Abstract
The mitochondrial theory of aging, a mainstream theory of aging which once included accumulation of mitochondrial DNA (mtDNA) damage by reactive oxygen species (ROS) as its cornerstone, has been increasingly losing ground and is undergoing extensive revision due to its inability to explain a growing body of emerging data. Concurrently, the notion of the central role for mtDNA in the aging process is being met with increased skepticism. Our progress in understanding the processes of mtDNA maintenance, repair, damage, and degradation in response to damage has largely refuted the view of mtDNA as being particularly susceptible to ROS-mediated mutagenesis due to its lack of “protective” histones and reduced complement of available DNA repair pathways. Recent research on mitochondrial ROS production has led to the appreciation that mitochondria, even in vitro, produce much less ROS than previously thought, automatically leading to a decreased expectation of physiologically achievable levels of mtDNA damage. New evidence suggests that both experimentally induced oxidative stress and radiation therapy result in very low levels of mtDNA mutagenesis. Recent advances provide evidence against the existence of the “vicious” cycle of mtDNA damage and ROS production. Meta-studies reveal no longevity benefit of increased antioxidant defenses. Simultaneously, exciting new observations from both comparative biology and experimental systems indicate that increased ROS production and oxidative damage to cellular macromolecules, including mtDNA, can be associated with extended longevity. A novel paradigm suggests that increased ROS production in aging may be the result of adaptive signaling rather than a detrimental byproduct of normal respiration that drives aging. Here, we review issues pertaining to the role of mtDNA in aging.
Keywords: Mitochondrial DNA, Reactive oxygen species, DNA damage, DNA repair, Somatic mtDNA mutations, Antioxidants, Reactive oxygen species signaling, Mitochondrial DNA degradation, Electron transport, Aging
Core tip: The notion of reactive oxygen species (ROS) -mediated accumulation of mutations in mitochondrial DNA (mtDNA) as a driving force behind aging is increasingly losing ground forcing a revision of the Mitochondrial Theory of Aging. While mitochondrial involvement remains in the center of attention of aging research, the focus is shifting from mtDNA mutations to mitochondrial physiology. The positive effect of increased ROS production on longevity is increasingly viewed as evidence that increased ROS production in aging may be adaptive rather than maladaptive. This novel paradigm explains failure of antioxidants to delay aging in clinical trials.
INTRODUCTION
While there is no universally accepted definition of the aging process, it is often defined as changes (mostly detrimental) that occur in organisms during their lifespan. Most researchers agree that aging is: (1) universal; (2) intrinsic (i.e., “built-in”); (3) progressive; (4) deleterious; and (5) irreversible. The universality of the aging process suggests the existence of an equally universal mechanism or mechanisms that govern it. Over time, different aging theories proposed a variety of such basic mechanisms. Perhaps the most popular of these theories was (and, arguably, remains) the Free Radical/Mitochondrial Theory of Aging (henceforth MTA) first proposed by Harman[1] in 1956. Initially, this theory simply postulated that aging results from the accumulation of oxygen free radical [reactive oxygen species (ROS)] damage to cellular components, including nucleic acids[1]. Over the years, the theory was refined by, first, the identification of mitochondria as both the source and the target of the ROS[2], and, then, the identification of mitochondrial DNA (mtDNA) as a tally-keeper for the damage. The latter concept was introduced by Fleming et al[3] and Miquel et al[4,5], and is of particular importance because it provided an answer to critics who questioned the capability of other mitochondrial macromolecules such as lipids, proteins, or RNA to accumulate longitudinal damage over an organism’s lifetime. Unlike damage to other macromolecules, damage to mtDNA can be converted to point mutations and deletions, which can be transmitted to and accumulated in daughter molecules through the process of replication, enabling deterioration of the integrity of hereditary information over time. It is this damage-sustaining capacity of mtDNA that makes it central to discussions of aging, and it is this property that will be the focus of the current review. Over the years, the MTA underwent many revisions to accommodate new experimental evidence, and thus, there are almost as many versions of it as there are investigators. As Jacobs observed more than a decade ago, “opponents of the hypothesis (MTA) tend to define it in such a narrow and extreme way that it is almost self-evidently falsified by generally accepted facts. Conversely, its proponents are liable to state the theory in such a vague and general way that it is virtually unfalsifiable experimentally”[6]. Here, we review our current knowledge of mtDNA maintenance as it pertains to the MTA, which consists of the following basic tenets: (1) Mitochondria are a significant source of ROS in the cell; (2) Mitochondrial ROS inflict damage on mtDNA; (3) Oxidative mtDNA damage results in mutations; (4) mtDNA mutations lead to the synthesis of defective polypeptide components of the electron transport chain (ETC); (5) Incorporation of these defective subunits into the ETC leads to a further increase in ROS production, initiating a “vicious” cycle of ROS production, mtDNA mutations, and mitochondrial dysfunction (Figure 1). This tenet appears to be the most controversial, and is no longer recognized as a part of the MTA by many researchers[7]; and (6) Eventually, mtDNA mutations, ROS production and cellular damage by ROS reach levels incompatible with life.
Figure 1.
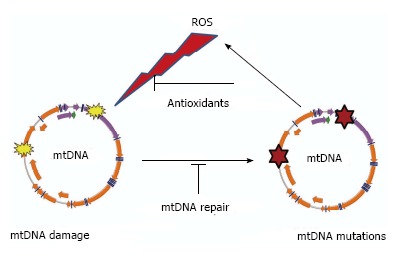
“Vicious cycle” of reactive oxygen species production, mitochondrial DNA damage, mitochondrial DNA mutagenesis and further reactive oxygen species production. The cycle implies an exponential growth of reactive oxygen species (ROS) production and mitochondrial DNA (mtDNA) mutagenesis.
Some recent experimental evidence has called into question the validity of the MTA, prompting its reevaluation (see e.g.,[7]). Here, we present a historical perspective of our views on the role of mtDNA in aging and update our earlier critical review of the topic[8].
MTDNA
MtDNA (Figure 2) in mammals is a circular molecule that encodes 37 genes, including 2 rRNAs, 22 tRNAs, and 13 polypeptides. All 13 polypeptides are components of the oxidative phosphorylation (OXPHOS) system. They are encoded using a non-standard genetic code, which requires its own translational machinery separate from that of the nucleus. Two rRNAs and 22 tRNAs involved in this mitochondrial protein synthesis are also encoded by mtDNA. Mitochondrial DNA is densely packed into nucleoids, each containing as few as 1-2 mtDNA molecules[9].
Figure 2.
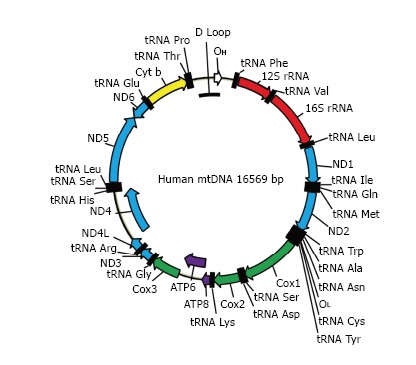
The map of human mitochondrial DNA. OH and OL: Origins of heavy and light strand replication, respectively; ND1-ND6: Subunits of NADH dehydrogenase (ETC complex I) subunits 1 through 6; COX1-COX3: Subunits of cytochrome oxidase subunits 1 through 3 (ETC complex IV); ATP6 and ATP8: Subunits 6 and 8 of mitochondrial ATPase (complex V); Cyt b: Cytochrome b (complex III); ETC: Electron transport chain.
A significant body of indirect evidence implicating mtDNA in longevity was contributed by studies on the inheritance patterns of longevity, which suggested possible cytoplasmic (mitochondrial) inheritance[10], and from studies which revealed the association of some mtDNA variations with longevity[11-14]. However, other studies indicate that these associations are weak[15]. The latest large-scale study on mtDNA and aging suggests that the relationship between mtDNA variants and longevity may be much more complex, and that while mutations in the OXPHOS complex I may beneficially affect longevity, the coincidence of mutations in complexes I and III as well as the simultaneous presence of mutations in complexes I and V are detrimental. These more complex relationships escape detection by haplogroup analysis and require sequencing of complete mitochondrial genomes[16]. Overall, these findings indirectly support the idea that mtDNA variations may contribute to longevity.
MITOCHONDRIA ARE A SIGNIFICANT SOURCE OF ROS IN CELLS
ROS generation by mitochondria
In the course of their migration through the respiratory chain, electrons can “escape” and participate in the single-electron reduction of oxygen resulting in the formation of the superoxide radical (O2•- Eq. 1). The detailed overview of this process is presented elsewhere[8,17]. While the exact magnitude of ROS production in vivo remains debatable, we and others repeatedly argued[8,17] that the values of 1%-2% of total oxygen consumption[18] frequently cited in the literature are not reflective of physiological conditions and that the real rates are much lower.
ROS are produced by multiple sites in mitochondria[19]. Sites other than complexes I and III are rarely mentioned in the context of aging. However, recent data suggest that some of these sites may have higher ROS production capacity than respiratory chain complex I, which is often viewed as a major source of matrix superoxide production[20]. Moreover, it was argued that the endoplasmic reticulum and peroxisomes have a greater capacity to produce ROS than mitochondria do[21]. Another important consideration is that O2•- produced by the mitochondrial respiratory chain inactivates aconitase, thus suppressing the Krebs cycle and reducing supply of NADH and FADH2 to the respiratory chain. This can reduce electron flow through ETC, lower the reduction of ETC complexes, and diminish the production of O2•-[22,23]. Thus, O2•- production by ETC may be regulated by a negative feedback loop. Finally, actively respiring mitochondria may consume more ROS than they are capable of producing[24].
Mitochondrial ROS neutralization
ETC-generated ROS are detoxified through a two-step process. First, O2•- is converted to H2O2 either spontaneously, or with the help of superoxide dismutases (Eq. 2). Two superoxide dismutases were described in mitochondria: SOD2 in the matrix and SOD1 in the intermembrane space. Interestingly, there is evidence of SOD1 activation by O2•-[25]. The relative stability and membrane permeability of H2O2 ensure its ready access to mtDNA, yet like O2•- this ROS is unable to efficiently react with DNA[8]. Only when H2O2 undergoes Fenton chemistry in the presence of transition metal ions (Eq. 3) is it converted to the extremely reactive hydroxyl radical. This ROS can efficiently damage mtDNA and other mitochondrial components[26,27]. At the second step, H2 O2 in the mitochondrial matrix is detoxified by peroxiredoxins III and V (PrxIII and PrxV, Eq. 4 and 5, respectively[28]) and by glutathione peroxidase 1 (GPx1, Eq. 6). Of the eight known GPx isoforms, this one is targeted to the mitochondrial matrix[29]. Another isoform, GPx4, is involved in detoxification of the mitochondrial membrane hydroperoxides[30] and is relevant due to the close association between mtDNA and the inner mitochondrial membrane. Prx III is about 30-fold more abundant in mitochondria than GPx 1[31]. It is generally believed that catalase does not localize to mitochondria[32]. Therefore, GPx 1, and Prx III and V appear to be the main contributors to H2O2 detoxification in the mitochondrial matrix.
O2 + e- → O2•- (Eq. 1)
2 O2•- + 2 H+ → H2O2 + O2 (Eq. 2)
Fe2+ + H2O2 → Fe3+ +•OH + OH- (Eq. 3)
H2O2 + 2PrxIII (SH)2 → 2H2O + PrxIII(SH)-S-S(SH)PrxIII (Eq. 4)
H2O2 + PrxV(SH)2 → 2H2O + PrxV(S-S) (Eq. 5)
H2O2 + 2GSH → GS-SG + 2H2O (Eq. 6)
Remarkably, the thioredoxin/peroxiredoxin system is capable of detoxifying extramitochondrial H2O2 in a respiration-dependent manner, providing evidence that mitochondrial OXPHOS is involved not only in the production of ROS, but also in their detoxification, and raising the question of whether mitochondria in vivo are a net source or a net sink of ROS[24].
MTDNA DAMAGE BY ROS
The reaction of O2•- with non-radicals is spin forbidden[33-37]. In biological systems, this means that the main reactions of O2•- are with itself (dismutation) or with another biological radical, such as nitric oxide. Therefore, direct reactions of O2•- with mtDNA are unlikely. This ROS is far more likely to undergo dismutation to H2O2 (Eq. 2). As indicated above, H2O2 in the presence of transition metal ions, in particular Fe2+ and Cu+, can undergo Fenton chemistry to form the extremely reactive •OH. Mitochondria are rich in iron, as many mitochondrial enzymes possess heme groups and iron-sulfur clusters in their active centers, and this abundance of iron may favor •OH production[38]. Therefore, it has been argued that mitochondria may be particularly susceptible to •OH -mediated oxidation, which plays a major role in DNA oxidation[39]. In this respect, it is important to note that mitochondrial iron is not free, but chelated (bound). Some experimental evidence does support the availability of chelated iron for Fenton-type reactions[40,41], and it is also true that iron chelators like desferrioxamine can efficiently suppress DNA mutagenesis by Fenton chemistry in vitro[42]. However, there is still a need for studies that could directly assess the ability of the iron bound in mitochondrial heme- and Fe-S proteins to promote generation of •OH.
Is mtDNA more sensitive to damage?
The mitochondrial genome accumulates germline mutations approximately one order of magnitude faster than nuclear DNA (nDNA)[43-45]. To evaluate relative accumulation of somatic mutations in nDNA vs mtDNA, we used 6 × 10-8 per nucleotide per cell division as an upper estimate for the rate of nDNA mutagenesis (8). Considering that the number of cells in the human body is 3.72 × 1013 (9), which roughly corresponds to 45 cell divisions starting with a fertilized egg, we arrive at 6 × 45 × 10-8 = 2.7 × 10-6 mutations per base pair for the somatic nDNA mutation burden in an aged human, provided that there is no further nDNA mutagenesis after reaching adulthood. The somatic mtDNA mutation burden has been recently estimated to be 1.9 × 10-5 (10), which is less than 1 order of magnitude higher than the 2.7 × 10-6 just calculated for nDNA. mtDNA is turned over with half-lives of 10-30 d in different tissues (11), and therefore the difference in the rates of spontaneous somatic mtDNA mutagenesis between mtDNA and nDNA on per doubling basis may be even smaller than 1 order of magnitude [because in a 70-year-old human mtDNA has replicated on average (assuming a half-life of 30 d) at least 12/2 × 70 + 45 = 465 times compared to 45 times for nDNA, not counting repair synthesis]. Therefore, somatic mutations may accumulate at the same per doubling rate in nDNA as they do in mtDNA, while the cumulative burden of mutations in mtDNA may be one order of magnitude higher than that in nDNA in a 70-year-old individual.
In the literature, three properties of the mitochondrial genome are frequently cited as responsible for this faster rate of mtDNA mutagenesis: (1) Its proximity to the source of ROS (ETC); (2) Its lack of “protective” histones; and (3) A limited repertoire of DNA repair pathways available in mitochondria.
It has been argued, however, that proximity to the source of ROS, by itself, is unable to explain the higher mutation rate of mtDNA, and that some available experimental evidence directly contradicts the notion of the protective role of histones[8]. Observations that mtDNA is covered by TFAM[46] and that at least some prominent oxidative DNA lesions are repaired more efficiently in mitochondria than they are in the nucleus[47] also contradict the above arguments.
Moreover, mitochondria evolved a unique way to deal with excessive or irreparable damage: a pathway for degradation or abandonment of damaged molecules (Figure 3)[48,49]. This pathway is enabled by the high redundancy of mtDNA (hundreds to thousands of copies per cell). MtDNA degradation has been reported in response to both oxidative stress[50-52] and to enzymatically-induced abasic sites[53]. It also has been suggested that substrates for the Nucleotide Excision Repair pathway, which has not been detected in mitochondria, are also mitigated through mtDNA turnover[54,55].
Figure 3.
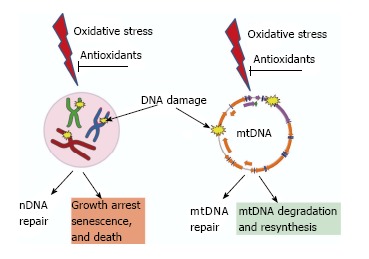
Consequences of unrepaired DNA damage in the nucleus and in the mitochondria. Oxidative damage induces lesions in both nDNA (left) and mtDNA (right). Both nuclei and mitochondria possess DNA repair systems to deal with these lesions. However, cellular consequences of unrepaired damage to nDNA and mtDNA are different. While persistent damage in nDNA results in the activation of cell cycle checkpoints, growth arrest, senescence and death. In contrast, mtDNA molecules with unrepairable damage are simply degraded and new molecules are synthesized using intact molecules as templates. This figure uses Servier elements available under Creative Commons license (155).
If three of the above mentioned rationales in support of mtDNA’s higher susceptibility to (oxidative) damage and mutagenesis are not satisfactorily supported by experimental evidence, what then is the basis for the frequently cited higher (compared to nDNA) susceptibility of mtDNA to oxidative stress? Here, one ought to make a distinction between damage to DNA bases-which may lead, upon replication, to point mutations- and damage to the sugar phosphate backbone. The first report comparing the content of the oxidative DNA base lesion, 8-oxodG, in nDNA vs mtDNA indicated that mtDNA may accumulate up to 15 times higher levels of this DNA oxidation product[56]. However, it was later established that this dramatic difference was a technical artefact[57]. Independent studies since confirmed that levels of 8-oxodG are similar in nDNA and mtDNA[58-60]. As far as sugar-phosphate backbone damage is concerned, Yakes and Van Houten[61] reported that in mouse embryonic fibroblasts exposed to H2O2, mtDNA accumulates more polymerase-blocking lesions than nDNA. These lesions are predominantly single- and double-strand breaks (SSB and DSB) as well as abasic sites with minor contribution from base modifications such as thymine glycol[50]. However, sugar-phosphate backbone damage may induce mtDNA turnover, thus preventing mutagenesis, rather than inducing it[48,50].
CAN MITOCHONDRIAL ROS INDUCE RELEVANT LEVELS OF MTDNA MUTATIONS?
Experimental evidence in support of the mutagenicity of mitochondrially produced ROS remains scarce. There are more studies attempting to assign oxidative stress as a cause of the observed mtDNA mutations than there are studies of mutations induced in mtDNA by experimental exposure of biological systems to oxidative stress. We were unable to detect a statistically significant increase in the level of mtDNA mutations in cells chronically treated with rotenone, which induces ROS production by inhibiting ETC complex I, and in cells repeatedly exposed to damaging levels of extracellular H2O2[50], which suggests that mtDNA is fairly resistant to ROS-induced mutagenesis. Similarly, recent studies indicate that mtDNA mutagenesis is not increased in flies with inactivated SOD and OGG1, an enzyme involved in the repair of oxidatively damaged DNA[62]. In aqueous environments, ionizing radiation induces DNA-damaging ROS: most importantly, the highly reactive •OH. With this in mind, Guo et al[63] evaluated 44 DNA blood samples from 18 mothers and 26 children. All mothers underwent radiation therapy for cancer in their childhood, and radiation doses to their ovaries were determined based on medical records and computational models. Sequencing of the entire mitochondrial genome in these patients revealed that the mother’s age at sample collection was positively correlated with mtDNA heteroplasmy, a condition in which the cell possesses more than one mtDNA variant (the mitochondrial equivalent of nuclear heterozygosity). However, Guo et al[63] failed to detect any significant difference in single nucleotide polymorphisms between mother and offspring. Also, there was no significant correlation between radiation dose to the ovaries and the level of heteroplasmic mtDNA mutations among mothers and children. Therefore, radiation therapy-induced ROS do not appear to contribute, in a substantial way, to mtDNA mutagenesis[63]. This finding is significant because radiation therapy, by design, produces levels of ROS that are much higher than those observed under physiological conditions and therefore have a higher potential to overwhelm cellular antioxidant defenses and produce oxidative damage.
PROPERTIES OF AGING-ASSOCIATED MTDNA MUTATIONS
It is of note that even though age-associated mtDNA mutations are randomly distributed around the genome, there is some bias for the type of mtDNA mutations observed in aging in mitotic vs post-mitotic tissues. In mitotic tissues, most common type of mtDNA mutations identified is base substitutions. In contrast, large-scale-deletions are more commonly identified in post-mitotic tissues[64]. Among point mutations in dividing cells, transitions dominate the spectrum (90%) with the remaining fraction of mutations almost equally divided between transversions and small deletions. The frequency of non-synonymous (65.4%) and frameshift/premature termination codons (16.5%) in aging cells is significantly elevated as compared with variants found in the general population (34% and 0.6%, respectively). Also, the predicted pathogenicity of aging-associated mtDNA mutations is higher than that of mutations in the general population[64]. This suggests that human somatic cells, unlike germline cells[65], lack mechanisms to protect them from the accumulation of deleterious mutations.
The advent of Next Generation Sequencing enabled cost-effective interrogation of large numbers of mtDNA bases for mutations. These analyses revealed a minimal contribution of G > T transversions to the spectrum of aging-associated mutations. G > T transversions can be induced by 8-oxodG, a frequently used measure of oxidative DNA damage. This has led some investigators to conclude that oxidative damage does not contribute to aging-associated mtDNA mutagenesis[64,66]. Some observations, however, caution against this interpretation: (1) The most frequent base substitution induced by oxidative stress is a G > A transition[67,68]. This is the most prominent base change observed in mtDNA from aged tissues[66]; (2) 8-oxodG in mammalian cells can also induce G > A transitions[69], and therefore the available evidence does not allow for the complete exclusion of the contribution of this lesion to mtDNA mutagenesis in aging; (3) Cumulative evidence suggests that oxidative stress can induce all possible base substitutions, both in vitro and in vivo[68], cautioning against basing a conclusion regarding the involvement of oxidative stress in age-related mtDNA mutagenesis solely on an increase in the frequency of G > T transversions. Therefore, in the absence of studies that determine the mutational signature of ROS in mtDNA, any mutation can be interpreted as resulting from oxidative stress. And, conversely, no particular mtDNA mutation can be used, with confidence, as evidence of oxidative stress; (4) It has been shown that oxidative DNA damage does not necessarily lead to an increase in G > T transversions. For example, in DNA oxidatively damaged in vitro and passed through bacterial cells, the frequency of G > C transversions was increased, whereas the frequency of G > T transversions was actually decreased as compared to that of untreated DNA[70]. In an almost identical experiment, the frequency of base substitutions at A/T pairs in oxidatively damaged DNA was elevated, whereas the frequency G > T transversions remained unchanged after passing damaged DNA through mammalian cells[42]; and (5) The specific spectrum of oxidative-damage induced DNA mutations is determined, to a great extent, by the particular properties of the experimental system used (reviewed in[67]). At present, we lack a precise understanding of how oxidative mtDNA lesions are processed by mitochondria to produce mutations. Therefore, no definitive conclusion regarding the contribution of oxidative stress to the spectrum of aging-associated mtDNA mutations can be drawn from the absence of an increase in G > T transversions.
WHAT IS THE FUNCTIONAL SIGNIFICANCE OF AGING-ASSOCIATED MTDNA MUTATIONS?
Given that mtDNA mutations accumulate with aging, are they a cause of (1) mitochondrial dysfunction and/or (2) aging? It is well established that mitochondrial function is only compromised when the fraction of cellular copies of a given mtDNA-encoded gene affected by a given mutation exceeds a certain threshold specific to the mutation (and tissue). This threshold phenomenon can be mediated, at least in part, by intra- and intermitochondrial complementation[71-73]. It is usually accepted that this threshold is 60% to 70% of mutant mtDNA in chronic progressive external ophthalmoplegia and may be close to 95% in the syndromes of mitochondrial encephalopathy, myopathy, lactic acidosis, and stroke-like episodes, and myoclonic epilepsy with ragged red fibers[74]. Therefore, generally, more than 60% of cellular copies for a given mitochondrial gene have to be affected by a pathogenic mutation in order to observe phenotypic manifestation of the mutation[75]. In aging, mtDNA mutations are random, which brings about two caveats. First, not all aging-associated mutations are detrimental. Because of the degeneracy of the genetic code, 25% of mutations will not alter the amino acid sequence of the encoded protein (68.8% of mtDNA encodes for proteins), and others, while causing an amino acid or nucleotide substitution, will not negatively affect the function of the encoded protein or RNA molecule. Second, these mutations are not localized to a particular gene, but rather are randomly distributed among 37 mitochondrially-encoded genes. This means that in order to affect 60% of cellular copies of the largest mtDNA-encoded polypeptide MT-ND5 (which spans 11% of the mitochondrial genome), each mtDNA molecule has to carry on average 0.6/0.11 = 5.45 mutations. For smaller genes, this number will be proportionally higher. Since there is no experimental evidence that supports a selective advantage for deleterious point mutations, both of these caveats suggest that the presence of several aging-associated mutations per mtDNA molecule is required before impairment of mitochondrial function can be observed. These levels are indeed achieved in tissues of mtDNA mutator mice[76,77], but not in naturally aged tissues of experimental animals or humans. Based on the reported frequency of mtDNA mutations, it can be calculated that in mice aged 24-33 mo, mutations affect as little as 20% of mtDNA molecules[78]. Similar calculations using reported values for humans aged 75-99 years[66] suggest that only about 32% of mtDNA molecules are affected by mutations. Therefore, it is highly unlikely that the relatively low mutation loads observed in naturally aged tissues[50,79,80] can account for the observed age-related measurable decline in mitochondrial function and, by extension, cause aging, provided that these mutations are maintained in a heteroplasmic state. Intriguingly, though, some studies indicate that the fraction of respiratory chain-deficient colonocytes in aging mammalian tissues increases after 35 years, and by 70 years of age, up to a third of colonocytes can be respiration-negative. This can be explained by a random genetic drift model. According to this model, multiple rounds of replication may result in the clonal expansion of random mtDNA molecules, leading to a loss of heteroplasmy[81]. In humans, this model predicts that clonal expansion may take decades to occur. Therefore, random drift may provide a satisfactory explanation for the mechanism of respiratory dysfunction observed in aged tissues provided that it can be demonstrated that cell types other than colon epithelium accumulate similar levels of clonally expanded mutations. The random genetic drift in colon epithelium, the tissue in which this phenomenon is best understood, however, appears to be highly heterogeneous, and its extent does not correlate well with chronological age between individuals. For example, a 75-year-old individual may have a lower percentage of respiration-deficient crypts than a 45-year-old[82]. This heterogeneity is inconsistent with the steady and relatively uniform process of aging, and, therefore, argues against random genetic drift being the sole or even a major driving force of aging. It is also unclear whether clonally expanded somatic mtDNA mutations can drive aging in short-lived species. For example, in human colon such mutations are not detectable until about 30 years of age[82]. Can clonally expanded mtDNA mutations explain aging in Caenorhabditis. elegans whose lifespan is only 2-3 wk? It is implausible that mtDNA in this organism turns over so much faster to allow for clonal expansion comparable to that observed in humans. Therefore, clonally expanded mtDNA mutations are more likely to be a contributing, rather than driving, factor of aging.
IS THERE EVIDENCE FOR THE EXISTENCE OF THE “VICIOUS” CYCLE?
As noted above, “vicious” cycle is the most contentious part of the MTA. The main premise of the “vicious” cycle hypothesis is the existence of a feed-forward cycle of ROS production and mtDNA mutation. That is: (1) increased ROS production in aging leads to increased mtDNA mutagenesis; and (2) increased mtDNA mutation loads result in increased mitochondrial dysfunction and ROS production. The first part of this premise appears intuitive and plausible. Indeed, no antioxidant defense or DNA repair system works with 100% efficiency, and an increase in ROS will inevitably lead to an increase in mtDNA damage and mutagenesis, however little. The second part of this premise, however, is more contentious. While observations in patients with mitochondrial disease may partially support the notion of increased ROS production in response to increased mtDNA mutation loads, these observations, paradoxically, also refute this notion. First, while some pathogenic mtDNA mutations result in increased ROS production[83,84], this is not a universal property of mutations in mtDNA. This point is best illustrated by observations made in “mito-mice” (mice that age prematurely due accumulation of random mtDNA mutations): these mice accumulate mtDNA mutation loads exceeding those observed in normal aging by more than one order of magnitude, and still this increase does not result in elevated levels of ROS production[76,77,85]. Thus, the majority of mtDNA point mutations will not affect mitochondrial ROS production regardless of their levels. Second, no accelerated aging or increase in mtDNA mutagenesis rates were reported in patients with mitochondrial diseases which are characterized by increased ROS production. Therefore, while increased ROS production is expected to increase the rate of mtDNA mutagenesis, this increase may not be physiologically relevant or experimentally detectable. This second point is relevant to the discussion above regarding threshold levels of mtDNA mutations.
Moraes et al[42] argued that if a “vicious” cycle played an important role in the accumulation of mtDNA deletions in somatic tissues, patients with compromised OXPHOS should accumulate mtDNA deletions at an accelerated rate. Their experiments did not support this prediction, leading Moraes et al[42] to the conclusion that a “vicious” cycle is not likely to play an important role in the accumulation of age-related mtDNA deletions[86].
To reconcile MTA with the new evidence, Gustavo Barja has put forward a new version of it that does not include the “vicious” cycle. Barja argues that the damage amplification step provided by the “vicious” cycle is unnecessary for the validity of the MTA[7].
ROS PRODUCTION AND LONGEVITY
It is predicted by the MTA that higher ROS production should lead to increased cellular oxidative stress, which should result in increased damage to cellular macromolecules including mtDNA, and ultimately lead to reduced longevity. Conversely, all other conditions being equal, lower ROS production and oxidative stress are expected to be associated with increased longevity. Since the principal contribution of the mtDNA to the aging process, within the framework of the MTA, is through the effects of mtDNA instability on cellular ROS production, it follows that an examination of the role of ROS in aging would be informative. Indeed, the lack of unequivocal evidence establishing a causative role for ROS in aging makes alterations in mtDNA, which are purportedly induced by ROS and contribute to aging by increasing ROS production, irrelevant.
Evidence from animal models
Early on, comparative biology studies established a positive correlation between body size and longevity. More detailed biochemical studies revealed an inverse correlation between mitochondrial ROS production and mtDNA damage on one hand and longevity on the other, across different biological taxa (reviewed in ref[7]), which is in agreement with the MTA. Unexpectedly, and conflicting with the predictions of the MTA, antioxidant defenses also correlated negatively with longevity[87]. Perhaps not surprisingly, an extension of this analysis to other species revealed that in many species, long lifespans defied explanation by the tenets of the MTA. One of the most striking examples in this category is that of the naked mole-rat. These animals, about the size of mice, live almost 8 times longer than mice[88,89]. Strikingly, these animals have very unremarkable antioxidant defenses: their glutathione peroxidase levels are 70 times lower than in mice, resembling those of knockout animals[88]. In the absence of compensatory upregulation of other antioxidant systems, this, predictably, leads to higher levels of oxidative damage in these animals: at least 10-fold higher levels of urinary isoprostanes (a marker of oxidative stress), eightfold increased levels of 8-oxodG (increased DNA damage) in the liver accompanied by reduced urinary excretion of 8-oxodG (reduced DNA repair), and high cellular (especially, mitochondrial) protein carbonyls were reported in this study[89]. The fact that naked mole-rats live longer than mice despite this increased oxidative burden (especially in mtDNA and mitochondrial proteins) strongly argues against the role of oxidative damage as a key determinant of longevity.
Another line of evidence against the MTA comes from studies on C. elegans. This organism has five genes encoding different isoforms of the SOD, an enzyme catalyzing the first step in the detoxification of superoxide (Eq. 2). Inactivation of the SOD isoforms in this organism either individually or in groups of three (including inactivation of all mitochondrial isoforms), failed to decrease the lifespan[90]. Instead, inactivation of sod-2 led to increased longevity, which was associated with increased oxidative damage to proteins. Moreover, an sod-2 mutation further increased lifespan of long-lived clk-1 mutants. Finally, the same group has recently inactivated all five sod genes in C. elegans and demonstrated that while animals completely lacking any SOD activity are more sensitive to multiple stressors, they have normal longevity[91]. Similarly, inactivation of the major mitochondrial antioxidant system by mutating Prx III (Eq. 4) decreased overall fitness in this organism, but failed to affect the lifespan[92].
In the fruit fly, somatic mtDNA mutagenesis was not affected by inactivation of SOD either alone, or in combination with OGG1, an enzyme involved in repair of oxidative DNA damage, even though lifespan was affected[62]. These observations suggest a minimal contribution of oxidative stress to age-related somatic mtDNA mutagenesis.
Mclk1+/- mice heterozygous for the key enzyme in the biosynthesis of ubiquinone, an electron transporter and mitochondrial membrane antioxidant, demonstrate extended longevity. This genetic defect is accompanied by an impairment of the ETC and by increased mitochondrial, but not cytoplasmic, oxidative stress[93]. Inactivation of the homologous gene clk-1 in C. elegans also resulted in increased longevity. This led the authors to hypothesize that an increase in the generation of mitochondrial ROS might accompany aging not because ROS play a causal role in this process but rather because ROS stimulate protective and restorative processes that help to counteract age-dependent damage[94,95].
Track record of antioxidant-based life-extending strategies
It is predicted by the MTA that reducing intracellular ROS production should reduce damage to macromolecules, including mtDNA, and ultimately increase longevity. As a result, numerous interventional studies have been performed in both vertebrate and invertebrate models. Treatments in these studies typically included either life-long supplementation with nonenzymatic antioxidants or genetic manipulation of intracellular levels of enzymatic antioxidants. These studies produced inconclusive results: while in some instances it was possible to achieve a modest increase in longevity, many studies revealed the lack of correlation, or even a negative correlation, between antioxidant defenses and lifespan (reviewed in ref[7]). In some instances, these studies produced different results in different species. For example, mitochondrial expression of catalase was reported to have no effect on the longevity of drosophila[96], but resulted in a modest (17%-21%) lifespan extension in mice[97]. In contrast, in C. elegans, a fivefold increase in longevity was reported for animals carrying two mutations (daf-2 and clk-1) in nDNA[98]. This suggests that nuclear genes play a pivotal role in determining longevity. To date, no manipulation of mtDNA or the systems involved in its replication, maintenance, or repair has produced comparable extension of the life-span.
Howes[99] reviewed the results of antioxidant studies which involved more than 550000 human subjects, and concluded that “not only have antioxidants failed to stop disease and aging but also they may cause harm and mortality, which precipitated the stoppage of several large studies”. Recent meta-studies support his findings: Bjelakovic et al[100] analyzed the results of 78 studies between 1977 and 2012, involving a total of 296707 participants, and concluded that antioxidant supplements neither reduce all-cause mortality nor extend lifespan, while some of them, such as beta carotene, vitamin E, and higher doses of vitamin A, may actually increase mortality[100]. The most direct interpretation of these findings in the context of the MTA as it pertains to mtDNA is that reduced oxidative damage to mtDNA does not extend longevity.
Caloric restriction (30%-40% reduction in caloric food intake without malnutrition) is frequently cited as the most reliable means of extending lifespan across diverse taxa and is frequently employed as a means to investigate the mechanisms of aging. Its effect is widely attributed to reduced ROS production and mtDNA damage[101]. However, in a recent survey of 41 laboratory mouse strains, 40% caloric restriction shortened lifespan in more strains than in which it lengthened it[102]. Similarly, a recent study by the National Institute of Aging revealed no beneficial effect of caloric restriction on longevity in primates[103,104].
CONCLUSION
Recently, there has been an emergence of experimental data challenging many aspects of the MTA as defined in the Introduction. This, in turn, has resulted in both a growing skepticism towards the role of mtDNA mutations in aging, and in the transformation of some of our views on mtDNA, ROS, and aging. Thus, the increased susceptibility of mtDNA to ROS-induced strand breaks (but not to oxidative base damage) is now viewed as a component of the mitochondria-specific mechanism for the maintenance of mtDNA integrity through abandonment and degradation of severely damaged mtDNA molecules, rather than as a mechanism for accelerated mtDNA mutagenesis (Figure 3). Also, we have begun to appreciate that increased ROS production in aging may represent evidence for adaptive signaling aimed at mitigating detrimental changes, rather than constituting an unwanted but unavoidable byproduct of respiration.
Even though its current status is controversial, it is the MTA that stimulated the research that advanced our understanding of aging and clarified the place of mtDNA in this process. While it is no longer plausible that mtDNA is either the sole or the main determinant of aging, epidemiological studies do still suggest a contribution of mtDNA variation to longevity[16]. Also, it is becoming increasingly obvious that maternally transmitted low levels of germline mtDNA mutations can have a significant impact on health and lifespan[105]. The random genetic drift theory[81] has the potential to reconcile the observed mitochondrial dysfunction in aged organs with the low average levels of mtDNA mutations in some tissues. These and other findings demonstrate that despite dramatic advances, our understanding of the role of mtDNA in aging remains incomplete. This incomplete understanding persists in large part due to our limited ability to manipulate mitochondria in a meaningful way. The lack of approaches to introduce defined base lesions into mtDNA impedes our progress in understanding the specifics of mitochondrial processing of oxidative DNA damage. This, in turn, limits our ability to deconvolute and interpret the spectrum of mtDNA mutations observed in aging.
In the near future there is great promise for further advances in our understanding of mtDNA’s contribution to aging. The advent of Duplex Sequencing methodology now makes it possible to determine the mutational signature of oxidative stress in mitochondria, which is one of the most important next steps in mtDNA research. The dire need for reliable markers of oxidative mtDNA damage is becoming increasingly obvious. Despite concerted efforts[106,107], detection of the widely used marker 8-oxodG remains variable between labs, which has resulted in contradictory reports: both a 20-fold increase[108] and no change[109] in 8-oxodG content in the mtDNA of OGG1 knockout animals have been reported. The development of methods for the determination of both the identity of mitochondrial ROS generated in vivo and the rates of their production would greatly aid in evaluating the interactions between mtDNA and ROS. Finally, a better understanding of the incidence, kinetics, and extent of random intracellular drift of mtDNA heteroplasmy in different tissues is needed for an accurate determination of its possible contribution to mitochondrial dysfunction in aging.
ACKNOWLEDGMENTS
The authors are grateful to Alexeeva O for critical reading of the manuscript.
Footnotes
Supported by The National Institutes of Health grants No. ES03456, PO1 HL66299, and No. OD010944
P- Reviewer: Lee HC S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Fleming JE, Miquel J, Cottrell SF, Yengoyan LS, Economos AC. Is cell aging caused by respiration-dependent injury to the mitochondrial genome? Gerontology. 1982;28:44–53. doi: 10.1159/000212510. [DOI] [PubMed] [Google Scholar]
- 4.Miquel J, Binnard R, Fleming JE. Role of metabolic rate and DNA-repair in Drosophila aging: implications for the mitochondrial mutation theory of aging. Exp Gerontol. 1983;18:167–171. doi: 10.1016/0531-5565(83)90009-8. [DOI] [PubMed] [Google Scholar]
- 5.Miquel J, Economos AC, Fleming J, Johnson JE. Mitochondrial role in cell aging. Exp Gerontol. 1980;15:575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs HT. The mitochondrial theory of aging: dead or alive? Aging Cell. 2003;2:11–17. doi: 10.1046/j.1474-9728.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 7.Barja G. Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts. Antioxid Redox Signal. 2013;19:1420–1445. doi: 10.1089/ars.2012.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexeyev MF. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009;276:5768–5787. doi: 10.1111/j.1742-4658.2009.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand FN, Kiely DK, Kannel WB, Myers RH. Family patterns of coronary heart disease mortality: the Framingham Longevity Study. J Clin Epidemiol. 1992;45:169–174. doi: 10.1016/0895-4356(92)90009-c. [DOI] [PubMed] [Google Scholar]
- 11.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova R, Lepage V, Charron D, Schächter F. Mitochondrial genotype associated with French Caucasian centenarians. Gerontology. 1998;44:349. doi: 10.1159/000022041. [DOI] [PubMed] [Google Scholar]
- 13.Bilal E, Rabadan R, Alexe G, Fuku N, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, et al. Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS One. 2008;3:e2421. doi: 10.1371/journal.pone.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexe G, Fuku N, Bilal E, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Bhanot G, et al. Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum Genet. 2007;121:347–356. doi: 10.1007/s00439-007-0330-6. [DOI] [PubMed] [Google Scholar]
- 15.Castri L, Melendez-Obando M, Villegas-Palma R, Barrantes R, Raventos H, Pereira R, Luiselli D, Pettener D, Madrigal L. Mitochondrial polymorphisms are associated both with increased and decreased longevity. Hum Hered. 2009;67:147–153. doi: 10.1159/000181152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raule N, Sevini F, Li S, Barbieri A, Tallaro F, Lomartire L, Vianello D, Montesanto A, Moilanen JS, Bezrukov V, et al. The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunits, not detected by haplogroup analysis, affects human longevity and is population specific. Aging Cell. 2014;13:401–407. doi: 10.1111/acel.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 20.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown GC, Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Gardner PR. Superoxide-driven aconitase FE-S center cycling. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 23.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 24.Drechsel DA, Patel M. Respiration-dependent H2O2 removal in brain mitochondria via the thioredoxin/peroxiredoxin system. J Biol Chem. 2010;285:27850–27858. doi: 10.1074/jbc.M110.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iñarrea P, Moini H, Han D, Rettori D, Aguiló I, Alava MA, Iturralde M, Cadenas E. Mitochondrial respiratory chain and thioredoxin reductase regulate intermembrane Cu,Zn-superoxide dismutase activity: implications for mitochondrial energy metabolism and apoptosis. Biochem J. 2007;405:173–179. doi: 10.1042/BJ20061809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henle ES, Luo Y, Gassmann W, Linn S. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxyguanosine family. J Biol Chem. 1996;271:21177–21186. [PubMed] [Google Scholar]
- 27.Henle ES, Luo Y, Linn S. Fe2+, Fe3+, and oxygen react with DNA-derived radicals formed during iron-mediated Fenton reactions. Biochemistry. 1996;35:12212–12219. doi: 10.1021/bi961235j. [DOI] [PubMed] [Google Scholar]
- 28.Rhee SG, Yang KS, Kang SW, Woo HA, Chang TS. Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid Redox Signal. 2005;7:619–626. doi: 10.1089/ars.2005.7.619. [DOI] [PubMed] [Google Scholar]
- 29.Merry TL, Tran M, Stathopoulos M, Wiede F, Fam BC, Dodd GT, Clarke I, Watt MJ, Andrikopoulos S, Tiganis T. High-fat-fed obese glutathione peroxidase 1-deficient mice exhibit defective insulin secretion but protection from hepatic steatosis and liver damage. Antioxid Redox Signal. 2014;20:2114–2129. doi: 10.1089/ars.2013.5428. [DOI] [PubMed] [Google Scholar]
- 30.Yoo SE, Chen L, Na R, Liu Y, Rios C, Van Remmen H, Richardson A, Ran Q. Gpx4 ablation in adult mice results in a lethal phenotype accompanied by neuronal loss in brain. Free Radic Biol Med. 2012;52:1820–1827. doi: 10.1016/j.freeradbiomed.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang TS, Cho CS, Park S, Yu S, Kang SW, Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J Biol Chem. 2004;279:41975–41984. doi: 10.1074/jbc.M407707200. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Kang YJ. Cellular and subcellular localization of catalase in the heart of transgenic mice. J Histochem Cytochem. 2000;48:585–594. doi: 10.1177/002215540004800502. [DOI] [PubMed] [Google Scholar]
- 33.Halliwell B, Aruoma OI. DNA damage by oxygen-derived species. Its mechanism and measurement in mammalian systems. FEBS Lett. 1991;281:9–19. doi: 10.1016/0014-5793(91)80347-6. [DOI] [PubMed] [Google Scholar]
- 34.Lesko SA, Lorentzen RJ, Ts’o PO. Role of superoxide in deoxyribonucleic acid strand scission. Biochemistry. 1980;19:3023–3028. doi: 10.1021/bi00554a029. [DOI] [PubMed] [Google Scholar]
- 35.Rowley DA, Halliwell B. DNA damage by superoxide-generating systems in relation to the mechanism of action of the anti-tumour antibiotic adriamycin. Biochim Biophys Acta. 1983;761:86–93. doi: 10.1016/0304-4165(83)90365-3. [DOI] [PubMed] [Google Scholar]
- 36.Blakely WF, Fuciarelli AF, Wegher BJ, Dizdaroglu M. Hydrogen peroxide-induced base damage in deoxyribonucleic acid. Radiat Res. 1990;121:338–343. [PubMed] [Google Scholar]
- 37.Brawn K, Fridovich I. DNA strand scission by enzymically generated oxygen radicals. Arch Biochem Biophys. 1981;206:414–419. doi: 10.1016/0003-9861(81)90108-9. [DOI] [PubMed] [Google Scholar]
- 38.Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Nucleic acid oxidation in Alzheimer disease. Free Radic Biol Med. 2008;44:1493–1505. doi: 10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Santos RX, Correia SC, Zhu X, Smith MA, Moreira PI, Castellani RJ, Nunomura A, Perry G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer’s disease. Antioxid Redox Signal. 2013;18:2444–2457. doi: 10.1089/ars.2012.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith JB, Cusumano JC, Babbs CF. Quantitative effects of iron chelators on hydroxyl radical production by the superoxide-driven fenton reaction. Free Radic Res Commun. 1990;8:101–106. doi: 10.3109/10715769009087980. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Abe Y, Kanagawa K, Shoji T, Mashino T, Mochizuki M, Tanaka M, Miyata N. Iron-chelating agents never suppress Fenton reaction but participate in quenching spin-trapped radicals. Anal Chim Acta. 2007;599:315–319. doi: 10.1016/j.aca.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Moraes EC, Keyse SM, Pidoux M, Tyrrell RM. The spectrum of mutations generated by passage of a hydrogen peroxide damaged shuttle vector plasmid through a mammalian host. Nucleic Acids Res. 1989;17:8301–8312. doi: 10.1093/nar/17.20.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatarenkov A, Avise JC. Rapid concerted evolution in animal mitochondrial DNA. Proc Biol Sci. 2007;274:1795–1798. doi: 10.1098/rspb.2007.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 46.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorslund T, Sunesen M, Bohr VA, Stevnsner T. Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst) 2002;1:261–273. doi: 10.1016/s1568-7864(02)00003-4. [DOI] [PubMed] [Google Scholar]
- 48.Bendich AJ. DNA abandonment and the mechanisms of uniparental inheritance of mitochondria and chloroplasts. Chromosome Res. 2013;21:287–296. doi: 10.1007/s10577-013-9349-9. [DOI] [PubMed] [Google Scholar]
- 49.Bendich AJ. Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol Direct. 2010;5:42. doi: 10.1186/1745-6150-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010;38:e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst) 2012;11:684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shokolenko IN, Wilson GL, Alexeyev MF. Persistent damage induces mitochondrial DNA degradation. DNA Repair (Amst) 2013;12:488–499. doi: 10.1016/j.dnarep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clayton DA, Doda JN, Friedberg EC. Absence of a pyrimidine dimer repair mechanism for mitochondrial DNA in mouse and human cells. Basic Life Sci. 1975;5B:589–591. doi: 10.1007/978-1-4684-2898-8_26. [DOI] [PubMed] [Google Scholar]
- 56.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci USA. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anson RM, Sentürker S, Dizdaroglu M, Bohr VA. Measurement of oxidatively induced base lesions in liver from Wistar rats of different ages. Free Radic Biol Med. 1999;27:456–462. doi: 10.1016/s0891-5849(99)00091-x. [DOI] [PubMed] [Google Scholar]
- 59.Anson RM, Hudson E, Bohr VA. Mitochondrial endogenous oxidative damage has been overestimated. FASEB J. 2000;14:355–360. doi: 10.1096/fasebj.14.2.355. [DOI] [PubMed] [Google Scholar]
- 60.Lim KS, Jeyaseelan K, Whiteman M, Jenner A, Halliwell B. Oxidative damage in mitochondrial DNA is not extensive. Ann N Y Acad Sci. 2005;1042:210–220. doi: 10.1196/annals.1338.023. [DOI] [PubMed] [Google Scholar]
- 61.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, Cardozo-Pelaez F, Pallanck LJ. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014;10:e1003974. doi: 10.1371/journal.pgen.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo Y, Cai Q, Samuels DC, Ye F, Long J, Li CI, Winther JF, Tawn EJ, Stovall M, Lähteenmäki P, et al. The use of next generation sequencing technology to study the effect of radiation therapy on mitochondrial DNA mutation. Mutat Res. 2012;744:154–160. doi: 10.1016/j.mrgentox.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greaves LC, Elson JL, Nooteboom M, Grady JP, Taylor GA, Taylor RW, Mathers JC, Kirkwood TB, Turnbull DM. Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet. 2012;8:e1003082. doi: 10.1371/journal.pgen.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Termini J. Hydroperoxide-induced DNA damage and mutations. Mutat Res. 2000;450:107–124. doi: 10.1016/s0027-5107(00)00019-1. [DOI] [PubMed] [Google Scholar]
- 68.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 69.Kamiya H, Miura K, Ishikawa H, Inoue H, Nishimura S, Ohtsuka E. c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res. 1992;52:3483–3485. [PubMed] [Google Scholar]
- 70.Akasaka S, Yamamoto K. Hydrogen peroxide induces G: C to T: A and G: C to C: G transversions in the supF gene of Escherichia coli. Mol Gen Genet. 1994;243:500–505. doi: 10.1007/BF00284197. [DOI] [PubMed] [Google Scholar]
- 71.Legros F, Malka F, Frachon P, Lombès A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 72.Nakada K, Sato A, Hayashi J. Mitochondrial functional complementation in mitochondrial DNA-based diseases. Int J Biochem Cell Biol. 2009;41:1907–1913. doi: 10.1016/j.biocel.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 73.Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- 74.Nonaka I. Mitochondrial diseases. Curr Opin Neurol Neurosurg. 1992;5:622–632. [PubMed] [Google Scholar]
- 75.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 77.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 78.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 79.Khrapko K, Kraytsberg Y, de Grey AD, Vijg J, Schon EA. Does premature aging of the mtDNA mutator mouse prove that mtDNA mutations are involved in natural aging? Aging Cell. 2006;5:279–282. doi: 10.1111/j.1474-9726.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- 80.Song X, Deng JH, Liu CJ, Bai Y. Specific point mutations may not accumulate with aging in the mouse mitochondrial DNA control region. Gene. 2005;350:193–199. doi: 10.1016/j.gene.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 81.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 84.Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, Wiedmann M, Manfredi G. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 85.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci USA. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tengan CH, Gabbai AA, Shanske S, Zeviani M, Moraes CT. Oxidative phosphorylation dysfunction does not increase the rate of accumulation of age-related mtDNA deletions in skeletal muscle. Mutat Res. 1997;379:1–11. doi: 10.1016/s0027-5107(97)00076-6. [DOI] [PubMed] [Google Scholar]
- 87.Pamplona R, Costantini D. Molecular and structural antioxidant defenses against oxidative stress in animals. Am J Physiol Regul Integr Comp Physiol. 2011;301:R843–R863. doi: 10.1152/ajpregu.00034.2011. [DOI] [PubMed] [Google Scholar]
- 88.Andziak B, O’Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–1212. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 89.Lewis KN, Andziak B, Yang T, Buffenstein R. The naked mole-rat response to oxidative stress: just deal with it. Antioxid Redox Signal. 2013;19:1388–1399. doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci USA. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ranjan M, Gruber J, Ng LF, Halliwell B. Repression of the mitochondrial peroxiredoxin antioxidant system does not shorten life span but causes reduced fitness in Caenorhabditis elegans. Free Radic Biol Med. 2013;63:381–389. doi: 10.1016/j.freeradbiomed.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 93.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J Biol Chem. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hekimi S. Enhanced immunity in slowly aging mutant mice with high mitochondrial oxidative stress. Oncoimmunology. 2013;2:e23793. doi: 10.4161/onci.23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mockett RJ, Bayne AC, Kwong LK, Orr WC, Sohal RS. Ectopic expression of catalase in Drosophila mitochondria increases stress resistance but not longevity. Free Radic Biol Med. 2003;34:207–217. doi: 10.1016/s0891-5849(02)01190-5. [DOI] [PubMed] [Google Scholar]
- 97.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 98.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 99.Howes RM. The free radical fantasy: a panoply of paradoxes. Ann N Y Acad Sci. 2006;1067:22–26. doi: 10.1196/annals.1354.004. [DOI] [PubMed] [Google Scholar]
- 100.Bjelakovic G, Nikolova D, Gluud C. Antioxidant supplements to prevent mortality. JAMA. 2013;310:1178–1179. doi: 10.1001/jama.2013.277028. [DOI] [PubMed] [Google Scholar]
- 101.Barja G. Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biol Rev Camb Philos Soc. 2004;79:235–251. doi: 10.1017/s1464793103006213. [DOI] [PubMed] [Google Scholar]
- 102.Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010;9:92–95. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Austad SN. Ageing: Mixed results for dieting monkeys. Nature. 2012;489:210–211. doi: 10.1038/nature11484. [DOI] [PubMed] [Google Scholar]
- 105.Ross JM, Stewart JB, Hagström E, Brené S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.European Standards Committee on Oxidative DNA Damage (ESCODD) Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic Biol Med. 2003;34:1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 107.Gedik CM, Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 108.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 109.Trapp C, McCullough AK, Epe B. The basal levels of 8-oxoG and other oxidative modifications in intact mitochondrial DNA are low even in repair-deficient (Ogg1(-/-)/Csb(-/-)) mice. Mutat Res. 2007;625:155–163. doi: 10.1016/j.mrfmmm.2007.06.006. [DOI] [PubMed] [Google Scholar]


