Abstract
Growing experimental evidences suggest that dimerization and oligomerization are important for G Protein-Coupled Receptors (GPCRs) function. The detailed structural information of dimeric/oligomeric GPCRs would be very important to understand their function. Although it is encouraging that recently several experimental GPCR structures in oligomeric forms have appeared, experimental determination of GPCR structures in oligomeric forms is still a big challenge, especially in mimicking the membrane environment. Therefore, development of computational approaches to predict dimerization of GPCRs will be highly valuable. In this review, we summarize computational approaches that have been developed and used for modeling of GPCR dimerization. In addition, we introduce a novel two-dimensional Brownian Dynamics based protein docking approach, which we have recently adapted, for GPCR dimer prediction.
Keywords: Membrane protein dimerization, Computer modeling, Protein docking, Brownian dynamics simulations, Molecular dynamics simulations, Coarse Grained MD simulations
Introduction
GPCRs constitute the largest family of cell surface receptors for a diverse range of ligands, including ions, hormones, neurotransmitters, odorants, tastants and light, and transduce signals to initiate cellular activities 1. GPCRs are important drug targets for various diseases. There are about 50-60% approved drugs eliciting their therapeutic effects by regulating GPCR activities 2. Growing experimental evidences suggest that GPCRs function either as homodimers, heterodimers or higher oligomers 3-5. This highlights the importance of research on prediction of the interface of dimerization. Experimental approaches, including cysteine crosslinking, coimmunoprecipitation, western blot analysis, fluorescence resonance energy transfer (FRET), and bioluminescence resonance energy transfer (BRET) were extensively applied to study dimerization and oligomerization of GPCRs, and provided convincing evidences of dimeric or oligomeric GPCR formations. However, it is very difficult to construct structure models of dimeric or oligomeric GPCRs in a detailed molecular level based on the results from these experimental approaches 6-11. Structural approaches, such as cryo-electron microscopy (cryo-EM) and atomic force microscopy (AFM) were used to obtain low resolution oligomeric structure information of rhodopsins. The projection structures of rhodopsins based on the two-dimensional crystals from cryo-EM12-14, and the organization of rohodopsin in native membranes from AFM14-16, provided valuable information of arrangement of rhodopsin in vivo. Molecular models of rohodopsin in oligomeric forms were proposed based on by fitting the crystal structure into the density maps from the AFM experiments17. Recent studies using direct biophysical techniques (Fluorescence Correlation Spectroscopy (FCS) with photon counting histogram (PCH) analysis and Total Internal Reflection Fluorescence (TIRF)) have provided the first conclusive demonstration of GPCR homodimers at the single molecule level 18,19.
Crystallography also contributed to our knowledge of the GPCR dimerization. Recently several GPCR structures in oligomeric forms have appeared, including Opsin (PDB: 3CAP) 20, CXCR4 chemokine receptor (PDB: 3ODU) 21, metarhodopsin II (PDB: 3PXO) 22, μ-opioid receptor (PDB: 4DKL) 23, κ-opioid receptor (PDB: 4DJH) 24, and β1-adrenergic receptor (PDB: 4GPO) 25. In addition to oligomer GPCR structures, even more monomer structures of GPCRs were obtained by crystallography. Emergence of the GPCR structures paved the way to conduct structure-based prediction of GPCR dimerization by computational methods, such as protein-protein docking, molecular dynamics (MD) simulations and coarse grained MD (CGMD) simulation. In addition, sequence-based approaches, as the other branch of computational methods, can be used to complement structure-based approaches to study the GPCR oligomerization. However, prediction of GPCR oligomer interfaces by computational approaches is still a very challenging task that is currently not fully resolved. The previous reviews 26,27 and books 5,28 well summarized the research area of GPCR oligomerization using computational approaches. Here we review some more recent progresses in this field.
Oligomeric Interface from GPCR Crystal Structures
Simply repeating asymmetric units from crystal structures may generate oligomeric forms of GPCR with one or more interfaces (Table 1 and Figure 1). Some of these interfaces might be artificial and not represent functional biological assembly. However, it still provides us a possible scenario of how GPCRs interact with each other.
Table 1.
Crystal Structure Packing Dimer Interface
| PDB id | Name Description | Resolution (Å) | Interface | Ref. |
|---|---|---|---|---|
| 1N3M | Rhodopsin, Semi-empirical model based on AFM | TM1/TM2/H8, TM4/TM5, TM1/TM2 - CL3 | Liang, 2003, 17 | |
| 2I35, 2I36 | Rhodopsin | 3.80, 4.10 | TM1/TM2/H8 | Salom, 2006, 29 |
| 2I37 | Photoactivated rhodopsin | 4.15 | TM1/TM2/H8 | Salom, 2006, 29 |
| 2RH1 | β2-AR | 2.40 | TM1 - H8 | Cherezov, 2007, 95 |
| 3CAP | Opsin | 2.90 | TM1/TM2/H8 | Park, 2008, 20 |
| 2Z73 | Squid Rhodopsin | 2.50 | TM4/TM5, TM5 | Murakami and Kouyama, 2008, 32 |
| 3ODU | CXCR4 | 2.50 | TM5/TM6 | Wu, 2010, 21 |
| 3OE(0, 6, 8, 9) | 2.90 - 3.20 | |||
| 3RZE | Histamine H1 receptor | 3.10 | TM4 | Shimamura, 2011, 33 |
| 3PXO | metarhodopsin II | 3.00 | TM1/TM2/H8 | Choe, 2011, 22 |
| 4DKL | HOR | 2.80 | TM1/TM2/H8, TM5/TM6 | Manglik, 2012, 23 |
| 4DJH | κOR | 2.90 | TM1/TM2/H8 | Wu, 2012, 24 |
| 4GPO | β1-AR | 3.50 | TM1/TM2/H8, TM4/TM5 | Huang, 2013, 25 |
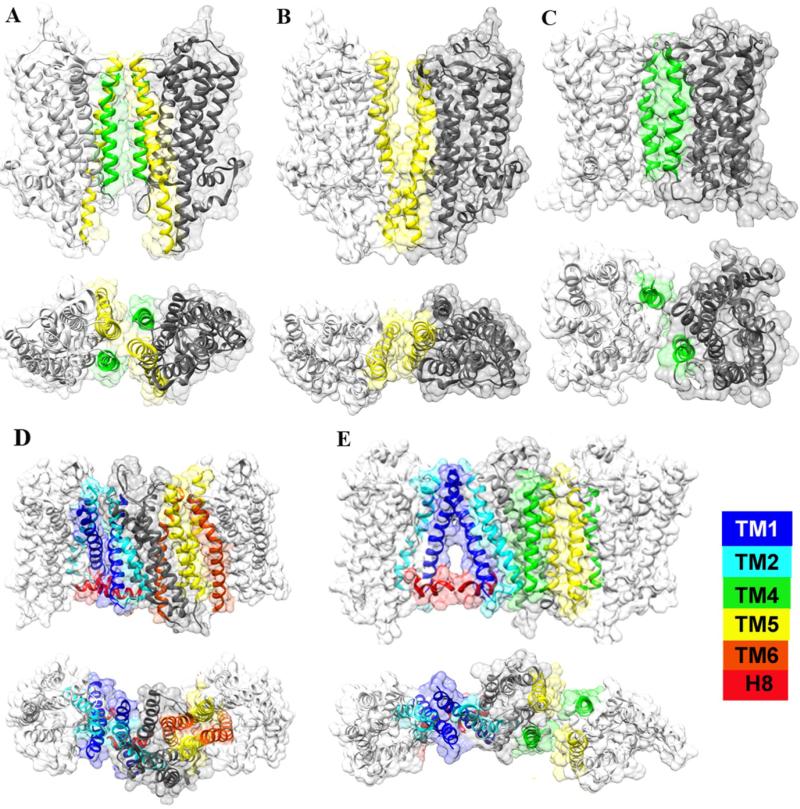
Fig. 1.
GPCR Oligomeric Interface. A. Squid rhodopsin, TM4/TM5-TM4/TM5 (PDBID:2Z73); B. Squid rhodopsin, TM5-TM5 (PDBID:2Z73); C. Histamine H1 receptor, TM4-TM4 (PDBID: 3RZE); D. μ-OR, TM1/TM2/H8-TM1/TM2/H8, TM5/TM6-TM5/TM6 (PDBID:4DKL); E. β1-AR homo oligomers, TM1/TM2/H8-TM1/TM2/H8 , TM4/TM5-TM4/TM5 (PDBID: 4GPO). (Color coded figures are available for the web version of this article)
The first semi-empirical model of the dimer of rhodopsin and opsin (PDB: 1N3M) established three interfaces, TM1/TM2/H8 - TM1/TM2/H8, TM4/TM5 - TM4/TM5 and TM1/TM2 - CL3 (cytoplasmic loop connecting helices V and VI) 17. Interfaces involving the TM1, TM2 and H8 are then repeatedly observed in the crystal structures of rhodopsin 29, opsin 30, metarhodopsin II 22, μOR 23, κOR 24, and β1-AR 25, indicating a relative conserved interface in various GPCRs. The interface in the 1N3M shows loose contact with a buried surface area (BSA) 146 Å2 while it has more contact in the crystal structures, e.g. 615 Å2 in the μOR structure (PDB: 4DKL). Considering the BSA, TM1/TM2/H8 is not the dominant interface in most cases. A recent computation study on rhodopsin indicates that the most stable dimers display a symmetric TM1/TM2/H8 interface though the interface has relatively small BSA 31. That study, on the one hand, highlights the significance of the TM1/TM2/H8 interface; but challenges the utility of the BSA as a predictor to measure the strength of membrane protein-protein interaction on the other hand 31. The TM4/TM5 interface appears similar in the three structures, Semi-empirical model 1N3M17, squid rhodopsin 2Z73 32 and β1-AR 4GPO 25. TM5 and TM6 are also found to form interface in the structures of CXCR4 21 and μOR 23. Yet they are quite different interfaces with distinct contact area. Additionally, the histamine H1 receptor structure has a unique TM4 - TM4 interface 33; one TM5 - TM5 interface is formed in squid rhodopsin structure 2Z73 32.
Sequence-based Approaches
Bioinformatics techniques can be used to predict dimer interfaces of GPCRs. These techniques include: Evolutionary Trace (ET) 34, Correlated Mutation Analysis (CMA) 35,36, Subtractive Correlated Mutations (SCM), and hidden-site class evolutionary model 37. Those sequence-based approaches have been extensively reviewed elsewhere 27,38-40. It has been pointed out that although sequence-based approaches have been successfully applied for prediction of GPCR dimerization, they could be more valuable when combined with other structural based approaches for their predictions 26, especially, as more high resolution crystal structures of GPCRs either in monomers or oligomers become available.
All-atom Molecular Dynamics Simulations
All-atom MD simulation is widely used to describe the dynamic behavior of biomacromolecules at atomic level. Increased availability of GPCR crystal structures facilitates performing reliable MD simulations for the study of many aspects of GPCR, including dimerization/oligomerization and relevant issues. The utilization of MD simulations on GPCRs has been recently reviewed 41. Recently a website pipeline, GPCRModSim (http://gpcr.usc.es) server, was developed to automatically construct a model of a GPCR based on available structure templates, and conduct all atom MD simulations on a single GPCR or a GPCR dimer 42. The server could significantly reduce the time and effort needed to perform modeling and simulating study on GPCRs.
To explore the role of the dimer form of rhodopsin in signal transduction, Neri et al. 43 conducted MD simulations based on its AFM dimer model 15. Rhodopsin, as the light detector, is bound with 11-cis retinal in the dark state. Photoinduced isomerization of the retinal from 11-cis to all-trans inside the binding pocket leads to a cascade of conformational changes related to the receptor activation and downstream signaling. In the simulation, a harmonic restraint was applied to enforce the isomerization process of the retinal in one subunit of the dimer, Rho*. Four independent MD simulations on the Rho*-Rho dimer indicated a tandem activation mechanism in which the light sensor Rho* induces conformational changes in the other monomer Rho where the “ionic lock” between R135 in TM3 and E247 in TM6 breaks triggering evolution toward the meta-II state. Dimer interface of the rhodopsin in this model involves TM4, TM5, part of TM3 and intracellular loop 2 (IL2). After isomerization, TM4* (TM4 in the Rho*) translated rigidly away from the dimer interface, resulting in a tilt of the C-terminal of TM3 (C-terminal of TM3 is anchored by the N-terminal of TM4*). The tilt of TM3 caused the breakage of the “ion lock” in the dark subunit Rho, which evolves towards an opsin-like active configuration.
100 ns MD simulations were conducted based on two available crystal structures of CXCR4 homodimer (PDB: 3ODU and 3OE0) 44. In the 3ODU and 3OE0, CXCR4 was co-crystallized with a small-molecular antagonist IT1t and with a cyclic peptide inhibitor CVX15, respectively. The dimerization interfaces involving TM5 and TM6 in both structures are essentially identical. The main difference between the two structures are in the intracellular side of the dimer where 3OE0 presents contacts that 3ODU does not have 21. In addition to the crystallized receptors which contain mutations to stabilize the dimer conformation (L1253.41W in the 3ODU and L1253.41W/ T2406.36P in the 3OE0), corresponding WT variants were also constructed to conduct MD simulations, which produced four comparable simulation systems. The buried surface area (BSA) of the dimerization interface increases in all systems from 10% (3OE0 WT) to 75% (3ODU mutant system) after 40 ns simulations, indicating the enlargement of the dimerization interfaces. And the relative approach of the protomers is mainly located in the intracellular side of the interface in all cases. On the other hand, the BSAs in the mutant systems increase more and the mutant systems reach the stable phase faster than their corresponding WT systems, supporting the stabilization effect of the mutations on the dimerization. Simulations on 3ODU also suggest that a relative rotation between the two protomers occurs, moving the TM4 from each of the two monomers close to each other. Such movement does not occur in the 3OE0-based simulations. In addition, a set of novel hydrogen bonds which stabilizes the intracellular contacts of the 3OE0-based structures are identified, involving Tyr1353.51 and His1403.56(IL2), Arg1464.35 and Ser2255.63 /Ser2285.66. These interactions could be further validated by experiment. A review concerning the CXCR4 structure-based studies is also available 45.
Heterodimeric complex of the metabotropic glutamate receptor type-2 (mGluR2) and 5HT2A serotoninergic receptor (2AR) has been reported to be a new target for the treatment of psychosis and shows distinct functional behavior from their monomer 46. Their signaling mechanism induced by antipsychotic drugs was also uncovered recently 47. No crystal structures of heteromeric GPCRs are available so far. Bruno et al 48 built homology models of the mGluR2 and 2AR, respectively, and constructed the heterodimeric complex. Sampling of dimer poses was conducted by Rosetta program. And the final complex model was selected by visual inspection because the scoring function was unable to correctly sort the interface disposition. Using experimental data as a guide, they selected a complex model with the TM4/TM5 interface which resembles the interface in the semi-empirical model of rhodopsin (PDB: 1N3M). A 40 ns MD simulation was then performed on the complex. Results displayed the formation of the dimerization interface between the two protomers that allosterically affects the shape of the binding pocket of the individual protomers.
Coarse Grained Molecular Dynamics Simulations on GPCRs
Compared to all-atom MD, the CGMD method is capable of simulation of larger systems and longer simulation time at the price of lower resolution and accuracy. CGMD thus has an advantage in simulating oligomerization of GPCR involving couples of receptors in the system. Periole et al. utilized large scale CGMD to study the self-assembly behavior of rhodopsin 31. Their simulation contains 64 receptors embedded in lipid bilayer at a protein-tolipid ratio of 1:100. Predominant dimer interface is a symmetric arrangement involving TM1/TM2 on extracellular side and amphipathic Helix8 on intracellular side. Additional interfaces TM5-TM5, TM4/TM5-TM4/TM5 are also observed in the simulations. All of these interfaces together with interfaces TM4-TM6 and TM4-TM4 are further investigated using umbrella sampling/potential of mean force (PMF) calculations. Free energy surface of PMFs confirms that the symmetric TM1/TM8/H8 interface leads to a stable dimer configuration with a deep free energy well around 13 kcal/mol and present no energy barrier for protomers binding with each other; In contrast, when forming TM4-TM6 and TM4-TM4 interfaces, an energy barrier 2.4−3.6 kcal/mol exist for binding. A model of the rows-of-dimers organization based on AFM images was also constructed in this study. According to this high protein density simulations, TM4, TM5 and TM6 may form interfaces related to high-order oligomerization of rhodopsin.
By conducting 18 microsecond CGMD simulations based on crystal structure 2RH1, Mondal et al. 49 demonstrated that spatial organization of the β2-AR receptor was dependent on the pattern of protein-membrane hydrophobic mismatch in the monomer. Oligomerization of the monomers at specific interfaces involving TM1, TM4 and TM5 eliminated the residual hydrophobic mismatch and thus reduced the energy penalty. Another study 50 also used CGMD simulations to study self-assembly of the β2-AR receptor based on the crystal structure 3SN6. 16 monomers of β2-AR inserted in DSPC lipid bilayer formed dimers in which 6 distinct dimeric units were observed and these dimers further assembled to form higher oligomeric clusters. TM1-TM1, H8-H8, TM1/TM5- TM1/TM5 and TM6-TM6 were involved in forming interfaces in which TM1/TM1 and H8/H8 form the most stable dimerization interface.
A recently published study investigated how cholesterols mediate the dimerization of β2-AR receptor 51 by CGMD simulations. Several GPCRs are shown to be influenced by cholesterol in the membrane 52-61. And the binding sites of cholesterol in various GPCRs were also investigated by simulations studies 62-64. In this CGMD simulation, the β2-AR receptors were immersed in POPC lipids in the presence of increasing concentrations of cholesterol: 0%, 9%, 30%, and 50%. Interestingly, in the absence of cholesterol dimer formed with the TM4/TM5-TM4/TM5 interface dominating during the simulation; with increasing concentration of cholesterol (9% and 30%), they observed interfaces involving the TM4/TM5 of one protomer and the TM1/TM2 of the other; when concentration of cholesterol reaches to 50%, the TM1/TM2-TM1/TM2 interface was formed between the receptors. This progressive change of dimer interface, according to the simulation, is due to cholesterol occupancy at TM4, which restricts its involvement in the dimer interface. Occupancy of the cholesterol binding site on TM4 is stochastic and dependent on the membrane cholesterol concentration.
A series of studies conducted by the Filizola group utilized a protocol of coarse-grained biased MD simulations to estimate dimerization free-energy in GPCRs 65-67. The free energy surface along the reaction coordinates (the distance between the center of mass (COM) of protomers or the rotational angle θ of interested TMs) was obtained and further used to calculate the dimerization constant KD and estimate the lifetime of the dimer. Simulations on the β1-AR and β2-AR homodimers indicated that both homodimers with TM1/H8 interface were more stable and long-lived than interfaces involving TM4/TM3. The β2-AR homodimer involving TM1/H8 interface can last minutes based on estimation derived from the calculated free energy. In contrast, both homodimers involving TM4/TM3 interface appeared transient with estimated lifetime shorter than milliseconds.
Protein-protein Docking
Compared to the MD simulations, the protein-protein docking method is able to generate possible dimer configurations with short time and low computational cost. The generated complexes provide structural information/residue interacting pairs that can be used to design experiments. Vice versa, preliminary experimental data was also utilized as constraint to filter the docking poses.
Based on the validation study by Kaczor et al. 68, several popular protein-protein docking programs widely used to predict structure of water-soluble protein complexes cannot be expected to give fairly good performance to transmembrane proteins, as the desolvation energy term incorporated in these docking algorithms and scoring functions are biased towards water-soluble proteins 68,69. They selected eleven transmembrane proteins, including ion channels, transporters and GPCRs (opsin, CXCR4 and κOR dimers), all with oligomer crystal structure available. Even for such redocking experiment, GPCR dimers are still tough targets and only low quality results were obtained. Analysis of all the cases indicted that the average docking success generally correlates to a ratio between protein interface and total surface (%IST). The GPCR dimers with low %IST (< 3.7%) were predicted by the average best RMSD in the range of 7-21 Å. Of those tested programs (ZDOCK, ClusPro, HEX, GRAMM-X, PatchDock, SymmDock and HADDOCK), GRAMM-X shows the best performance, as it is the only tool that uses additional information, such as an evolutionary conservation term in the scoring function. In addition to the small area of interface, interfaces of GPCRs do not have obvious geometric complementarity that usually exists in the protein-protein interface. That is another reason causing the difficulty of predicting GPCR interfaces compared to the other transmembrane proteins tested.
All default settings of the docking programs were used in this study, in order to test the usability of the programs to transmembrane proteins. Since program parameters are derived from water-soluble proteins it is not surprising to get low-quality results under such conditions.
Another study conducted by Casciari et al. presented a practical procedure of membrane protein docking 69. The approach consists of rigid-body docking samplings, membrane topology-based filtering and cluster analysis. The rigid-body docking is performed by ZDOCK, starting with one monomer against the other. Each docking run for one monomer is followed by the filtering and clustering until the whole oligomer model is constructed. For all these systems, predictions lead to native-like structures with Cα-RMSDs lower than 2.5 Å from the native oligomer. Two strategies adopted in this approach deserve to be noticed: neglecting the desolvation term as it has not been parameterized to the membrane environment; and removing the solutions that have inappropriate orientation with respect to the membrane, namely, membrane topology-based filtering. It was reported that >94% solutions within the 4000 structures generated in each docking run can be discarded by the filtering step, which ‘concentrates’ the correct poses. Usually only a few groups can be obtained by the subsequent cluster analysis, with the best solutions in the main cluster, further emphasizing the goodness of the membrane topology-based filter. This approach was then used to predict the dimerization of the lutropin receptor 70, neurotensin 1 receptor 71, A2A adenosine receptor 72, and thromboxane A2 receptor 73. In the docking studies of neurotensin 1 receptor and A2A adenosine receptor, a possible dimer interface involving the CL3 was found: CL3-CL2 in the homodimer of neurotensin 1 receptor and CL3-H8 in the homodimer of A2A receptor. Several lines of experimental evidence have pointed out that cytoplasmic loops and C-terminus may play important roles in GPCR heteromerization; e.g. heteromers of D5-D2 dopamine receptors, adenosine A2A-D2, cannabinoid CB1-A2A, CB1-D2, and A2A-CB1-D2, can be formed in the cell by means of electrostatic interactions between cytoplasmic loops and C-terminus 74,75. Yet crystal structures of GPCRs usually miss N- and C-terminus and cytoplasmic loops (especially lack of CL3 which is replaced by fused T4 lysozyme). Computational modeling and simulation methods should take more care of these domains for understanding the molecular mechanism of the GPCR heteromerization.
Two-dimensional Brownian Dynamics (2D-BD) Simulations
The docking feature of Brownian Dynamics (BD) method76 can be used to predict protein-protein interactions interactions 77-83. Previously, we successfully predicted the interactions between potassium channels and various scorpion toxins using the BD simulations. We found that the interactions between potassium channels and scorpion toxins are mainly from electrostatics. Although the original MacroDox (BD program) considers electrostatic interactions only between the two proteins, the program was able to predict the channel-toxin complexes correctly 84,85. For example, the complex of potassium channel (KcsA) and scorpion toxin (Lq2) we predicted is very similar to the complex solved later by NMR experiments 84,86. However, in general, protein docking methods need to include other short-range interaction terms for scoring and ranking of the results correctly. Therefore, we adapted the original MacroDox program to include additional Van der Waals, and desolvation energy terms87. The performance of the adapted MacroDox program was evaluated by prediction of a test set of protein complexes, for which the crystal structures are available, and obtained very encouraging results 88.
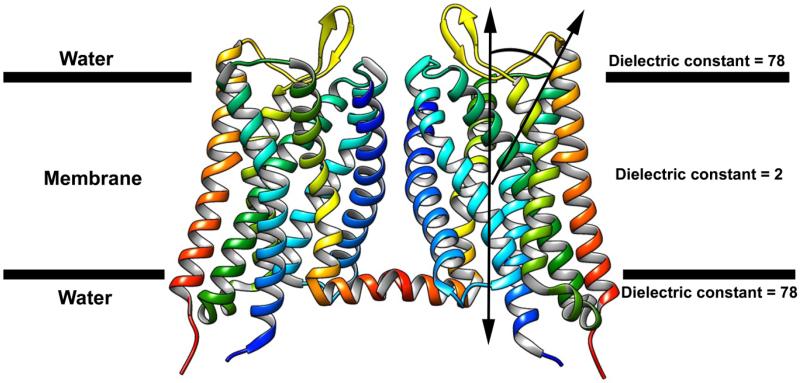
Like most docking programs, the BD program is not suitable to simulate the association of membrane proteins, which are in a special hybrid of a water/membrane environment (Fig. 2). In addition, sampling the entire 3D space for membrane proteins is computationally wasteful, and inefficient, since the movement of membrane proteins is restricted in a nearly planar environment of the membrane (2D). To predict dimerization of membrane proteins, we have adapted the BD program (MacroDox) to include a hybrid electrostatic potential map of membrane and water for electrostatic interaction calculations, and restriction of sampling within 2D space. We added post-docking refinement accounting for flexibility of proteins and rescored. Our adapted 2D- BD program was successfully used to predict the dimerization of the Outer Membrane Phospholipase A (OMPLA) and glycophorin A (GPA) 89.
Fig. 2.
Hybrid electrostatic potential map of membrane and water for electrostatic interaction calculations for 2D-BD simulations. The arrows present additional relaxations of motions perpendicular to the membrane and rotation out of the membrane, as a suitable approximation or simulations of membrane proteins that translate and rotate within the two-dimensional membrane.
Current work employed the 2D BD simulation to predict the oligimerization interfaces of the thyroid-stimulation hormone receptor's transmembrane domain [TRANSMEMBRANE DOMAINS OF ATTRACTION ON THE TSH RECEPTOR, Rauf Latif, M. Rejwan Ali, Mihaly Mezei, and Terry F Davies to be submitted]. The BD simulations produced two distinct clusters suggesting contacts between helices 1 and 4, and between helices 2 and 5. These contacts were subsequently confirmed by mutating six residues that were in contact in according to the BD simulation to cysteines and performing crosslinking experiments.
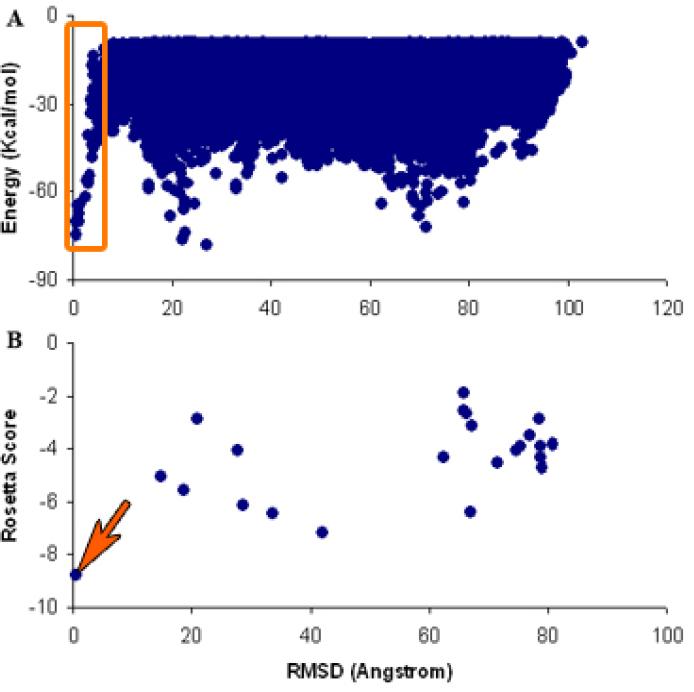
To test the performance of our 2D-BD approach, supplemented with local energy minimization, on GPCRs, we performed a re-docking simulation experiment on the newly solved β1-adrenergic receptor oligomeric form crystal structure (PDB: 4GPO) 25. We performed 1,000,000 2D-BD runs, and obtained 37,920 compact dimer complexes. Fig. 3 shows the center of mass distribution of Monomer II around the Monomer I of β1-adrenergic receptor. The complex structures were subjected to rigid body energy minimization, and ranked by interaction energies. The top 200 lowest interaction energy complex structures were selected for clustering analysis (cutoff = 5Å), and 23 clusters were obtained. The lowest energy structures were selected from each cluster as representative structures, and followed by further energy minimization using the Rosetta program 90, which takes into account protein side chain flexibility. The root-mean-square deviation (RMSD) of α carbon atoms between the predicted and crystal structures were calculated, and the plot of interaction energy (Rosetta score (Rscore)) vs. RMSD is shown in Fig. 4. From Fig. 4A, we can see that the 2D-BD approach is able to sample the near crystal structure (RMSD = 0) efficiently, which requires a van der Waals energy term to be presented during the sampling stage (the nearest structure from the crystal structure sampled without the van der Waals term is larger than 9 Å, data not shown). Fig. 4B shows that after application of further local energy minimization using the Rosetta program, the best score (Rscore) predicted structure is very close to the crystal structure (RMSD = 0.49 Å). The predicted structure with best score superimposed on the crystal structure is shown in Fig. 5.
Fig. 3.
The center of mass distribution of Monomer II (gray balls) around Monomer I of β1-adrenergic receptor from 2D-BD simulations (Left: top view, Right: side view). The center of mass of crystal structure of Monomer II is shown as a black ball.
Fig. 4.
Interaction energy (score) vs. RMSD between the predicted and crystal structures. (A) Structures from BD simulations followed by rigid body energy minimization. The sampled complexes near to crystal dimer are marked with an orange rectangular box. (B) 23 reprehensive structures of the cluster from the top 200 lowest energy predicted structures. The lowest energy predicted complex is marked with an orange arrow (RMSD = 0.49 Å from the crystal structure).
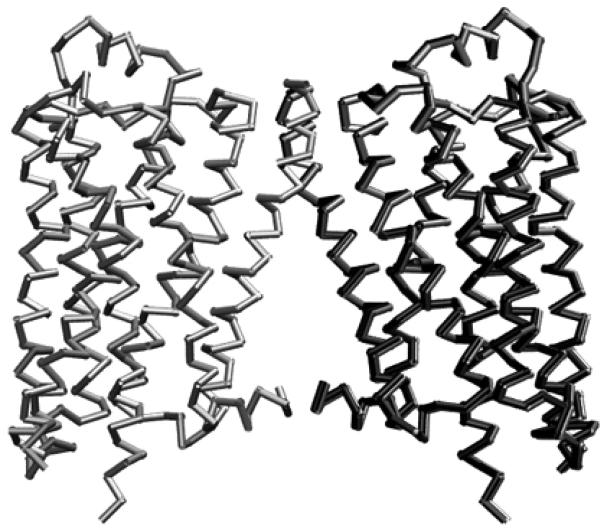
Fig. 5.
Comparison of crystal structure and 2D-BD predicted structure. The BD predicted Monomer II (right, in black) is very close to that of the crystal structure (right, in gray), and Monomer I (left) in colored in gray (RMSD: 0.49 Å)
Future Perspective
Not just for GPCRs, many other membrane proteins also function as dimers or oligomers, such as protein-tyrosine kinase receptors, cytokine receptors, antigen receptors, tumor necrosis receptors, protein-serine/threonine kinase receptors, and ion channels 6,91-94. Dimerization is also involved in the regulation of a large number of biological processes. It is of high importance to develop docking approaches suitable for predicting dimerization of membrane proteins, including sampling in the membrane environment, and improved scoring functions for membrane proteins.
The recent emergence of a number of GPCR crystal structures in oligomer forms provides an opportunity to evaluate the performance of new docking approaches. The goal will be to predict dimer structures correctly from monomer forms of GPCRs based on efficient sampling and scoring of protein docking approaches. Docking approaches can be used in combination with CGMD or all-atom MD simulations to take account induced-fit effects of GPCR dimerization. Using computational approaches alone to predict dimerization of GPCRs is a challenging task. In combination with experimental approaches, the computational approaches could become more powerful and useful. For example, cysteine crosslinking results between the dimer interfaces 11 could be used as distance restraints to reduce the sampling space of docking simulations dramatically. The predictions from computational approaches need to be tested and validated by experimental approaches.
Table 2.
Computational modeling methods employed for studies of GPCR oligomers
| GPCR | Type of Oligomerization |
Interface studied | Methodology | Ref |
|---|---|---|---|---|
| Rhodopsin | Dimerization | TM4/ TM5/part of TM3/CL2 | All-atom MD | 43 |
| CXCR4 | Dimerization | TM5/TM6 | All-atom MD | 44 |
| mGluR2 and 2AR | Heterodimerization | TM4/TM5 | All-atom MD | 48 |
| Rhodopsin | Oligomerization | TM1/TM2/H8, TM5, TM4/TM5, TM4, TM4-TM6 | CGMD, umbrella sampling/PMF | 31 |
| β2-AR | Oligomerization | TM1, TM5, TM4/TM5 | CGMD | 49 |
| β2-AR | Oligomerization | TM1, H8, TM1/TM5, TM6 | CGMD | 50 |
| β2-AR | Dimerization | TM4/TM5, TM1/TM2-TM4/TM5, TM1/TM2 | CGMD | 51 |
| δ-OR | Dimerization | TM4 | CGMD, umbrella sampling | 65 |
| δ-OR | Dimerization | TM4, TM4/TM5 | CGMD, well-tempered metadynamics simulation, umbrella sampling | 66 |
| β1-AR, β2-AR | Dimerization | TM1/H8, TM4/TM3 | CGMD well-tempered metadynamics simulation, umbrella sampling | 67 |
| Opsin | Dimerization | TM1/TM2/H8 | Re-docking to test programs of ZDOCK, ClusPro, HEX, GRAMM-X, PatchDock, SymmDock, and HADDOCK | 68 |
| CXCR4 | Dimerization | TM5/TM6* | Re-docking to test programs of ZDOCK, ClusPro, HEX, GRAMM-X, PatchDock, SymmDock, and HADDOCK | 68 |
| κOR | Dimerization | TM1/TM2/H8 | Re-docking to test programs of ZDOCK, ClusPro, HEX, GRAMM-X, PatchDock, SymmDock, and HADDOCK | 68 |
| Lutropin receptor | Dimerization | TM4-TM4, TM4-TM6, TM5-TM6 | ZDOCK plus membrane topology-based filtering | 70 |
| Neurotensin 1 receptor | Dimerization | TM1/EL1-TM2/TM4/EL1, TM4-TM4/EL2-EL2 | ZDOCK plus membrane topology-based filtering, Free Energy Estimations | 71 |
| A2A adenosine receptor | Dimerization | TM1-TM1/TM2-TM2,TM1-TM4/TM2-TM2, TM6-TM6/TM6-TM7/H8-CL3 | ZDOCK plus membrane topology-based filtering | 72 |
| Thromboxane A2 receptor | Dimerization | TM1-TM1, TM1-TM2/EL1, H8-H8 | ZDOCK plus membrane topology-based filtering | 73 |
| Thyroid-stimulation hormone receptor | Dimerization | TM1-TM5/TM2-TM5 | 2D BD simulation | To be submitted |
| β1-AR | Dimerization | TMi/TM2/H8 | Re-docking to test method of 2D BD simulation | This review |
CXCR4 forms TM5/TM6 interface. It was written as the TM4/TM5 interface in the Table 4 of ref 68, which probably was a typo.
Acknowledgement
This work was supported by the National Institute Health Grants DC008996 (M.C.) and S10RR027411 (M.C.), and the Center for High Performance Computing (CHiPC) and Institute of Structural Biology and Drug Discovery, at Virginia Commonwealth University (VCU).
References
- 1.Ellis C. The state of GPCR research in 2004. Nat. Rev. Drug Discov. 2004;3(7):575, 577–575, 626. doi: 10.1038/nrd1458. [DOI] [PubMed] [Google Scholar]
- 2.Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr. Med. Chem. 2000;7(9):861–888. doi: 10.2174/0929867003374534. [DOI] [PubMed] [Google Scholar]
- 3.Pin JP, Neubig R, Bouvier M, Devi L, Filizola M, Javitch JA, Lohse MJ, Milligan G, Palczewski K, Parmentier M, Spedding M. International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the recognition and nomenclature of G protein-coupled receptor heteromultimers. Pharmacol. Rev. 2007;59(1):5–13. doi: 10.1124/pr.59.1.5. [DOI] [PubMed] [Google Scholar]
- 4.Ferre S, Baler R, Bouvier M, Caron MG, Devi LA, Durroux T, Fuxe K, George SR, Javitch JA, Lohse MJ, Mackie K, Milligan G, Pfleger KD, Pin JP, Volkow ND, Waldhoer M, Woods AS, Franco R. Building a new conceptual framework for receptor heteromers. Nat. Chem. Biol. 2009;5(3):131–134. doi: 10.1038/nchembio0309-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filizola M. Advances in Experimental Medicine and Biology: G Protein-Coupled Receptors - Modeling and Simulation, Part II GPCRs in Motion: Insights from Simulations. 2014;796:37–125. [Google Scholar]
- 6.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U. S. A. 2000;97(7):3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angers S, Salahpour A, Bouvier M. Dimerization: an emerging concept for G protein- coupled receptor ontogeny and function. Annu. Rev. Pharmacol. Toxicol. 2002;42:409–435. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 8.Cheng ZJ, Miller LJ. Agonist-dependent dissociation of oligomeric complexes of G protein-coupled cholecystokinin receptors demonstrated in living cells using bioluminescence resonance energy transfer. J. Biol. Chem. 2001;276(51):48040–48047. doi: 10.1074/jbc.M105668200. [DOI] [PubMed] [Google Scholar]
- 9.McVey M, Ramsay D, Kellett E, Rees S, Wilson S, Pope AJ, Milligan G. Monitoring receptor oligomerization using time-resolved fluorescence resonance energy transfer and bioluminescence resonance energy transfer. The human delta -opioid receptor displays constitutive oligomerization at the cell surface, which is not regulated by receptor occupancy. J. Biol. Chem. 2001;276(17):14092–14099. doi: 10.1074/jbc.M008902200. [DOI] [PubMed] [Google Scholar]
- 10.Dinger MC, Bader JE, Kobor AD, Kretzschmar AK, Beck-Sickinger AG. Homodimerization of neuropeptide y receptors investigated by fluorescence resonance energy transfer in living cells. J. Biol. Chem. 2003;278(12):10562–10571. doi: 10.1074/jbc.M205747200. [DOI] [PubMed] [Google Scholar]
- 11.Guo W, Shi L, Filizola M, Weinstein H, Javitch JA. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. Sci. U. S. A. 2005;102(48):17495–17500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schertler GF, Hargrave PA. Projection structure of frog rhodopsin in two crystal forms. Proc. Natl. Acad. Sci. U. S. A. 1995;92(25):11578–11582. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies A, Schertler GF, Gowen BE, Saibil HR. Projection structure of an invertebrate rhodopsin. J. Struct. Biol. 1996;117(1):36–44. doi: 10.1006/jsbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- 14.Davies A, Gowen BE, Krebs AM, Schertler GF, Saibil HR. Three-dimensional structure of an invertebrate rhodopsin and basis for ordered alignment in the photoreceptor membrane. J. Mol. Biol. 2001;314(3):455–463. doi: 10.1006/jmbi.2001.5167. [DOI] [PubMed] [Google Scholar]
- 15.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic- force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421(6919):127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 16.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564(3):281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J. Biol. Chem. 2003;278(24):21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrick-Davis K, Grinde E, Cowan A, Mazurkiewicz JE. Fluorescence correlation spectroscopy analysis of serotonin, adrenergic, muscarinic, and dopamine receptor dimerization: the oligomer number puzzle. Mol. Pharmacol. 2013;84(4):630–642. doi: 10.1124/mol.113.087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai RS, Kusumi A. Single-molecule imaging revealed dynamic GPCR dimerization. Curr. Opin. Cell Biol. 2014;27:78–86. doi: 10.1016/j.ceb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471(7340):651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 23.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485(7398):321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485(7398):327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Chen S, Zhang JJ, Huang XY. Crystal structure of oligomeric beta1- adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 2013;20(4):419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson LM, Taddese B, Wall ID, Reynolds CA. Bioinformatics and molecular modelling approaches to GPCR oligomerization. Curr. Opin. Pharmacol. 2010;10(1):30–37. doi: 10.1016/j.coph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Selent J, Kaczor AA. Oligomerization of G protein-coupled receptors: computational methods. Curr. Med. Chem. 2011;18(30):4588–4605. doi: 10.2174/092986711797379320. [DOI] [PubMed] [Google Scholar]
- 28.Conn PM. Receptor-Receptor Interactions. Chapter 4 Simulating G Protein-Coupled Receptors in Native-Like Membranes: From Monomers to Oligomers. 2013;117:63–90. doi: 10.1016/B978-0-12-408143-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 29.Salom D, Lodowski DT, Stenkamp RE, Le T,I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2006;103(44):16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 31.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J. Am. Chem. Soc. 2012;134(26):10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453(7193):363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 33.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, Kobayashi T, Stevens RC, Iwata S. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475(7354):65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257(2):342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 35.Gobel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18(4):309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 36.Gouldson PR, Dean MK, Snell CR, Bywater RP, Gkoutos G, Reynolds CA. Lipid-facing correlated mutations and dimerization in G-protein coupled receptors. Protein Eng. 2001;14(10):759–767. doi: 10.1093/protein/14.10.759. [DOI] [PubMed] [Google Scholar]
- 37.Soyer OS, Dimmic MW, Neubig RR, Goldstein RA. Dimerization in aminergic G-protein-coupled receptors: application of a hidden-site class model of evolution. Biochemistry. 2003;42(49):14522–14531. doi: 10.1021/bi035097r. [DOI] [PubMed] [Google Scholar]
- 38.Filizola M, Weinstein H. The study of G-protein coupled receptor oligomerization with computational modeling and bioinformatics. FEBS J. 2005;272(12):2926–2938. doi: 10.1111/j.1742-4658.2005.04730.x. [DOI] [PubMed] [Google Scholar]
- 39.Vohra S, Chintapalli SV, Illingworth CJ, Reeves PJ, Mullineaux PM, Clark HS, Dean MK, Upton GJ, Reynolds CA. Computational studies of Family A and Family B GPCRs. Biochem. Soc. Trans. 2007;35(Pt 4):749–754. doi: 10.1042/BST0350749. [DOI] [PubMed] [Google Scholar]
- 40.Reggio PH. Computational methods in drug design: modeling G protein-coupled receptor monomers, dimers, and oligomers. AAPS. J. 2006;8(2):E322–E336. doi: 10.1007/BF02854903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanni S, Rothlisberger U. A closer look into G protein coupled receptor activation: X- ray crystallography and long-scale molecular dynamics simulations. Curr. Med. Chem. 2012;19(8):1135–1145. doi: 10.2174/092986712799320493. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez-de-Teran H, Bello X, Rodriguez D. Characterization of the dynamic events of GPCRs by automated computational simulations. Biochem. Soc. Trans. 2013;41(1):205–212. doi: 10.1042/BST20120287. [DOI] [PubMed] [Google Scholar]
- 43.Neri M, Vanni S, Tavernelli I, Rothlisberger U. Role of aggregation in rhodopsin signal transduction. Biochemistry. 2010;49(23):4827–4832. doi: 10.1021/bi100478j. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez D, Gutierrez-de-Teran H. Characterization of the homodimerization interface and functional hotspots of the CXCR4 chemokine receptor. Proteins. 2012;80(8):1919–1928. doi: 10.1002/prot.24099. [DOI] [PubMed] [Google Scholar]
- 45.Zhu L, Zhao Q, Wu B. Structure-based studies of chemokine receptors. Curr. Opin. Struct. Biol. 2013;23(4):539–546. doi: 10.1016/j.sbi.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Maeso J, Ang RL, Yuen T, Chan P, Weisstaub NV, Lopez-Gimenez JF, Zhou M, Okawa Y, Callado LF, Milligan G, Gingrich JA, Filizola M, Meana JJ, Sealfon SC. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature. 2008;452(7183):93–97. doi: 10.1038/nature06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fribourg M, Moreno JL, Holloway T, Provasi D, Baki L, Mahajan R, Park G, Adney SK, Hatcher C, Eltit JM, Ruta JD, Albizu L, Li Z, Umali A, Shim J, Fabiato A, MacKerell AD, Jr., Brezina V, Sealfon SC, Filizola M, Gonzalez- Maeso J, Logothetis DE. Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell. 2011;147(5):1011–1023. doi: 10.1016/j.cell.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruno A, Guadix AE, Costantino G. Molecular dynamics simulation of the heterodimeric mGluR2/5HT(2A) complex. An atomistic resolution study of a potential new target in psychiatric conditions. J. Chem. Inf. Model. 2009;49(6):1602–1616. doi: 10.1021/ci900067g. [DOI] [PubMed] [Google Scholar]
- 49.Mondal S, Johnston JM, Wang H, Khelashvili G, Filizola M, Weinstein H. Membrane driven spatial organization of GPCRs. Sci. Rep. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh A, Sonavane U, Joshi R. Multiscale modelling to understand the self-assembly mechanism of human beta2-adrenergic receptor in lipid bilayer. Comput. Biol. Chem. 2014;48:29–39. doi: 10.1016/j.compbiolchem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Prasanna X, Chattopadhyay A, Sengupta D. Cholesterol modulates the dimer interface of the beta(2)-adrenergic receptor via cholesterol occupancy sites. Biophys. J. 2014;106(6):1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818(2):234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao Z, Kobilka B. Using synthetic lipids to stabilize purified beta2 adrenoceptor in detergent micelles. Anal. Biochem. 2005;343(2):344–346. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Paila YD, Jindal E, Goswami SK, Chattopadhyay A. Cholesterol depletion enhances adrenergic signaling in cardiac myocytes. Biochim. Biophys. Acta. 2011;1808(1):461–465. doi: 10.1016/j.bbamem.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Pontier SM, Percherancier Y, Galandrin S, Breit A, Gales C, Bouvier M. Cholesterol-dependent separation of the beta2-adrenergic receptor from its partners determines signaling efficacy: insight into nanoscale organization of signal transduction. J. Biol. Chem. 2008;283(36):24659–24672. doi: 10.1074/jbc.M800778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pucadyil TJ, Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin(1A) receptors from bovine hippocampus. Biochim. Biophys. Acta. 2004;1663(1-2):188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Pucadyil TJ, Chattopadhyay A. Cholesterol depletion induces dynamic confinement of the G-protein coupled serotonin(1A) receptor in the plasma membrane of living cells. Biochim. Biophys. Acta. 2007;1768(3):655–668. doi: 10.1016/j.bbamem.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Saxena R, Chattopadhyay A. Membrane cholesterol stabilizes the human serotonin(1A) receptor. Biochim. Biophys. Acta. 2012;1818(12):2936–2942. doi: 10.1016/j.bbamem.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 59.Jafurulla M, Chattopadhyay A. Membrane lipids in the function of serotonin and adrenergic receptors. Curr. Med. Chem. 2013;20(1):47–55. [PubMed] [Google Scholar]
- 60.Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 2011;21(6):802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Paila YD, Chattopadhyay A. Membrane cholesterol in the function and organization of G-protein coupled receptors. Subcell. Biochem. 2010;51:439–466. doi: 10.1007/978-90-481-8622-8_16. [DOI] [PubMed] [Google Scholar]
- 62.Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J. Am. Chem. Soc. 2012;134(40):16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sengupta D, Chattopadhyay A. Identification of cholesterol binding sites in the serotonin1A receptor. J. Phys. Chem. B. 2012;116(43):12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- 64.Cang X, Du Y, Mao Y, Wang Y, Yang H, Jiang H. Mapping the functional binding sites of cholesterol in beta2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B. 2013;117(4):1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 65.Provasi D, Johnston JM, Filizola M. Lessons from free energy simulations of delta- opioid receptor homodimers involving the fourth transmembrane helix. Biochemistry. 2010;49(31):6771–6776. doi: 10.1021/bi100686t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnston JM, Aburi M, Provasi D, Bortolato A, Urizar E, Lambert NA, Javitch JA, Filizola M. Making structural sense of dimerization interfaces of delta opioid receptor homodimers. Biochemistry. 2011;50(10):1682–1690. doi: 10.1021/bi101474v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnston JM, Wang H, Provasi D, Filizola M. Assessing the relative stability of dimer interfaces in g protein-coupled receptors. PLoS. Comput. Biol. 2012;8(8):e1002649. doi: 10.1371/journal.pcbi.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaczor AA, Selent J, Sanz F, Pastor M. Modeling Complexes of Transmembrane Proteins: Systematic Analysis of Protein-Protein Docking Tools. Mol. Inf. 2013;32:717–733. doi: 10.1002/minf.201200150. [DOI] [PubMed] [Google Scholar]
- 69.Casciari D, Seeber M, Fanelli F. Quaternary structure predictions of transmembrane proteins starting from the monomer: a docking-based approach. BMC. Bioinformatics. 2006;7:340. doi: 10.1186/1471-2105-7-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fanelli F. Dimerization of the lutropin receptor: insights from computational modeling. Mol. Cell Endocrinol. 2007;260-262:59–64. doi: 10.1016/j.mce.2005.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casciari D, Dell'Orco D, Fanelli F. Homodimerization of neurotensin 1 receptor involves helices 1, 2, and 4: insights from quaternary structure predictions and dimerization free energy estimations. J. Chem. Inf. Model. 2008;48(8):1669–1678. doi: 10.1021/ci800048d. [DOI] [PubMed] [Google Scholar]
- 72.Fanelli F, Felline A. Dimerization and ligand binding affect the structure network of A(2A) adenosine receptor. Biochim. Biophys. Acta. 2011;1808(5):1256–1266. doi: 10.1016/j.bbamem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 73.Fanelli F, Mauri M, Capra V, Raimondi F, Guzzi F, Ambrosio M, Rovati GE, Parenti M. Light on the structure of thromboxane A(2)receptor heterodimers. Cell Mol. Life Sci. 2011;68(18):3109–3120. doi: 10.1007/s00018-010-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O'Dowd BF, Nguyen T, Ji X, George SR. D5 dopamine receptor carboxyl tail involved in D5-D2 heteromer formation. Biochem. Biophys. Res. Commun. 2013;431(3):586–589. doi: 10.1016/j.bbrc.2012.12.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Navarro G, Ferre S, Cordomi A, Moreno E, Mallol J, Casado V, Cortes A, Hoffmann H, Ortiz J, Canela EI, Lluis C, Pardo L, Franco R, Woods AS. Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers. J. Biol. Chem. 2010;285(35):27346–27359. doi: 10.1074/jbc.M110.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossky PJ, Doll JD, Friedman HL. Brownian dynamics as smart Monte Carlo simulation. Journal of Chemical Physics. 1978;69(10):14628–14633. [Google Scholar]
- 77.Ouporov IV, Knull HR, Thomasson KA. Brownian dynamics simulations of interactions between aldolase and G- or F-actin. Biophys. J. 1999;76(1 Pt 1):17–27. doi: 10.1016/S0006-3495(99)77174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearson DC, Jr., Gross EL. Brownian dynamics study of the interaction between plastocyanin and cytochrome f. Biophys. J. 1998;75(6):2698–2711. doi: 10.1016/S0006-3495(98)77714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lowe SL, Adrian C, Ouporov IV, Waingeh VF, Thomasson KA. Brownian dynamics simulations of glycolytic enzyme subsets with F-actin. Biopolymers. 2003;70(4):456–470. doi: 10.1002/bip.10530. [DOI] [PubMed] [Google Scholar]
- 80.Motiejunas D, Gabdoulline R, Wang T, Feldman-Salit A, Johann T, Winn PJ, Wade RC. Protein-protein docking by simulating the process of association subject to biochemical constraints. Proteins. 2008;71(4):1955–1969. doi: 10.1002/prot.21867. [DOI] [PubMed] [Google Scholar]
- 81.Ouporov IV, Knull HR, Huber A, Thomasson KA. Brownian dynamics simulations of aldolase binding glyceraldehyde 3-phosphate dehydrogenase and the possibility of substrate channeling. Biophys. J. 2001;80(6):2527–2535. doi: 10.1016/S0006-3495(01)76224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elcock AH, Sept D, McCammon JA. Computer simulation of protein-protein interactions. Journal of Physical Chemistry B. 2001;105(8):1504–1518. [Google Scholar]
- 83.Haddadian EJ, Gross EL. Brownian dynamics study of cytochrome f interactions with cytochrome c6 and plastocyanin in Chlamydomonas reinhardtii plastocyanin, and cytochrome c6 mutants. Biophys. J. 2005;88(3):2323–2339. doi: 10.1529/biophysj.104.053561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui M, Shen J, Briggs JM, Luo X, Tan X, Jiang H, Chen K, Ji R. Brownian dynamics simulations of interaction between scorpion toxin Lq2 and potassium ion channel. Biophys. J. 2001;80(4):1659–1669. doi: 10.1016/S0006-3495(01)76138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui M, Shen J, Briggs JM, Fu W, Wu J, Zhang Y, Luo X, Chi Z, Ji R, Jiang H, Chen K. Brownian dynamics simulations of the recognition of the scorpion toxin P05 with the small-conductance calcium-activated potassium channels. J. Mol. Biol. 2002;318(2):417–428. doi: 10.1016/S0022-2836(02)00095-5. [DOI] [PubMed] [Google Scholar]
- 86.Yu L, Sun C, Song D, Shen J, Xu N, Gunasekera A, Hajduk PJ, Olejniczak ET. Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex. Biochemistry. 2005;44(48):15834–15841. doi: 10.1021/bi051656d. [DOI] [PubMed] [Google Scholar]
- 87.Cui M, Mezei M, Osman R. Prediction of protein loop structures using a local move Monte Carlo approach and a grid-based force field. Protein Eng Des Sel. 2008;21(12):729–735. doi: 10.1093/protein/gzn056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meng XY, Zhang HX, Mezei M, Cui M. Predicting Protein Interactions by Brownian Dynamics Simulations. Journal of Biomedicine and Biotechnology. 2012;2012 doi: 10.1155/2012/121034. Article ID 121034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui M, Mezei M, Osman R. Modeling dimerizations of transmembrane proteins using Brownian dynamics simulations. J. Comput. Aided Mol. Des. 2008;22(8):553–561. doi: 10.1007/s10822-008-9198-3. [DOI] [PubMed] [Google Scholar]
- 90.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 91.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80(2):213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 92.Milligan G. Oligomerisation of G-protein-coupled receptors. J. Cell Sci. 2001;114(Pt 7):1265–1271. doi: 10.1242/jcs.114.7.1265. [DOI] [PubMed] [Google Scholar]
- 93.Rios CD, Jordan BA, Gomes I, Devi LA. G-protein-coupled receptor dimerization: modulation of receptor function. Pharmacol. Ther. 2001;92(2-3):71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 94.George SR, O'Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discov. 2002;1(10):808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 95.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]