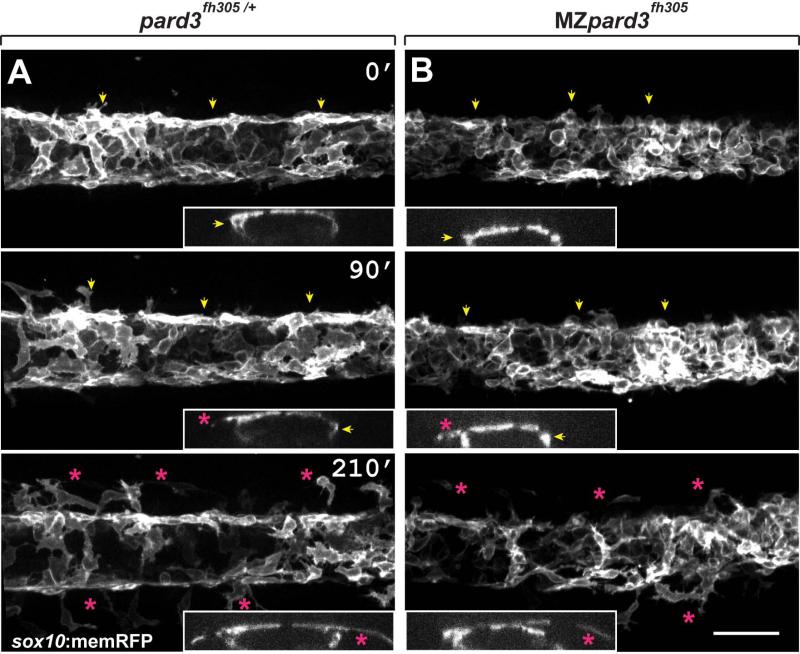
Schwann cells are specified from neural crest cells, which arise by delamination of neuroepithelial cells from dorsal neural tube. Delamination can occur via several processes including asymmetric division, force generation, and down-regulation of cellular adhesion complexes (Ahlstrom and Erickson, 2009; Berndt et al., 2008; Clay and Halloran, 2010; Theveneau and Mayor, 2012). Pard3 mediates the formation and maintenance of apical cellular adhesion complexes within mouse and chick neuroepithelial cells (Afonso and Henrique, 2006; Takekuni et al., 2003) and, in zebrafish, Pard3 localizes along the apical domain of pre-migratory neuroepithelial cells (Clay and Halloran, 2013). Therefore, we hypothesized that Pard3 mediates the timing of trunk neural crest cell delamination. If so, absence of Pard3 might result in premature neural crest cell delamination and migration. To test this hypothesis, we introduced the transgene Tg(sox10:memRFP) (Kucenas et al., 2008), which marks neural crest cells with membrane tethered RFP, into MZpard3fh305 embryos and used time-lapse microscopy to analyze neural crest exit from the dorsal neural tube. As previously described (Raible and Eisen, 1996; Raible et al., 1992), beginning at 18 hours post fertilization (hpf) cells exited from the dorsal neural tube in control embryos (n=7 embryos) and migrated away from the dorsal midline in an anterior-to-posterior progression (Fig. 2A; Supplementary Movie S1). The timing of neural crest cell delamination and initiation of migration was similar in MZpard3fh305 embryos (n=9 embryos) to that of control embryos (Fig. 2B). Additionally, wild-type and MZpard3fh305 embryos formed similar numbers of sox10:mRFP+ trunk neural crest cells (wild-type, 14.53 ± 0.29 cells/80 μM; mutant, 14.13 ± 0.64 cells/80 μM; n=15 embryos for each genotype; p = 0.5). Consistent with this, we found no difference in cell death between cells/ dorsal controls and mutants using anti-Caspase 3 labeling (control, 2.53 ± 0.37 region, n=15 embryos; mutant, 2.15 0.32 cells/ dorsal region, n=20 embryos; p = 0.56). We conclude that Pard3 function within neuroepithelial cells is not required to regulate neural crest cell formation, survival and delamination from the neural tube.
Fig. 2.
Schwann cells exit the dorsal neural tube on schedule in pard3 mutants. Panels show representative frames from time-lapse movies of trunk neural crest cells between 18-24 hpf. Views are from dorsal with anterior to the left. Insets display z-plane orthogonal views of the migrating neural crest for each frame. Numbers denote time elapsed, in minutes, from start of imaging. Embryos are siblings and imaged sequentially in the same chamber. A: Initiation of neural crest migration in a control pard3fh305/+ embryo. Neural crest cells are marked by sox10:memRFP expression. Arrows mark migration on the medial pathway between neural tube and somites and asterisks mark migration on the lateral pathway, across the dorsolateral surface of somites. B: Migration of neural crest in a MZpard3fh305 mutant embryo. The onset and pattern of migration is similar to the control. Scale bar = 50 μM.
Pard3 Promotes Contact Mediated Inhibition But Does Not Drive Neural Crest Cell Migration
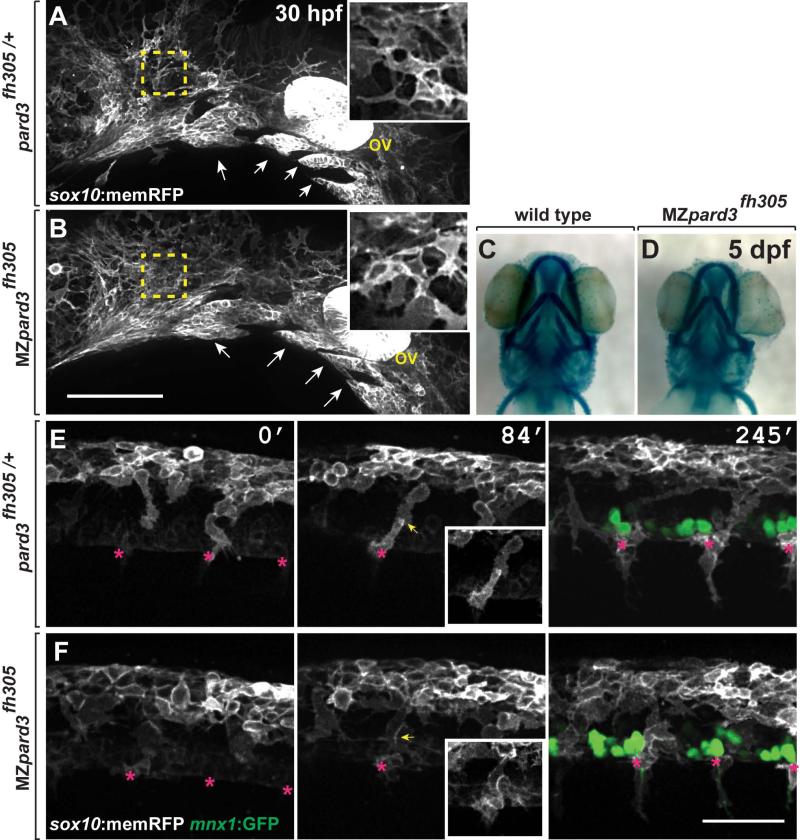
Subsequent to delamination, neural crest cells migrate along discrete pathways to populate peripheral tissues. Cranial neural crest cells exit the neuroepithelium and migrate toward the eye and pharyngeal arches forming distinct streams (Knight and Schilling, 2006; Schilling and Kimmel, 1994). Similarly, trunk neural crest forms columns or streams of cells as they migrate into the periphery. Recent data produced using pard3 antisense morpholino oligonucleotides in zebrafish indicated that loss of Pard3 function inhibited both cranial and trunk neural crest migration (Moore et al., 2013). However, cranial and trunk neural crest migration appeared normal in MZpard3fh305 mutant embryos. In particular, cranial neural crest cells formed distinct streams as they migrated from the neuroepithelium at 30 hpf, similar to control (Fig. 3A,B) and formed normal jaw structures by 5 dpf (Fig. 3C,D). Trunk neural crest cells also migrated to their motor axon targets in control and mutant embryos (Fig. 3E,F; Supplementary Movie S2). Therefore, our genetic analysis indicates that Pard3 function is not necessary to drive neural crest cell migration.
Fig. 3.
pard3 is not essential for neural crest migration. A,B: Representative images of cranial neural crest marked by Tg(sox10:memRFP) reporter expression in 30 hpf control pard3fh305/+ (A) and MZpard3fh305 mutant (B) embryos. View is from lateral with anterior to the left and dorsal up. Arrows indicate migrating groups of neural crest cells. In some regions, indicated by the outlined boxes and magnified in the insets, neural crest cells appear more tightly packed in mutant than in control. ov, otic vesicle. C,D: Representative images of jaw structures at 5 dpf in wild-type (C) and MZpard3fh305 mutant (B) larvae. The jaw in the mutant larva appears similar to that of wild type. E,F: Representative frames captured from time-lapse movies of trunk neural crest marked by Tg(sox10:memRFP) reporter expression. Motor neurons and axons are marked by Tg(mnx1:GFP) expression (green) in the final frames of the sequences. Images are from lateral with anterior to the left and dorsal up. Numbers indicate time, in minutes, elapsed from beginning of imaging. Images were captured from 18-24 hpf. In the control embryo (E), neural crest cells form streams (asterisks) with little overlap at points of contact (arrows). By contrast, neural crest cells appear less organized and overlap more in the MZpard3fh305 mutant embryo (F). Scale bar for E,F = 100 μM.
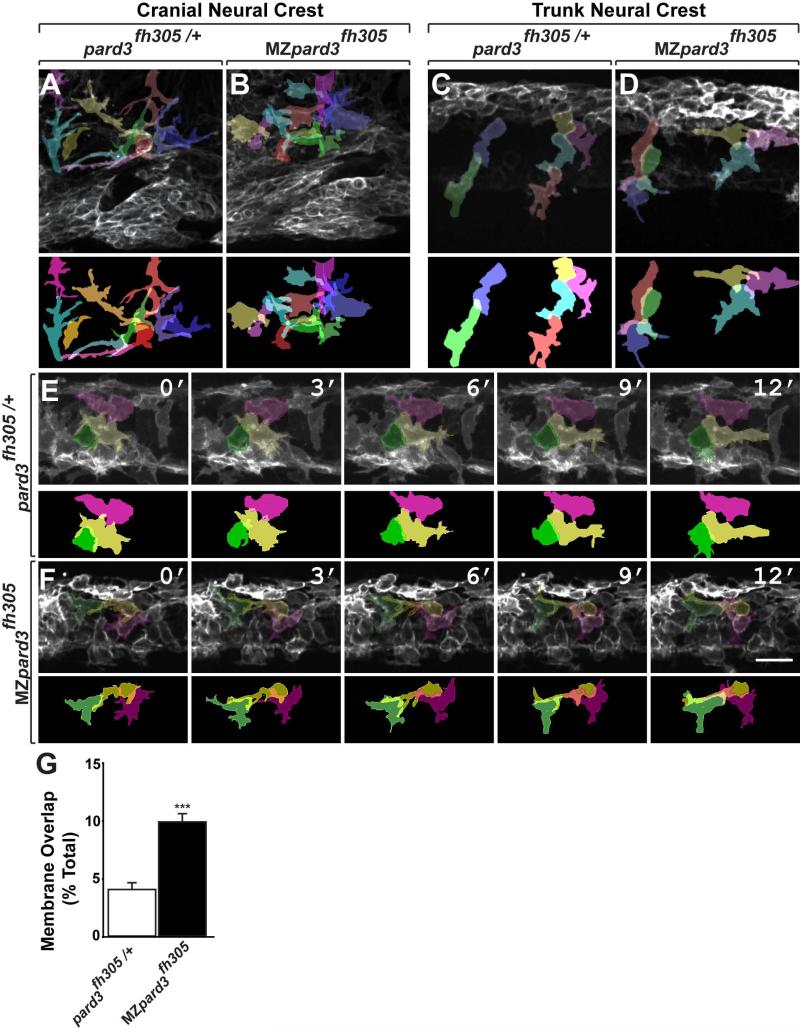
Although neural crest cells arrived at their normal destinations in MZpard3fh305 mutant embryos, we noted differences in organization of the neural crest population. In control embryos many migrating neural crest cells appeared to be loosely associated, with few contacts between them (Fig. 3A,E). By contrast, in MZpard3fh305 mutant embryos migrating neural crest cells appeared to have more area of contact between them (Fig. 3B,F; Supplementary Movie S1). We therefore examined confocal image stacks to assess the amount of membrane contact between neighboring cells. For analysis of cranial neural crest we focused on cells located outside of branchial arch streams. This confirmed that in control embryos neighboring cranial neural crest cells had only a few, thin membrane contacts (Fig. 4A) but that neural crest cells in MZpard3fh305 mutant embryos had substantial amounts of overlapping membrane (Fig. 4B). This increase in the amount of membrane contact did not result from an increase in cell density in mutant animals (control, 51.75 ± 0.68 cells/volume; mutant, 52.67 ± 0.81 cells/volume; n=5 embryos for each genotype; p = 0.4). In the trunk, neural crest cells in control embryos were elongated in the direction of migration with neighbor cell contacts limited to a single point, indicative of chain migration (Fig. 4C; Supplementary Movie S2). By contrast, trunk neural crest cells of MZpard3fh305 mutant embryos were less elongated and had more area of neighbor cell contact (Fig. 4D). To further validate this observation we performed time-lapse imaging and examined cell-cell interactions by analyzing membrane overlap. To determine the percentage of membrane overlap we traced individual cell membranes over a period of 1 hour during neural crest cell exit from the dorsal neural tube and examined membrane overlap following contact. In control embryos neighboring neural crest cells collapsed their membrane projections following contact and restricted membrane overlap either by altering their migratory path or limiting process extension along a neighboring cell (Fig. 4E, n= 24 cells). By contrast, in MZpard3fh305 embryos neural crest cells frequently extended membrane processes across neighboring cells and maintained their migratory direction over and along neighboring cells despite contact (Fig. 4F, n= 30 cells). We quantified these behaviors by measuring the amount of overlap between adjacent cells, which confirmed that adjacent neural crest cells had more area of overlapping membrane in mutant embryos relative to control (Fig. 4G).
Fig. 4.
Pard3 mediates contact inhibition between neural crest cells. A-F: Individual neural crest cell tracings represented as colored objects. A,B: Representative images of cranial neural crest cell migration in control (A) and mutant (B) embryos. In some regions, neural crest cells appear more tightly packed in mutant than in control. C,D: Representative images of trunk neural crest cells in control (C) and mutant (D) embryos; lateral view. Neural crest cells form chains elongated in the direction of migration in the control embryo (C). By contrast, neural crest cells appear less elongated and to be less well organized as chains in the mutant (D). E,F: Representative frames captured from time-lapse movies of trunk neural crest cell delamination in control (E) and mutant (F) embryos. Views are from dorsal with anterior to the left. Numbers denote time elapsed, in minutes, from start of imaging. Embryos are siblings and imaged sequentially in the same chamber. Neural crest cells in control embryos (n=30) retract cell membrane upon contact with neighboring cells. By contrast, neural crest cells in mutant embryos (n=24) fail to retract cell membrane and maintain cell overlap (F). G: Quantification of trunk neural crest cell membrane overlap during delamination (pard3fh305/+, n= 3 embryos, 43 cells; MZpard3fh305, n= 4 embryos, 47 cells; *** p = 0.0001; Student's t-test). Scale bar, 25uM.
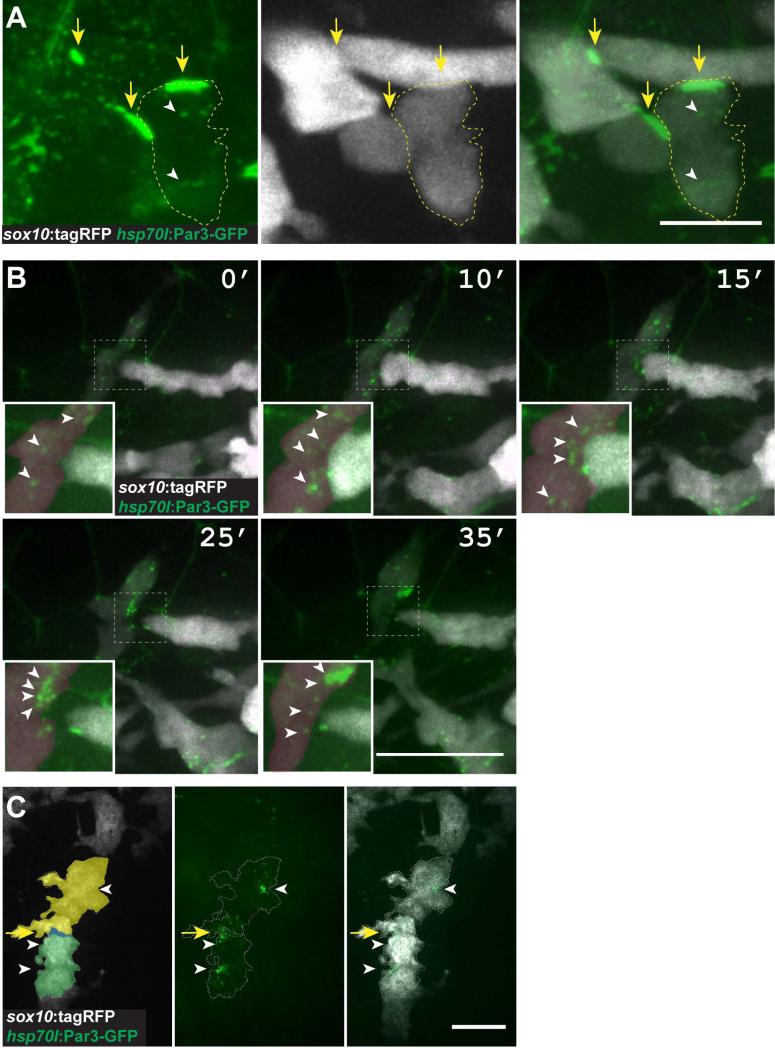
Migrating populations of cells often engage in contact inhibition of locomotion (CIL), which is characterized by the collapse of membrane projections upon contact with another cell and alteration of migration (Mayor and Carmona-Fontaine, 2010). The overlapping nature of neural crest cells in MZpard3fh305 mutant embryos is consistent with the loss of CIL. However, Pard3 function during CIL is unclear. To determine if Pard3 is localized to the site of cell-cell contact in migrating neural crest cells and therefore a candidate for regulation of CIL, we induced Pard3-GFP expression using the Tg(hsp70l:pard3-GFP) transgene. Pard3-GFP was evident as small aggregates within cells (Fig. 5A), similar to previously reported overexpression results (Buckley et al., 2013; Mizuno et al., 2003). At 20 hpf, Pard3-GFP was localized along contacting membranes of neighboring neural crest cells, consistent with the possibility that Pard3 regulates CIL (Fig. 5A). To investigate this further, we performed time-lapse imaging. This revealed that as neighboring cells came in contact, Pard3-GFP became localized to the membrane at the contact point. This was followed by retraction of membrane protrusions and dispersion of the Pard3-GFP that had been concentrated at the point of contact (Fig. 5B; Supplementary Movie S3). Concentrated Pard3-GFP was also associated with the points of contact between Schwann cells as they migrated along motor axons (Fig. 5C). Together with our observations that neighboring neural crest cells have more membrane contact in the absence of Pard3 function, these data support the idea that localization of Pard3 to points of cell contact promotes CIL. However, in contrast to previous conclusions (Moore et al., 2013) our data do not support the idea that Pard3-mediated CIL is a principal driver of neural crest cell migration.
Fig. 5.
Pard3 localizes at transient points of contact between neural crest cells. A: Image of neural crest cells marked by sox10:tagRFP (white) and Pard3-GFP (green) (24 hpf; lateral view of trunk). Pard3-GFP is concentrated at points of contact between cells (arrows) and forms puncta within cells (arrowheads) . B: Images captured from time-lapse movies of neural crest cells (24 hpf; dorsal view of trunk). Numbers indicate time elapsed since beginning of image sequence. Image sequence shows one neural crest cell (white) extending to contact another (red) followed by process withdrawal. Pard3-GFP clusters at the point of contact and then disperses following withdrawal (insets, arrowheads). C: Representative image of Pard3-GFP localization within migrating Schwann cells (20 hpf; lateral view of the trunk) Scale bars, A = 10 μM B = 25 μM C= 15 μM.
pard3 mutant Schwann cells myelinate motor axons
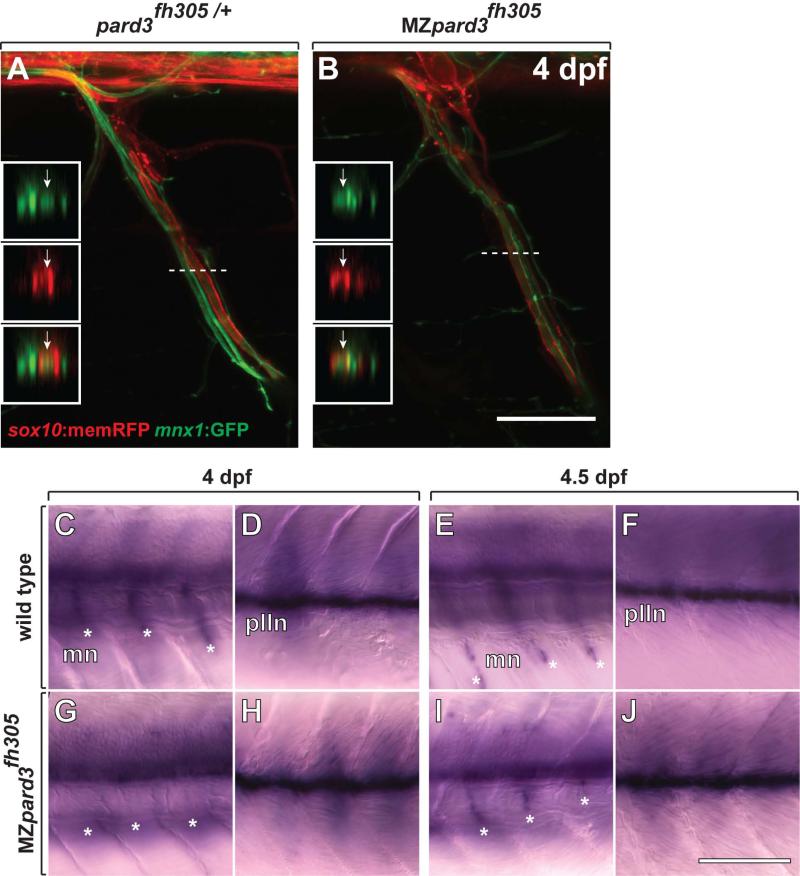
Upon association with peripheral axons, neural crest-derived Schwann cells change shape to ensheath axons with myelin membrane. Ensheathing Schwann cells have features of apical-basal polarity, with adaxonal and abaxonal membrane having characteristics of apical and basal membrane, respectively (Özçelik et al., 2010). Previous work implicated roles for Pard3 in both axon wrapping and myelination by Schwann cells. In particular, our own analysis using pard3 antisense morpholino oligonucleotides suggested that loss of pard3 function interferes with Schwann cell wrapping and myelination (Tep et al., 2012) and Pard3 siRNA knockdown in cultured Schwann cells blocked myelination (Chan et al., 2006). We therefore reinvestigated Schwann cell wrapping and myelination in vivo in the absence of both maternal and zygotic pard3 function. To do so we created Tg(sox10:memRFP);Tg(mnx1:GFP); MZpard3fh305 larvae, in which Schwann cells express RFP and motor axons are marked by GFP expression. At 4 dpf Schwann cells were elongated and tightly wrapped around motor axons in both control and mutant larvae, with no apparent differences in morphology (Fig. 6A,B). Therefore, we now conclude that Pard3 is not necessary for axon wrapping. Next, we investigated myelination by using in situ RNA hybridization to detect expression of mbp, which encodes a major myelin protein. At 4 dpf, Schwann cells along the anterior trunk motor roots and posterior lateral line nerve (plln) expressed mbp (Fig. 6C,D). In contrast, Schwann cells in MZpard3fh305 larvae lacked expression of mbp along the motor roots, although mbp expression was present along the plln and in the central nervous system (Fig. 6G,H). To determine if mbp expression was only delayed, we next performed in situ RNA hybridization using 4.5 dpf larvae. In this case, mbp expression at motor nerves of MZpard3fh305 larvae was similar to that of control larvae (Fig. 6E,F,I,J). These data indicate that Pard3 is not necessary for myelin gene expression in vivo but that it may promote the timing of the myelination program.
Fig. 6.
Schwann cells wrap motor axons but delay mbp expression in the absence of pard3 function. A,B: Representative images of control pard3fh305/+ (A) and MZpard3fh305 mutant (B) embryos, focused on a single motor root. Schwann cells are marked by sox10:memRFP (red) and motor axons are marked by mnx1:GFP (green). Schwann cells ensheath motor axons similarly in both control and mutant. Boxes show orthogonal views of motor nerves at the level of the dashed line. Arrows indicate ensheathed motor axons. C-J: Representative images of mbp RNA expression in wild-type and MZpard3fh305 mutant larvae. Views are from lateral of the trunk with anterior to the left and dorsal up. At 4 dpf, mbp expression is evident at motor nerves (mn, asterisks) (C) and the posterior lateral line nerve (pLLn) (D) of a wild-type larva. By contrast, no motor nerve expression is evident at motor nerves of a MZpard3fh305 mutant larva (E) although pLLn expression appears normal (F). At 4.5 dpf, mbp expression is similar in wild-type and MZpard3fh305 mutant larvae at both motor nerves and the pLLn (G-J). Scale bar = 15 μM (A,B), 100 μM (C-J).
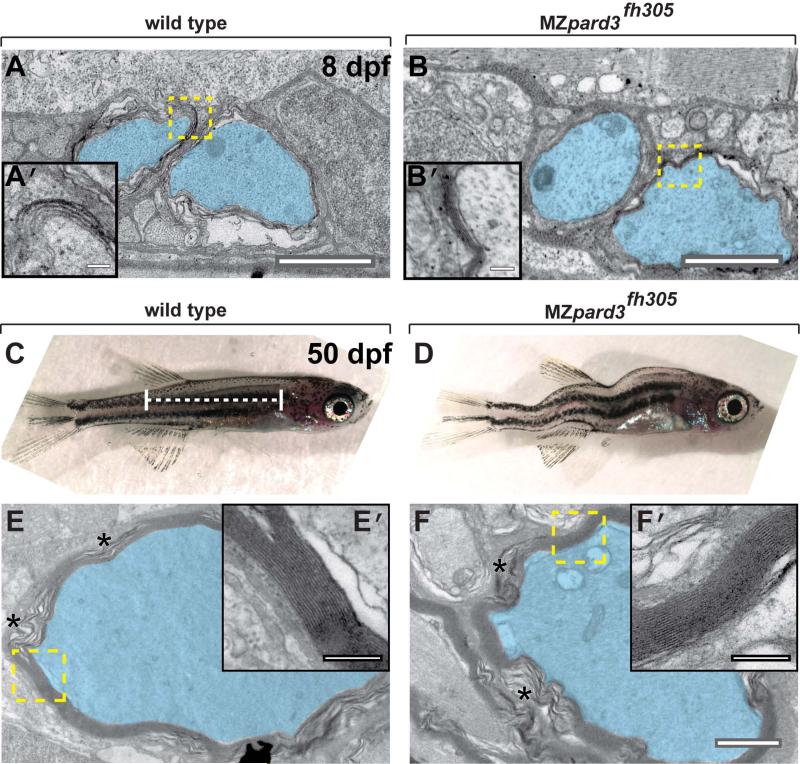
Additionally, we used electron microscopy to assess myelin formation. At 8 dpf, myelin was evident, although poorly compacted, in both control and MZpard3fh305 mutant larvae (Fig. 7A,B). To determine if myelin could become compacted in the absence of Pard3, we examined myelination in the small fraction of MZpard3fh305 mutant larvae that survived past larval stage (Fig. 7C,D). At 50 dpf, motor axons were surrounded by multiple layers of compacted myelin membrane in both wild-type and MZpard3fh305 mutant fish (Fig. 7E,F). We conclude that Pard3 is not essential for myelination, although it promotes its timely initiation during development.
Fig. 7.
pard3 is not essential for Schwann cell myelination. A-B: Transmission electron micrographs of coronal sections through the trunk region of 8 dpf wild-type (A) and MZpard3fh305 mutant (B) larvae. Motor axons are pseudocolored blue. Areas indicated by dashed boxes are shown at higher magnification in insets. Multiple layers of myelin membrane are evident in both wild type and mutant. C,D: Lateral views of 50 dpf wild-type (C) and MZpard3fh305 mutant (D) zebrafish. Dashed white line indicates region of coronal sections obtained for electron microscopy. MZpard3fh305 mutants are shortened and display severe body deformation, manifesting as a variable curved body axis. E-F: Transmission electron micrographs of coronal sections through the trunk region of 50 dpf fish. Areas indicated by dashed boxes are shown at higher magnfication in insets. Myelin ultrastructure is similar in wild type (E, E’) and MZpard3fh305 mutant (F, F’). Scale bars, A,B,C,D,1 μM; A’,B’,C’,D’, 0.25 μM.
DISCUSSION
Cells fated to become myelinating Schwann cells undergo a stepwise sequence of events during development including delamination from the neuroepithelium, directed migration into peripheral tissue, axon ensheathment and synthesis of myelin membrane. At each step these cells have a distinct polarity. Prior to delamination they have the apical-basal polarity characteristic of neuroepithelial cells, during migration they have distinct leading and trailing edges and during axon wrapping and myelination features of apical-basal polarity again become evident. These dynamic developmental steps imply dynamic functions of proteins that contribute to cell polarity. However, few studies have investigated the role of any polarity proteins during the entire Schwann cell progression in vivo. In this work we tested a hypothesis that the Par complex protein Pard3 regulates polarity necessary for directed migration, axon ensheathment and myelination by Schwann cells.
Genes that encode Par proteins, including Pard3, were first identified using forward genetic screens for embryonically lethal mutations that disrupt early developmental patterning events in nematodes (Kemphues et al., 1988) The fly ortholog of pard3, known as bazooka, was similarly identified in a forward screen for lethal mutations that altered larval cuticle pattern (Nusslein-Volhard et al., 1984). Mouse embryos also die midway through development, at least in part due to abnormal cardiac development (Hirose et al., 2006). We were therefore surprised that homozygous pard3 mutant zebrafish did not have dramatic morphological defects as embryos or larvae and that they survived to adulthood. The morphological defects of embryos and larvae produced by homozygous mutant adults was more severe than that of those produced by heterozygous animals and these individuals rarely survived past larval stage, indicating that maternal contribution of pard3 function is sufficient to sustain nearly normal development.
Pard3 localizes to tight junctions at the apical/lateral boundary of vertebrate epithelia (Izumi et al., 1998) and promotes tight junction assembly (Chen and Macara, 2005) Neural crest cells undergo an epithelial to mesenchymal transition (EMT) as they delaminate from the neuroepithelium, which is accompanied by disassembly of cellular adhesions such as tight junctions (Hay, 1995; Powell et al., 2013). Down-regulation of some tight junction components coincides with neural tube closure and neural crest delamination (Sauka-Spengler and Bronner-Fraser, 2008) and knock-down of the tight junction protein Cingulin enlarged the migratory neural crest population (Wu et al., 2011). We therefore considered the possibility that Pard3 similarly influences neural crest delamination. However, no abnormalities in the timing of neural crest delamination nor of the size of the neural crest population were evident in MZpard3fh305 mutant embryos, indicating that the timely onset of neural crest migration in zebrafish does not require modulation of Pard3 function.
As epithelial cells transform to migratory cells following EMT, apical-basal polarity changes to front-rear polarity (Nelson, 2009). Pard3 might contribute to front-rear polarity to facilitate directed migration, because knocking down Pard3 function impaired chemotaxis of isolated keratinocytes (Pegtel et al., 2007). However, most neural crest cells do not migrate as individuals, but as cohorts of cells that engage in transient contacts with one another. Upon contact, neural crest cells withdraw protrusions and frequently change directions (Carmona-Fontaine et al., 2008), a behavior known as contact inhibition of locomotion (Abercrombie and Heaysman, 1953). CIL has been implicated as an important driver of neural crest migration (Carmona-Fontaine et al., 2008; Matthews et al., 2008; Theveneau and Mayor, 2010; Theveneau et al., 2013). Recent data provided evidence that Pard3 localizes to the point of contact between neural crest cells and that an approximately 50% reduction of Pard3 levels by antisense morpholino oligonucleotide injection in frog and zebrafish embryos blocked CIL and migration of cranial and trunk neural crest (Moore et al., 2013). Our data, drawn from analysis of embryos lacking both maternal and zygotic Pard3 functions, are in good agreement with the conclusion that Pard3 mediates CIL because neural crest cells had substantially increased amounts of overlapping membrane. However, our data do not support the conclusion that Pard3-mediated CIL is an important driver of neural crest migration. Although both cranial and trunk neural crest cells were abnormally organized in mutant embryos, they nevertheless migrated normally. In particular, formation of the jaw, which requires long-distance migration of many cranial neural crest cells, was patterned normally in Pard3 mutant embryos. One possible explanation for these different observations is that use of morpholino oligonucleotides might delay neural crest migration through non-specific effects on development.
Neural crest-derived Schwann cells undergo one final reorganization of polarity upon completion of migration by transforming front-rear polarity back to apical-basal polarity (Özçelik et al., 2010). Consequently, the Schwann cell membrane in contact with the axon, known as the adaxonal membrane, has characteristics of apical membrane whereas the outer abaxonal membrane has features of basolateral membrane. This organization could have at least two important consequences for Schwann cell differentiation and myelination. First, juxtaposition of abaxonal-apical membrane to the axon could place molecules that facilitate axon ensheathment. Consistent with this, we previously concluded that loss of Pard3 function interfered with the ability of Schwann cells to tightly wrap motor axons (Tep et al., 2012). However, we now think that effect was likely an artifact of morpholino oligonucleotide injection because in this study we found no evidence of abnormal ensheathment. Second, juxtaposition of abaxonal-apical membrane to the axon could localize molecules that convey signals from axons to Schwann cells that promote myelination. Consistent with this possibility, Pard3 localized to abaxonal membrane and recruited p75 neurotrophin receptor, which can convey myelin-promoting signals (Chan et al., 2006). Knock down of Pard3 in cultured Schwann cells using short-hairpin RNA blocked expression of MBP, indicating that Schwann cell polarity is a critical feature of myelination (Chan et al., 2006). However, our in vivo analysis also fail to fully support this conclusion. Although mbp transcription was delayed in larvae lacking maternal and zygotic Pard3, the delay was slight and mature myelin appeared to be fully formed and compacted. Multiple signal transduction mechanisms promote Schwann cell myelination (Glenn and Talbot, 2013; Pereira et al., 2012; Salzer, 2012) and we speculate that these function normally even in the absence of Pard3.
In summary, our data support the idea that regulation of polarity contributes to migration and differentiation of neural crest and its derivative Schwann cells. However, our data point to a more nuanced role for polarity mechanisms than suggested by data derived from in vitro models and use of knock down methods for loss of function studies. We propose that Pard3 plays a modulatory role by organizing some of the many signaling mechanisms that influence neural crest migration and Schwann cell differentiation.
EXPERIMENTAL PROCEDURES
Zebrafish Husbandry
The animal work in this study was approved by the Institutional Animal Care and Use Committee at the University of Colorado School of Medicine. Embryos were produced by pair-wise mating and kept at 28.5°C in egg water or embryo medium. Embryos were staged to hours post fertilization (hpf) or days post fertilization (dpf) according to established zebrafish guidelines (Kimmel et al., 1995). Homozygous zygotic mutants for the pard3fh305 allele were created by pair-wise mating of pard3fh305/+ adults. Homozygous maternal zygotic mutants were created by pair-wise mating of pard3fh305/fh305 female and pard3fh305/+ male adults. The experiments conducted in this study used the following strains of zebrafish: Tg(sox10:memRFP)vu234 (Kucenas et al., 2008), Tg(hsp70l:pard3-EGFP)co14 (Hudish et al., 2013), Tg(mnx:GFP)ml2 (Flanagan-Steet et al., 2005) and Tg(sox10:tagRFP)co26.
Generation of Tg(Sox10(7.2):tagRFP)co26
The Tg(Sox10(7.2):tagRFP) construct was generated using Gateway (Invitrogen) recombination and inserted into a Tol2 Kit destination vector for microinjection (Kwan et al., 2007). Microinjected embryos were screened and transgene positive embryos were raised. Adults were mated with AB fish and progeny were screened for tagRFP expression and positive embryos were raised to establish transgenic lines.
pard3fh305 Genotyping
Embryos and larvae were genotyped using primers FWD: 5’-ATTGGCTTCAGCAGTTTTAAGAAA-3’ and REV: 5’-ATGATTGGCACTGAGTGAAGAAC-3’ and PCR to amplify a 155 base-pair (bp) product for. The PCR products were digested with HpyCH4IV (New England Bioscience). The wild-type allele remains undigested and mutant allele is digested into 87 bp and 68 bp. The complete protocol is available at the Zebrafish International Resource Center (http://zebrafish.org).
Time-lapse Imaging
At 18 hpf, embryos were embedded in low melting point agarose and mounted in a heated chamber (28.5°C) of a motorized stage. Z-stack images were obtained every 5-7 minutes from 18 to 25 hpf using a PerkinElmer UltraVIEW VoX Confocal System coupled with a Zeiss Axio Observer inverted compound microscope fitted with a 20X objective. Using Volocity software (PerkinElmer, Waltham, MA, USA) images were processed using deconvolution and contrast enhancement. Four-dimensional volumes were assembled and exported as QuickTime movie files.
Tg(hsp70l:pard3-EGFP) Heat Shock Procedure
Either non-transgenic pard3fh305/+ or Tg(hsp70l:pard3-EGFP);pard3fh305/+ males were mated to pard3fh305/fh305 females and the resulting embryos were spilt, with half untreated and half heat shocked for 30 minutes at 38°C in approximately 100 mililiters of egg water at 24, 48, and 72 hpf. Transgene expression was confirmed by screening for GFP. For rescue analysis, Tg(hsp70l:pard3-EGFP)+ larvae from non-heat shocked control and heat shocked groups were scored for the severity of the body deformation (Normal, Mild, or Severe) at 4 and 5 dpf. Larvae that had a straight body axis and inflated swim bladders were scored as normal. Larvae with body axis deformations and lacking inflated swim bladders were scored as severe. Larvae with subtle body deformations and slight or fully inflated swim bladders were scored as mild and genotyped to confirm the MZpard3fh305 mutant genotype.
Cell Membrane Overlap Analysis
Individual cells within 40 micron Z-stack volumes were traced using Volocity software v6.1.1. Total area for individual cells and cell overlap were determined using the Volocity. Statistical analysis of membrane overlap data was performed using Prism 6 (GraphPad) and two-tailed Student's t-test.
Whole Mount Cartilage Staining
5 dpf larvae were anesthetized and fixed overnight in 4% paraformaldehyde solution (PFA). Cartilage was stained with Alcian blue as previously described (Walker and Kimmel, 2007). Larvae were mounted in glycerol and bright-field images were taken using a Leica stereoscope.
In situ RNA Hybridization
Whole-mount in situ RNA hybridization was performed as described (Thisse and Thisse, 2008) to detect mbp (Brösamle and Halpern, 2002). Following hybridization, tissues were fixed with 4% paraformaldehyde, equilibrated in 70% glycerol and mounted on glass coverslips for whole-mount imaging. Images were collected using a Zeiss Axio Observer equipped with DIC optics, Retiga Exi digital color camera and Volocity software. All images were imported into Adobe Photoshop software and image processing was limited to changes in levels, contrast, brightness and cropping.
Transmission Electron Microscopy
Juvenile fish at 8 and 50 dpf were anesthetized with tricaine, placed on ice, and fixed in a solution of 2% glutaraldehyde, 4% paraformaldehyde and 0.1 M sodium cacodylate, pH 7.4. Membranes were enhanced using either secondary fixation with OsO4, uranyl acetate, and imidazole, or secondary fixation using OsO4-TCH-OsO4. Electron micrographs were collected using a FEI Techai G2 BioTwin microscope, transferred to Adobe Photoshop and image processing was limited to contrast and cropping.
Quantification of Cranial and Trunk Neural Crest Cells
Tg(sox10:memRFP);MZpard3fh305 and Tg(sox10:memRFP); pard3fh305/+ embryos were fixed at 24 hpf in 4% paraformaldehyde for 1 hour at room temperature and subsequently stained with DAPI (New England Bioscience; 1:1000) for 15 minutes at room temperature. Embryos were mounted in 1% low melting point agarose and imaged using a PerkinElmer UltraVIEW VoX Confocal System coupled with a Zeiss Axio Observer inverted compound microscope fitted with a 40x oil or 63X water objective. Using Volocity software (PerkinElmer, Waltham, MA, USA) three-dimensional volumes were assembled and regions of interest were defined for cell count analysis; dorsal trunk neural crest: 80 μM length, 20 μM width, 10 μM depth; cranial neural crest: 116 μM height, 116 μM width, 20 μM depth located posterior to the eye. DAPI positive cells within a defined region of interest were counted and data was compared using Student's t-test in the GraphPad Prism software (Version 6).
Quantification of Trunk Neural Crest Cell Survival
MZpard3fh305 and pard3fh305/+ embryos were assayed for Caspase 3 expression (BD Pharmingen, 1:500) at 20 hpf as previously described (Sorrells et al., 2013). Embryos were mounted in 1% low melting point agarose and imaged using a PerkinElmer UltraVIEW VoX Confocal System coupled with a Zeiss Axio Observer inverted compound microscope fitted with a 20x objective. Using Volocity software (PerkinElmer, Waltham, MA, USA) three-dimensional volumes were assembled and a 250 μM length, 50 μM width, 20 μM depth region of interest was defined for cell count analysis. Caspase 3-positive cells within a defined region of interest were counted and data were compared using Student's t-test in the GraphPad Prism software (Version 6).
Supplementary Material
Movie 1. In vivo time-lapse movie of zebrafish neural crest cell delamination. Dorsal view of Tg(sox10:mRFP); pard3fh305-/+ (left) and Tg(sox10:mRFP); MZpard3fh305 (right) with anterior to the top. Note neural crest cells in control embryos (left) limit membrane overlap during interactions (arrow). By contrast, mutant neural crest cells (right) display extensive membrane overlap during interactions (arrow). Despite differences in the interactions, both control and mutant neural crest cells migrate away from the dorsal neural tube in an anterior to posterior fashion over time. 5 minutes / frame; 4 frames / second.
Movie 2. In vivo time-lapse movie of zebrafish neural crest cell migration to motor exit point. Lateral view of Tg(sox10:mRFP); pard3fh305−/+ (left) and Tg(sox10:mRFP); MZpard3fh305 (right) with anterior to the left. Neural crest cells in control embryo (left) migrate in a chain-like fashion and limit membrane overlap (arrow) during migration to the motor axon exit point. In contrast, mutant neural crest cells (right) migrate along and over each other, exhibiting extensive membrane overlap during interactions (arrow).
Movie 3. In vivo time-lapse movie of Pard3-GFP localization within zebrafish neural crest cells during cell-cell contact. Pard3-GFP is localized within delaminating trunk neural crest cells to sites of cell-cell contact following collision. 5 minutes / frame; 1 frame / second.
Key findings.
Pard3 localizes to points of transient contact between migrating neural crest cells
Pard3 mediates contact inhibition between neural crest cells but does not drive neural crest migration
Pard3 promotes the timing of myelin gene expression by Schwann cells but is not essential for axon wrapping or myelination
ACKNOWLEDGMENTS
We thank members of the Appel lab for valuable discussions and Dorothy Dill for helping with the electron microscopy procedure. This work was supported by NIH NS062717 and a gift from the Gates Frontiers Fund. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility is supported by NIH grant P30 NS048154. The pard3fh305 mutant was identified by TILLING with the support of NIH grant NIH HD076585 to C.B.M.
Grant sponsor and numbers: NIH NS062717 (B.A.) and NIH HD076585 (C.B.M.)
REFERENCES
- Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Experimental Cell Research. 1953;5:111–131. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- Afonso C, Henrique D. PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. Journal of Cell Science. 2006;119:4293–4304. doi: 10.1242/jcs.03170. [DOI] [PubMed] [Google Scholar]
- Ahlstrom JD, Erickson CA. The neural crest epithelial-mesenchymal transition in 4D: a ‘tail’ of multiple non-obligatory cellular mechanisms. Development. 2009;136:1801–1812. doi: 10.1242/dev.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Developmental Biology. 2008;324:236–244. doi: 10.1016/j.ydbio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C. ErbB receptors and the development of the nervous system. Experimental Cell Research. 2009;315:611–618. doi: 10.1016/j.yexcr.2008.10.035. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser ME, LeDouarin NM. Development and evolution of the neural crest: an overview. Developmental Biology. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Buckley CE, Ren X, Ward LC, Girdler GC, Araya C, Green MJ, Clark BS, Link BA, Clarke JDW. Mirror-symmetric microtubule assembly and cell interactions drive lumen formation in the zebrafish neural rod. The EMBO Journal. 2013;32:30–44. doi: 10.1038/emboj.2012.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK, Cayouette M. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science. 2006;314:832–836. doi: 10.1126/science.1134069. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang M. The Par3/Par6/aPKC complex and epithelial cell polarity. Experimental Cell Research. 2013;319:1357–1364. doi: 10.1016/j.yexcr.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nature Cell Biology. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Clay MR, Halloran MC. Control of neural crest cell behavior and migration: Insights from live imaging. celladhesion. 2010;4:586–594. doi: 10.4161/cam.4.4.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay MR, Halloran MC. Rho activation is apically restricted by Arhgap1 in neural crest cells and drives epithelial-to-mesenchymal transition. Development. 2013;140:3198–3209. doi: 10.1242/dev.095448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Polarity proteins in glial cell functions. Current Opinion in Neurobiology. 2008;18:488–494. doi: 10.1016/j.conb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B. A 90 rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development. 2003;130:3767–3780. doi: 10.1242/dev.00603. [DOI] [PubMed] [Google Scholar]
- Glenn TD, Talbot WS. Signals regulating myelination in peripheral nerves and the Schwann cell response to injury. Current Opinion in Neurobiology. 2013;23:1041–1048. doi: 10.1016/j.conb.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Developmental Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED. An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Hirose T, Karasawa M, Sugitani Y, Fujisawa M, Akimoto K, Ohno S, Noda T. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 2006;133:1389–1398. doi: 10.1242/dev.02294. [DOI] [PubMed] [Google Scholar]
- Hudish LI, Blasky AJ, Appel B. miR-219 Regulates Neural Precursor Differentiation by Direct Inhibition of Apical Par Polarity Proteins. Developmental Cell. 2013;27:387–398. doi: 10.1016/j.devcel.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. The Journal of Cell Biology. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. CELL. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knight RD, Schilling TF. Cranial neural crest and development of the head skeleton. Adv Exp Med Biol. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park H-C, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat Neurosci. 2008;11:143–151. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lewallen KA, Shen Y-AA, la Torre De AR, Ng BK, Meijer D, Chan JR. Assessing the role of the cadherin/catenin complex at the Schwann cell-axon interface and in the initiation of myelination. Journal of Neuroscience. 2011;31:3032–3043. doi: 10.1523/JNEUROSCI.4345-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews HK, Marchant L, Carmona-Fontaine C, Kuriyama S, Larraín J, Holt MR, Parsons M, Mayor R. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development. 2008;135:1771–1780. doi: 10.1242/dev.017350. [DOI] [PubMed] [Google Scholar]
- Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends in Cell Biology. 2010;20:319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Suzuki A, Hirose T, Kitamura K, Kutsuzawa K, Futaki M, Amano Y, Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- Moore R, Theveneau E, Pozzi S, Alexandre P, Richardson J, Merks A, Parsons M, Kashef J, Linker C, Mayor R. Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion. Development. 2013;140:4763–4775. doi: 10.1242/dev.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011;138:799–809. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harbor Perspectives in Biology. 2009;1:a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster. Dev Genes Evol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- Özçelik M, Cotter L, Jacob C, Pereira JA, Relvas JB, Suter U, Tricaud N. Pals1 is a major regulator of the epithelial-like polarization and the extension of the myelin sheath in peripheral nerves. Journal of Neuroscience. 2010;30:4120–4131. doi: 10.1523/JNEUROSCI.5185-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Ellenbroek SIJ, Mertens AEE, van der Kammen RA, de Rooij J, Collard JG. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends in Neurosciences. 2012;35:123–134. doi: 10.1016/j.tins.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Powell DR, Blasky AJ, Britt SG, Artinger KB. Riding the crest of the wave: parallels between the neural crest and cancer in epithelial-to-mesenchymal transition and migration. Wiley Interdiscip Rev Syst Biol Med. 2013;5:511–522. doi: 10.1002/wsbm.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Regulative interactions in zebrafish neural crest. Development. 1996;122:501–507. doi: 10.1242/dev.122.2.501. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Salzer JL. Axonal regulation of Schwann cell ensheathment and myelination. J Peripher Nerv Syst. 2012;17(Suppl 3):14–19. doi: 10.1111/j.1529-8027.2012.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser ME. A gene regulatory network orchestrates neural crest formation. Nature Reviews Molecular Cell Biology. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Shoji W, Yee CS, Kuwada JY. Zebrafish semaphorin Z1a collapses specific growth cones and alters their pathway in vivo. Development. 1998;125:1275–1283. doi: 10.1242/dev.125.7.1275. [DOI] [PubMed] [Google Scholar]
- Sorrells S, Toruno C, Stewart RA, Jette C. Analysis of apoptosis in zebrafish embryos by whole-mount immunofluorescence to detect activated Caspase 3. J Vis Exp. 2013:e51060. doi: 10.3791/51060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekuni K, Ikeda W, Fujito T, Morimoto K, Takeuchi M, Monden M, Takai Y. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem. 2003;278:5497–5500. doi: 10.1074/jbc.C200707200. [DOI] [PubMed] [Google Scholar]
- Tep C, Kim ML, Opincariu LI, Limpert AS, Chan JR, Appel B, Carter BD, Yoon SO. Brain-derived neurotrophic factor (BDNF) induces polarized signaling of small GTPase (Rac1) protein at the onset of Schwann cell myelination through partitioning-defective 3 (Par3) protein. Journal of Biological Chemistry. 2012;287:1600–1608. doi: 10.1074/jbc.M111.312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Integrating chemotaxis and contact-inhibition during collective cell migration: Small GTPases at work. Small GTPases. 2010;1:113–117. doi: 10.4161/sgtp.1.2.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Developmental Biology. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nature Cell Biology. 2013;15:763–772. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Thompson BJ. Cell polarity: models and mechanisms from yeast, worms and flies. Development. 2013;140:13–21. doi: 10.1242/dev.083634. [DOI] [PubMed] [Google Scholar]
- Trotha von JW, Campos-Ortega JA, Reugels AM. Apical localization of ASIP/PAR-3:EGFP in zebrafish neuroepithelial cells involves the oligomerization domain CR1, the PDZ domains, and the C-terminal portion of the protein. Dev Dyn. 2006;235:967–977. doi: 10.1002/dvdy.20715. [DOI] [PubMed] [Google Scholar]
- Walker M, Kimmel C. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wei X, Cheng Y, Luo Y, Shi X, Nelson S, Hyde DR. The zebrafish Pard3 ortholog is required for separation of the eye fields and retinal lamination. Developmental Biology. 2004;269:286–301. doi: 10.1016/j.ydbio.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Alonso MBD, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, Wilton DK, Al-Shawi R, Simons P, Shen J, Guillemot F, Radtke F, Meijer D, Feltri ML, Wrabetz L, Mirsky R, Jessen KR. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhoo A, Sommer L. Development of the Schwann cell lineage: from the neural crest to the myelinated nerve. Glia. 2008;56:1481–1490. doi: 10.1002/glia.20723. [DOI] [PubMed] [Google Scholar]
- Wu C-Y, Jhingory S, Taneyhill LA. The tight junction scaffolding protein cingulin regulates neural crest cell migration. Dev Dyn. 2011;240:2309–2323. doi: 10.1002/dvdy.22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. In vivo time-lapse movie of zebrafish neural crest cell delamination. Dorsal view of Tg(sox10:mRFP); pard3fh305-/+ (left) and Tg(sox10:mRFP); MZpard3fh305 (right) with anterior to the top. Note neural crest cells in control embryos (left) limit membrane overlap during interactions (arrow). By contrast, mutant neural crest cells (right) display extensive membrane overlap during interactions (arrow). Despite differences in the interactions, both control and mutant neural crest cells migrate away from the dorsal neural tube in an anterior to posterior fashion over time. 5 minutes / frame; 4 frames / second.
Movie 2. In vivo time-lapse movie of zebrafish neural crest cell migration to motor exit point. Lateral view of Tg(sox10:mRFP); pard3fh305−/+ (left) and Tg(sox10:mRFP); MZpard3fh305 (right) with anterior to the left. Neural crest cells in control embryo (left) migrate in a chain-like fashion and limit membrane overlap (arrow) during migration to the motor axon exit point. In contrast, mutant neural crest cells (right) migrate along and over each other, exhibiting extensive membrane overlap during interactions (arrow).
Movie 3. In vivo time-lapse movie of Pard3-GFP localization within zebrafish neural crest cells during cell-cell contact. Pard3-GFP is localized within delaminating trunk neural crest cells to sites of cell-cell contact following collision. 5 minutes / frame; 1 frame / second.