Summary
The Calcyclin-Binding Protein/Siah-1-Interacting Protein (CacyBP/SIP) is highly expressed in the brain and was shown to regulate the β-catenin-driven transcription in thymocytes. Therefore, it was investigated whether in brain cells CacyBP/SIP might play a role as a transcriptional regulator. In BDNF- or forskolin-stimulated rat primary cortical neurons, overexpression of CacyBP/SIP enhanced transcriptional activity of the cAMP-response element (CRE). In addition, overexpressed CacyBP/SIP enhanced BDNF-mediated activation of the Nuclear Factor of Activated T-cells (NFAT) but not the Serum Response Element (SRE). These stimulatory effects required an intact C-terminal domain of CacyBP/SIP. Moreover, in C6 rat glioma cells, the overexpressed CacyBP/SIP enhanced activation of CRE- or NFAT- following forskolin- or serum stimulation, respectively. Conversely, knockdown of endogenous CacyBP/SIP reduced activation of CRE- and NFAT but not SRE. Taken together, these results indicate that CacyBP/SIP is a novel regulator of CRE- and NFAT-driven transcription.
Keywords: CacyBP/SIP, CRE-driven transcription, Brain development
Introduction
Transcription factors are key mediators of long-term responses to a variety of extracellular signals. For instance, neurotransmitters and neurotrophins including glutamate and BDNF regulate brain neuron morphogenesis, survival and synaptic plasticity by activating Extracellular signal-Regulated Kinase-1/2 (ERK1/2) (Adams and Sweatt 2002; Hetman and Gozdz 2004; Miller and Kaplan 2003; Parrish et al. 2007). In turn, ERK1/2 carries out these long-term responses by stimulating such transcription factors as cAMP Response Element Binding protein (CREB), Serum Response Factor (SRF) and the Nuclear Factor of Activated T-cells (NFAT) (Knoll and Nordheim 2009; Lonze and Ginty 2002; Vashishta et al. 2009). Although a great progress has been achieved in defining the mechanisms of ERK1/2-dependent transcriptional activation, the negative regulation of ERK1/2-driven transcription has been relatively understudied.
CacyBP/SIP has been initially isolated as a protein that interacts with calcyclin/S100A6 (Filipek and Kuznicki 1998) and later as a binding partner of a human Siah-1 (Matsuzawa and Reed 2001). Siah-1 is an E3 ubiquitin ligase that is induced by p53 in response to DNA damage (Amson et al. 1996; Matsuzawa et al. 1998). CacyBP/SIP helps to assemble multiprotein E3 ubiquitin ligase containing Siah-1, Skp1 and the F-box protein Ebi (Matsuzawa and Reed 2001). It has been proposed that the latter protein targets the complex towards β-catenin that is then polyubiquitinated and degraded. By inhibiting the pro-proliferative transcription that is mediated by β-catenin, CacyBP/SIP contributes to the p53-dependent cell cycle arrest following genotoxic stress and is essential for mouse thymocyte differentiation (Fukushima et al. 2006).
In addition to serving as an adaptor protein for the E3 ligase, CacyBP/SIP also exhibits phosphatase activity toward ERK1/2 and tau (Kilanczyk et al. 2011; Wasik and Filipek 2013). Moreover, CacyBP/SIP directly bound to ERK2 (Kilanczyk et al. 2009). This interaction was mediated by the C-terminal domain of CacyBP/SIP. Within this domain, the conserved residue E217 (amino acid numbering for mouse CacyBP/SIP) was required for ERK2 binding. When overexpressed in NB2a cells, the wild type but not the E217K mutant variant of CacyBP/SIP reduced the ERK2-mediated activation of the transcription factor Elk1 (Kilanczyk et al. 2009). Together with the Serum Response Factor (SRF), Elk1 activates the SRE-driven transcription in an ERK1/2-dependent manner (Knoll and Nordheim 2009). Therefore, in addition to inhibiting the β-catenin-mediated transcription, CacyBP/SIP may negatively regulate transcription factors that are activated by ERK1/2. Moreover, such an inhibitory interaction may require binding of CacyBP/SIP to ERK1/2.
Although CacyBP/SIP is abundantly expressed in the brain including forebrain neurons (Jastrzebska et al. 2000), its significance for brain development and/or function is unclear. Therefore, the current study has been initiated to evaluate whether CacyBP/SIP regulates ERK-dependent activation of transcription factors that have well established role in the brain.
Materials and Methods
Materials
All reagents were obtained from Sigma, EMD or other vendors, as indicated. The following plasmids have been previously described: CRE-Luc (CRE-luciferase reporter) and EF1αLacZ (expression vector for β-galatosidase, β-gal) (Impey et al. 1998), NFAT-Luc (NFAT-luciferase reporter) (Vashishta et al. 2009), SRE-Luc (SRE- luciferase reporter) (Wang and Prywes 2000), expression vectors for CacyBP/SIP-EGFP, CacyBP/SIP-E217K-EGFP and their empty cloning vector (pEGFP-C1) (Wolozin 2014); pSuper-based small hairpin RNA (shRNA) construct targeting GFP (Boehrs et al. 2007).
Cell Culture and Transfection
Cortical neurons were prepared from newborn Sprague-Dawley rats at postnatal day 1 as previously described (Habas et al. 2006). Briefly, the culture medium was Basal Medium Eagle (BME) supplemented with 10% heat-inactivated bovine calf serum (Hyclone, Logan, UT), 35 mM glucose, 1 mM L-glutamine, 100 U/mL of penicillin and 0.1 mg/mL streptomycin. Cytosine arabinoside (2.5 μM) was added on the second day after seeding (day in vitro 2, DIV2) to inhibit proliferation of non-neuronal cells. Neurons were transfected on DIV4 using Lipofectamine2000 (Invitrogen) as described previously (Hetman et al. 2002). Two days after transfections neurons were treated with 10 ng/ml BDNF diluted in phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) or 10 μM forskolin diluted in water. C6 astrocytic glioma cells were cultured in growth medium containing DMEM, 10% FBS, 1 mM L-glutamine, 100 U/mL of penicillin and 0.1 mg/mL streptomycin. C6 cells were transfected using the Lipofectamine2000 reagent according to the manufacturer's protocol. One day after transfection cells were placed in serum-free media for 12h and then stimulated by adding medium containing 10% fetal bovine serum (FBS) with or without 10 μM forskolin.
Immunocytochemistry
For GFP immunostaining a standard immunofluorescence protocol was followed using a rabbit anti-GFP antibody (MBL) diluted 1:500 (Hetman et al. 1999).
Luciferase reporter gene assays
Luciferase and β-gal activities were assayed using standard kits (Promega). Transcriptional activity was determined as a luciferase activity normalized to β-gal activity. For each condition, three sister cultures were used per experiment. The baseline activities were determined for each transfection condition in non-stimulated cells. In each experiment, stimulation responses were calculated as fold of the baseline for each plasmid combination. Such baseline-normalized values from at least three independent experiments were then compared using ANOVA and posthoc tests.
Generation of shRNA expression constructs
To generate CacyBP/SIP shRNA constructs, the rat CacyBP/SIP mRNA (gene bank accession Number NP_001004208.1) sequence was analyzed using shRNA design software (http://www.genelink.com/sirna/shrnai.asp/). One sequence corresponding to nucleotides 280-299 was selected. Oligonucleotide sequence: GATCCCCGGAACAAGATGCAGCAGAATTCAAGAGATTCTGCTGCATCTTGTTCCTTTTTA was designed together with their complementary counterparts, annealed and subcloned into a pSUPER vector digested with BglII and HindIII (OligoEngine, Seattle, WA, USA).
SDS-PAGE and Western blotting
A standard protocol for both SDS-PAGE and Western blot was applied. Mouse anti-CacyBP/SIP (Abcam) and mouse anti-GAPDH antibody (Alexis Biochemicals) were used at 1:1000 and 1:3000 dilutions, respectively. Densitometry analysis was used to evaluate the relative expression of CacyBP/SIP.
Results
Transcription modulation by CacyBP/SIP in primary neurons
In BDNF- or forskolin-treated rat cortical neurons, activation of CRE-driven transcription requires ERK1/2 (D'Amico et al. 2000). To determine effects of CacyBP/SIP overexpression on transcriptional responses downstream of ERK1/2, neurons were transfected with expression vectors for CacyBP/SIP-EGFP or CacyBP/SIP-E217K-EGFP together with a CRE-driven luciferase reporter construct. In initial experiments, equal expression and similar cellular localization of CacyBP/SIP-EGFP and CacyBP/SIP-E217K-EGFP was confirmed by immunofluorescence study. Both proteins appeared to accumulate in the perikarial cytosol and neurites resembling neuronal distribution of endogenous CacyBP/SIP (Fig. 1A, upper panel). Such a pattern did not change in response to BDNF stimulation (Fig. 1A, lower panel).
Figure 1. Effects of CacyBP/SIP overexpression on ERK1/2-mediated transcriptional responses in primary neurons.
DIV4 cortical neurons were transfected with the cDNA expression vector for CacyBP/SIP-EGFP (WT) or its E217K mutant (E217K) together with the transfection marker plasmid EF1αLacZ (to express β-gal) and the luciferase reporter constructs as indicated (0.5μg+0.2μg+0.2μg plasmid DNA/5×105 neurons, respectively). As a control an empty expression vector EGFP-C1 (Vector) was used. In A, neurons received only CacyBP/SIP-EGFP constructs as indicated. At 24h post transfection, neurons were stimulated with 10 ng/ml BDNF (A, B, D and E) or 10 μM forskolin (C) for 8h. A, Immunofluorescence using anti GFP antibody revealed similar expression levels and localization of CacyBP/SIP-GFP and its E217K mutant irrespective of BDNF stimulation; scale bar=20 μm. B-E, After stimulations, activities of luciferase and β-gal were determined and compared to unstimulated cells. CacyBP/SIP-EGFP selectively increased CRE- and NFAT- but not SRE-mediated transcriptional responses despite their similarly high sensitivity to the ERK1/2 pathway inhibitor U0126 (data not shown). Data represent 12 sister cultures from four independent experiments, error bars are SEM; ***p<0.001.
In unstimulated neurons, overexpression of CacyBP/SIP significantly increased the basal acitivity of CRE as compared to empty vector-transfected cells (5.7±1.2 vs. 1.0±0.05, p<0.001); no significant effects on baseline CRE were observed with the E217K mutant. Moreover, the wild type by not the mutant CacyBP/SIP increased CRE activation in response to BDNF or forskolin (Fig. 1B, C). In empty vector-transfected neurons, an 8h treatment with 10 ng/ml BDNF or 10 μM forskolin increased CRE activity 3.9- or 8.5 fold of non-stimulated controls, respectively. In CacyBP/SIP overexpressing neurons, CRE activation by BDNF or forskolin increased to 12- or 30 fold of non-stimulated controls, respectively. In contrast, overexpression of the E217K mutant did not alter CRE responses to either stimulus. Similar effects of overexpressed CacyBP/SIP were observed on activation of NFAT-driven transcription in response to BDNF (Fig. 1D). However, basal activity of NFAT was not affected by the wild type or the E217K mutant form of CacyBP/SIP (2±0.61 or 1.5±69 vs. 1±0.1, respectively, p>0.05). Thus, in neurons, the ERK1/2-dependent activation of CRE- or NFAT-driven transcription was enhanced by CacyBP/SIP. Such a modulation required intact C-terminal domain of CacyBP/SIP.
In addition to CRE or NFAT, SRE-mediated transcription is another well-established downstream target of the BDNF-ERK1/2 signaling pathway in neurons. However, overexpression of CacyBP/SIP or its E217K mutant did not modulate activation of SRE by BDNF (Fig. 1E). Likewise, in the absence of stimulation, baseline activity of SRE was unaffected by either construct (0.91±0.14 or 1.26±0.36 vs. 1±0.14, respectively, p>0.05). Importantly, similar requirement of ERK1/2 for stimulation of all three transcriptional systems was confirmed using the specific ERK1/2 inhibitor U0126 (data not shown) (Kalita et al., 2006; Vashishta et al. 2009). Therefore, CacyBP/SIP did not uniformly affect all ERK1/2-dependent transcription.
Transcription modulation by CacyBP/SIP in C6 astrocytic glioma cells
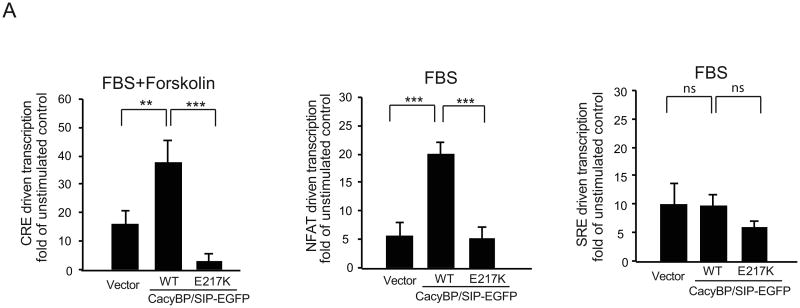
To investigate whether regulatory influence of CacyBP/SIP over CRE and NFAT is present in proliferating cells of brain origin, rat C6 astrocytic glioma cells were used. In unstimulated C6 cells, overexpression of CacyBP/SIP or its E217K mutant had no significant effects on basal activities of CRE or NFAT. For instance, in CacyBP/SIP overexpressing cells, basal CRE or NFAT activity was 1.6±0.34- or 1.9±0.43 fold of vector controls, respectively (p>0.05). However, CacyBP/SIP enhanced CRE- and NFAT- responses to activating stimuli including serum+forskolin (CRE) or serum (NFAT) (Fig. 2A, B). Such a modulatory action was observed, similarly as it was for neurons, only for the wild type CacyBP/SIP but not for its E217K mutant. Conversely, neither basal SRE activity nor its stimulation by serum was significantly affected by CacyBP/SIP (Fig. 2C and data not shown).
Figure 2. Overexpression of CacyBP/SIP selectively enhances CRE- and NFAT–driven transcription in C6 astrocytic glioma cells.
Cells were transfected as described for Figure 1. At 48h post transfection cell were placed for 12h in serum free media followed by 8h stimulations with 10% FBS±10μM forskolin as indicated. CacyBP/SIP-EGFP enhanced activation of CRE- (A) and NFAT- (B) but not SRE- (C) driven transcription. Data represent 9 sister cultures from three independent experiments, error bars are SEM; ***p<0.001, **p<0.01.
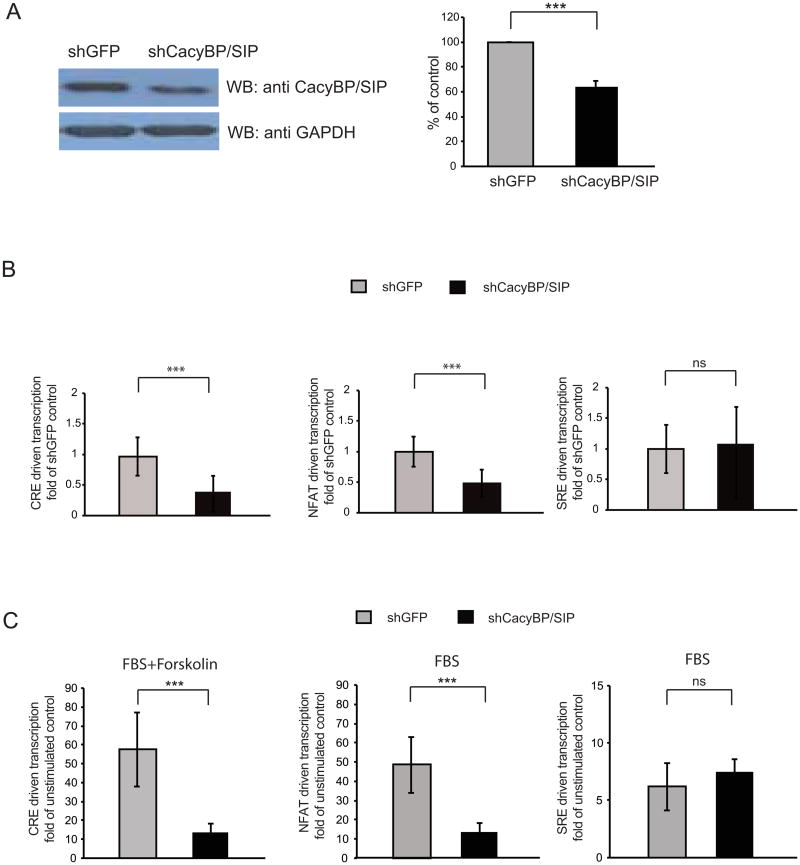
To determine the role of endogenous CacyBP/SIP in transcriptional regulation, an shRNA expression vector was prepared to target rat CacyBP/SIP (shCacyBP/SIP). When transfected into the C6 cells the shCacyBP/SIP reduced expression of endogenous CacyBP/SIP (Fig. 3A). In the absence of stimulation, knock down of CacyBP/SIP lowered basal activities of CRE and NFAT but not SRE as compared to control shRNA-transfected cells (Fig. 3B). Moreover, shCacyBP/SIP reduced activation of CRE- or NFAT-driven transcription in response to serum+forskolin or serum, respectively (Fig. 3C). However, it did not affect stimulation of SRE in response to serum (Fig. 3C). Taken together, these results indicate that both endogenous and overexpressed CacyBP/SIP regulate CRE and NFAT-driven transcription.
Figure 3. Knockdown of CacyBP/SIP reduces stimulation of CRE- and NFAT-driven transcription.
C6 cells were transfected with an expression vector for shRNA targeting rat CacyBP/SIP or a control shRNA against GFP (0.5 μg plasmid DNA/5×105 cells). In B-D, EF1αLacZ and the luciferase reporter constructs were also added as indicated (0.2μg+0.2μg plasmid DNA/5×105 cells, respectively). A, Expression of endogenous CacyBP/SIP was determined by Western blotting. To monitor total protein content in each lane the blot was re-probed for GAPDH. The graph represents averages of optical density ratios of CacyBP/SIP to GAPDH bands from three independent experiments; error bars are SEM; ***, p<0.001. Note that as transfection efficiency was estimated at no more than 50%, the extent of the knockdown is underrepresented by these data. B-C, Effects of CacyBP/SIP knockdown on CRE-, NFAT- and SRE-driven transcription was determined by luciferase reporter assays as indicated. In B, effects on basal transcriptional activities are presented as folds of values from shGFP-transfected cells; in C, stimulations were as described for Fig. 2. Data represent 9 sister cultures from three independent experiments; error bars are SEM; ***p<0.001.
Discussion
In this work we demonstrate that in rat primary cortical neurons and C6 astrocytic glioma cells, CacyBP/SIP does not uniformly affect transcription factors that are targets of ERK1/2 signaling. CRE- and NFAT-driven transcription was enhanced while SRE-driven transcription was unaffected. In addition, in C6 cells, an shRNA-based evidence was obtained that endogenous CacyBP/SIP is required for CRE-and NFAT activation by forskolin and/or serum. Finally, effects on CRE- and NFAT-driven transcription were dependent on the intact C-terminal domain of CacyBP/SIP.
All findings presented in this work suggest that CacyBP/SIP may work as a positive regulator of some but not all ERK1/2-driven transcription pathways including CRE and NFAT. Such effects may be mediated by either direct or indirect interactions between CacyBP/SIP and the components of the affected transcriptional systems. In the case of β-catenin-driven transcription, CacyBP/SIP is as an adaptor protein that facilitates β-catenin degradation by its Siah-1-mediated ubiquitination (Matsuzawa and Reed 2001). A similar mechanism that operates by destabilizing transcriptional activators or repressors may modulate CRE- and/or NFAT-driven transcription (Healey et al. 2013; Liu et al. 2009; Sen and Snyder 2011). Alternatively, CacyBP/SIP-facilitated degradation of β-catenin may affect CRE and/or NFAT. Finally, one can also consider a possibility that the adaptor functions of CacyBP/SIP may be used not only for the regulated proteasomal degradation but also for assembly of signaling complexes transducing extracellular signals into the nucleus in a proteasome-independent manner. Additional studies are needed to define mechanisms of transcriptional effects of CacyBP/SIP.
We report that overexpression or knockdown of CacyBP/SIP does not affect SRE. Our prior observations in neuroblastoma cells suggested that overexpressed CacyBP/SIP antagonized ERK1/2-dependent transcriptional activity of the overexpressed Gal4-Elk1 fusion transcription factor on the Gal4-luciferase reporter construct (Kilanczyk et al. 2009). As Elk1 contributes to regulation of SRE our current findings appear to contradict those initial observations. However, SRE is also regulated by SRF and the SRF interacting factors including MKL1 and MKL2 (Knoll and Nordheim 2009). In addition, the Gal4-Elk1/Gal4-luciferase system that was used in our previous work was designed to probe the transactivation capacity of Elk1 without taking into account modulation of its DNA binding or activity of other factors that co- regulate SRE. Hence, even if Elk-1-mediated transactivation is reduced by CacyBP/SIP, increased activity of other SRE regulators such SRF or MKL1/2 may result in normal SRE activity.
In our hands, integrity of the C-terminal domain of CacyBP/SIP, which was shown to interact with ERK2, was essential for transcriptional effects of CacyBP/SIP since its E217K mutant had no effect on CRE- or NFAT-driven transcription. Such a mutation was also shown to prevent effects of CacyBP/SIP on β-catenin levels (Kilanczyk et al. 2009; Matsuzawa and Reed 2001). While these interactions may contribute to the observed effects on the ERK1/2-regulated transcriptional pathways one can also consider alternative interactions with the C-terminal domain of CacyBP/SIP or structural changes within this domain that may contribute to these effects.
Findings presented in this work suggest that CacyBP/SIP is a novel regulator of NFAT. Critical involvement of NFAT in development and functions of the immune system is well established (Muller and Rao 2010). In addition, NFAT-driven transcription is required for heart development, angiogenesis, axonal growth, peripheral nerve myelination, and glutamate-mediated neuronal survival (Graef et al. 2003; Kao et al. 2009; Nilsson et al. 2008; Schulz and Yutzey 2004; Vashishta et al. 2009). NFAT also contributes to pathologies such as heart hypertrophy, cancer metastases or astroglia re-activation in degenerating brain (Abdul et al. 2009; Muller and Rao 2010; Nilsson et al. 2008; Sama et al. 2008). As a new addition to the list of NFAT regulators, CacyBP/SIP may affect these important processes. Such a claim is supported by overlapping effects of CacyBP/SIP- and NFATc3- gene knock outs on mouse thymocyte development (Cante-Barrett et al. 2007; Fukushima et al. 2006). Taken together, our results identify CacyBP/SIP as a novel positive regulator of CRE- and NFAT-driven transcription. Several questions are raised by such a conclusion including those about (i) mechanism of the CacyBP/SIP-mediated transcriptional regulation, (ii) identity of the CRE- or NFAT target genes that are regulated by CacyBP/SIP, and, (iii) biological significance of this regulation. Given recognized importance of CRE or NFAT and scarcity of data on functions of CacyBP/SIP, future studies towards answering these questions will be of high significance.
Acknowledgments
This work was supported by grants from NIH (NS073584-01 and 8P30GM103507, to MH), NSF (IOS1021860, to MH), The Polish National Science Center (NCN-NZ3/004250, to AF), EMBO (short-term travel fellowship to EK) as well as statutory funds from the Nencki Institute of Experimental Biology and the Commonwealth of Kentucky Challenge for Excellence Fund. The authors wish to thank Ms. Jing-Juan Zheng for excellent technical assistance and Dr. W. Leśniak for helpful discussion during preparation of this manuscript.
Abbreviations
- BDNF
Brain-Derived Neurotrophic Factor
- CacyBP/SIP
Calcyclin-binding protein/Siah-1-interacting protein
- CRE
cAMP Response Element
- ERK1/2
Extracellular signal Regulated Kinases 1 and 2
- NFAT
Nuclear Factor of Activated T-cells
- GAPDH
Glyceraldehyde 3-Phosphate Dehydrogenase
- PAGE
Polyacrylamide Gel Electrophoresis
- PBS
Phosphate Buffered Saline
- SDS
Sodium Dodecyl Sulfate
- SRF
Serum Response Factor
Footnotes
Conflict of Interests: The authors have no conflict of interests.
References
- Abdul HM, Sama MA, Furman JL, Mathis DM, Beckett TL, Weidner AM, Patel ES, Baig I, Murphy MP, LeVine H, 3rd, Kraner SD, Norris CM. Cognitive decline in Alzheimer's disease is associated with selective changes in calcineurin/NFAT signaling. J Neurosci. 2009;29(41):12957–12969. doi: 10.1523/JNEUROSCI.1064-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JP, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu Rev Pharmacol Toxicol. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Amson RB, Nemani M, Roperch JP, Israeli D, Bougueleret L, Le Gall I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, Piouffre L, Prieur S, Susini L, Alvaro V, Millasseau P, Guidicelli C, Bui H, Massart C, Cazes L, Dufour F, Bruzzoni-Giovanelli H, Owadi H, Hennion C, Charpak G, Telerman A, et al. Isolation of 10 differentially expressed cDNAs in p53-induced apoptosis: activation of the vertebrate homologue of the drosophila seven in absentia gene. Proc Natl Acad Sci USA. 1996;93(9):3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehrs JK, He J, Halaby MJ, Yang DQ. Constitutive expression and cytoplasmic compartmentalization of ATM protein in differentiated human neuron-like SH-SY5Y cells. J Neurochem. 2007;100(2):337–345. doi: 10.1111/j.1471-4159.2006.04254.x. [DOI] [PubMed] [Google Scholar]
- Cante-Barrett K, Winslow MM, Crabtree GR. Selective role of NFATc3 in positive selection of thymocytes. J Immunol. 2007;179(1):103–110. doi: 10.4049/jimmunol.179.1.103. [DOI] [PubMed] [Google Scholar]
- D'Amico M, Hulit J, Amanatullah DF, Zafonte BT, Albanese C, Bouzahzah B, Fu M, Augenlicht LH, Donehower LA, Takemaru K, Moon RT, Davis R, Lisanti MP, Shtutman M, Zhurinsky J, Ben-Ze'ev A, Troussard AA, Dedhar S, Pestell RG. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J Biol Chem. 2000;275(42):32649–32657. doi: 10.1074/jbc.M000643200. [DOI] [PubMed] [Google Scholar]
- Filipek A, Kuznicki J. Molecular cloning and expression of a mouse brain cDNA encoding a novel protein target of calcyclin. J Neurochem. 1998;70(5):1793–1798. doi: 10.1046/j.1471-4159.1998.70051793.x. [DOI] [PubMed] [Google Scholar]
- Fukushima T, Zapata JM, Singha NC, Thomas M, Kress CL, Krajewska M, Krajewski S, Ronai Z, Reed JC, Matsuzawa S. Critical function for SIP, a ubiquitin E3 ligase component of the beta-catenin degradation pathway, for thymocyte development and G1 checkpoint. Immunity. 2006;24(1):29–39. doi: 10.1016/j.immuni.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113(5):657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Habas A, Kharebava G, Szatmari E, Hetman M. NMDA neuroprotection against a phosphatidylinositol-3 kinase inhibitor, LY294002 by NR2B-mediated suppression of glycogen synthase kinase-3beta-induced apoptosis. J Neurochem. 2006;96(2):335–348. doi: 10.1111/j.1471-4159.2005.03543.x. [DOI] [PubMed] [Google Scholar]
- Healey M, Crow MS, Molina CA. Ras-induced melanoma transformation is associated with the proteasomal degradation of the transcriptional repressor ICER. Molecular Carcinogenesis. 2013;52(9):692–704. doi: 10.1002/mc.21908. [DOI] [PubMed] [Google Scholar]
- Hetman M, Gozdz A. Role of extracellular signal regulated kinases 1 and 2 in neuronal survival. Eur J Biochem. 2004;271(11):2050–2055. doi: 10.1111/j.1432-1033.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- Hetman M, Hsuan SL, Habas A, Higgins MJ, Xia Z. ERK1/2 Antagonizes Glycogen Synthase Kinase-3beta -induced Apoptosis in Cortical Neurons. J Biol Chem. 2002;277(51):49577–49584. doi: 10.1074/jbc.M111227200. [DOI] [PubMed] [Google Scholar]
- Hetman M, Kanning K, Smith-Cavanaugh JE, Xia Z. Neuroprotection by Brain-Derived Neurotrophic Factor Is Mediated by Extracellular-Signal-Regulated Kinase and Phosphatidylinositol-3 Kinase. J Biol Chem. 1999;274:22569–22580. doi: 10.1074/jbc.274.32.22569. [DOI] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci. 1998;1(7):595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Filipek A, Nowicka D, Kaczmarek L, Kuznicki J. Calcyclin (S100A6) binding protein (CacyBP) is highly expressed in brain neurons. J Histochem Cytochem. 2000;48(9):1195–1202. doi: 10.1177/002215540004800903. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323(5914):651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilanczyk E, Filipek S, Filipek A. ERK1/2 is dephosphorylated by a novel phosphatase--CacyBP/SIP. Biochem Biophys Res Com. 2011;404(1):179–183. doi: 10.1016/j.bbrc.2010.11.088. [DOI] [PubMed] [Google Scholar]
- Kilanczyk E, Filipek S, Jastrzebska B, Filipek A. CacyBP/SIP binds ERK1/2 and affects transcriptional activity of Elk-1. Biochem Biophys Res Com. 2009;380(1):54–59. doi: 10.1016/j.bbrc.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Knoll B, Nordheim A. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 2009;32(8):432–442. doi: 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. Faseb J. 2009;23(10):3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. The EMBO Journal. 1998;17(10):2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa SI, Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Molecular Cell. 2001;7(5):915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- Miller FD, Kaplan DR. Signaling mechanisms underlying dendrite formation. Curr Opin Neurobiol. 2003;13(3):391–398. doi: 10.1016/s0959-4388(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nature reviews Immunology. 2010;10(9):645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- Nilsson LM, Nilsson-Ohman J, Zetterqvist AV, Gomez MF. Nuclear factor of activated T-cells transcription factors in the vasculature: the good guys or the bad guys? Curr Opin Lipidol. 2008;19(5):483–490. doi: 10.1097/MOL.0b013e32830dd545. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Sama MA, Mathis DM, Furman JL, Abdul HM, Artiushin IA, Kraner SD, Norris CM. Interleukin-1beta-dependent signaling between astrocytes and neurons depends critically on astrocytic calcineurin/NFAT activity. J Biol Chem. 2008;283(32):21953–21964. doi: 10.1074/jbc.M800148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz RA, Yutzey KE. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266(1):1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Sen N, Snyder SH. Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc Natl Acad Sci USA. 2011;108(50):20178–20183. doi: 10.1073/pnas.1117820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishta A, Habas A, Pruunsild P, Zheng JJ, Timmusk T, Hetman M. Nuclear factor of activated T-cells isoform c4 (NFATc4/NFAT3) as a mediator of antiapoptotic transcription in NMDA receptor-stimulated cortical neurons. J Neurosci. 2009;29(48):15331–15340. doi: 10.1523/JNEUROSCI.4873-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Prywes R. Activation of the c-fos enhancer by the erk MAP kinase pathway through two sequence elements: the c-fos AP-1 and p62TCF sites. Oncogene. 2000;19(11):1379–1385. doi: 10.1038/sj.onc.1203443. [DOI] [PubMed] [Google Scholar]
- Wasik U, Filipek A. The CacyBP/SIP protein is sumoylated in neuroblastoma NB2a cells. Neurochem Res. 2013;38(11):2427–2432. doi: 10.1007/s11064-013-1155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discovery Medicine. 2014;17(91):47–52. [PMC free article] [PubMed] [Google Scholar]