Abstract
Background
Specific molecules involved in early inductive signaling from anterior neural tissue to the placodal ectoderm to establish a lens-forming bias, as well as their regulatory factors, remain largely unknown. In this study we sought to identify and characterize these molecules.
Results
Using an expression cloning strategy to isolate genes with lens-inducing activity, we identified the transcriptional cofactor ldb1. This, together with evidence for its nuclear dependence, suggests its role as a regulatory factor, not a direct signaling molecule. We propose that ldb1 mediates induction of early lens genes in our functional assay by transcriptional activation of lens-inducing signals. Gain-of-function assays demonstrate that the inductive activity of the anterior neural plate on head ectodermal structures can be augmented by ldb1. Loss-of-function assays show that knockdown of ldb1 leads to decreased expression of early lens and retinal markers and subsequently to defects in eye development.
Conclusions
The functional cloning, expression pattern, overexpression, and knockdown data show that an ldb1-regulated mechanism acts as an early signal for Xenopus lens induction.
Keywords: induction, lens, placodes, Xenopus, expression cloning, ldb1
INTRODUCTION
Embryonic lens induction in vertebrates is a complex process involving patterning events that take place over several stages in development. Early determination events in the Xenopus lens induction process, beginning in gastrula stages (Grainger 1992), have been studied extensively by embryological manipulation (reviewed by Baker and Bronner-Fraser, 2001). During gastrulation, ectoderm transiently becomes able to respond to lens inductive signaling. This is the period of lens competence (Servetnick and Grainger, 1991). The ectoderm adjacent to the forming neural plate then becomes biased (partially specified) toward the lens fate (Grainger et al., 1997) and may in fact be biased simultaneously toward multiple placodal fates (Jacobson, 1966). As the neural tube closes and the optic vesicles approach the overlying ectoderm (stage 19; Nieuwkoop and Faber, 1994), lens specification occurs (the ectoderm has sufficient information to begin differentiation when isolated in culture; Henry and Grainger, 1990).
Many molecules involved in lens inductive responses have been identified, though few genes are exclusive to the presumptive lens ectoderm (PLE) at early stages of determination. One such exclusive gene is foxe3. foxe3 mRNA is present in the anterior neural plate of stage 15 embryos and as development proceeds becomes limited to the PLE and lens epithelium (Kenyon et al., 1999), rendering it a useful PLE marker. Deletion analysis of the foxe3 enhancer region has revealed lens-specific regulation by rbpj and otx2, and demonstrated a role for notch signaling in lens induction (Ogino et al., 2008). Other signaling molecules (bmp4, bmp7, fgf8) are thought to be involved in lens specification and differentiation, and several transcription factors (e.g. maf and sox genes, among others more broadly expressed) are activated in the responding lens ectoderm at these stages (reviewed by Ogino and Yasuda, 2000 and Schlosser, 2006). However, little is understood about the transcriptional activation of early signals from and within the anterior neural plate that produce the lens-forming bias in the presumptive lens ectoderm.
As the neural plate forms, several genes that may be associated with the acquisition of lens-forming bias become activated in the anterior placodal region, such as otx2, pax6 (Zygar et al., 1998), and foxe3 (Kenyon et al., 1999). Following neural tube closure and optic vesicle contact, other genes become activated in the PLE (such as sox3 [Schlosser and Ahrens, 2004], maf-B and nrl-maf [Ogino and Yasuda, 1996]) and are associated with lens determination. The lens ectoderm thickens to form a placode at stage 25 and begins differentiation (Fini et al., 1997), as indicated by the expression of lens crystallins.
There is evidence that the early sensory epithelia and cement gland are determined by signals from the anterior neural plate (Jacobson, 1963a,b; Gallagher et al., 1996; Drysdale and Elinson, 1993). In addition, many genes expressed in the PLE are also present in other developing head structures. otx2 is expressed in the lens, olfactory epithelium, and cement gland. pax6, foxe3, and sox3 are all expressed in the lens and olfactory epithelium. Several other genes are expressed in some or all of the head placodes and cement gland. The dlx gene family shows early expression in the olfactory epithelium and cement gland, as well as later expression in the thickened otic placode, eya1 genes are shared by the lens, olfactory and otic placodes, and six genes are expressed widely in the olfactory, lens, otic, and other placodes (reviewed by Schlosser 2006). Because of the similarity of the early steps in their development as well as shared early gene expression, these structures may be induced by a common mechanism; a panplacodal primordium model has been proposed (Schlosser and Ahrens, 2004).
The common element of the anterior neural plate in lens and other placodal inductions suggests that early inducers, common or lens-specific, are predominantly produced in anterior neural tissue (Schlosser, 2006). The anterior neural plate has been shown to provide the primary lens inducer (Henry and Grainger, 1990), to establish lens bias in the head ectoderm (Grainger et al., 1997), and to activate early markers of the lens (Zygar et al., 1998). The activation of otx2, pax6, and foxe3 expression in lens-competent ectoderm provides a molecular assay for induction and allows the observation of early steps in determination.
We sought to identify early lens-inducing factors from Xenopus laevis neural plate stage embryos, with the focus on the anterior neural plate as the signaling source and on genes expressed early in the PLE as the inductive response. We used the Xenopus oocyte to express injected RNAs encoding putative signals, placed lens-competent animal caps on the oocytes, and assayed for an inductive response in these caps. We identified a gene that elicits a foxe3 response in the animal cap, the previously cloned ldb1 (Agulnick et al., 1996), which encodes a nuclear protein. ldb1 expression is able to recapitulate the early phases of induction of the lens, nose, and cement gland and can contribute to the ability of the neural plate to induce early markers of sensory determination. We propose that it acts upstream of the signals responsible for the induction of anterior sensory structures.
In order to better establish a role for ldb1 in the endogenous placodal induction process, we used antisense morpholino oligonucleotides (MO) directed against ldb1 mRNA to reduce the levels of ldb1 protein during early development. This MO-mediated depletion of ldb1 resulted in decreased expression of several markers of retina and lens providing further support for involvement of an ldb1-mediated event in lens formation.
RESULTS
Isolation of a clone mediating lens-inductive activity
To test the activity of inducing molecules and to screen for an early lens-inducing molecule, we developed an oocyte-animal cap assay system and isolated a clone by sib selection of a neurula cDNA library (see Experimental Procedures). RNA enriched in poly(A)+ transcripts from dorsalized stage 14 embryos was injected into oocytes, which were cultured and recombined with lens-competent ectoderm (as per Lustig and Kirschner 1995). After culturing the ectoderm to the equivalent of tailbud stages (24-30), we assayed the ability of transcripts to induce expression of foxe3 (Kenyon et al., 1999) in responding ectoderm. Approximately 10% of cases were positive for foxe3 expression when pooled transcripts of groups of 105 clones were tested. In subsequent experiments, 10 groups of pooled transcripts derived from the previous positive group were tested; the pool with the highest level of response in each experiment (10% to 36%) was selected for use in the next series. A single clone with foxe3 inducing activity was isolated and confirmed with 50/179 positive cases (28%).
Sequence analysis of the clone of interest revealed that the 750 bp sequence is 100% identical at the nucleic acid level with the previously identified ldb1, a LIM domain-homeodomain transcription factor (LIM-HD)-binding factor (Agulnick et al., 1996, GenBank accession #U74360); we therefore called the positive clone Δldb1. Its appearance on Northern hybridizations of 750-800 nucleotides is likely due to polyadenylation, as the AAUAAA polyadenylation hexanucleotide is present in the Δldb1 3’ UTR. Further analysis indicated that the partial clone corresponds to the 3’ terminal third of the ldb1 coding region (amino acids 244-376) plus the 3’ untranslated region. Δldb1 also overlaps with regions shown to be sufficient for the function of ldb1 family members (Fig. 1A). An in-frame ATG present just 5’ to the putative nuclear localization signal (Agulnick et al., 1996) is the presumed start site for translation of the C-terminal 131 amino acids of Δldb1. The level of identity suggests that it is a truncation of ldb1 and not a different gene.
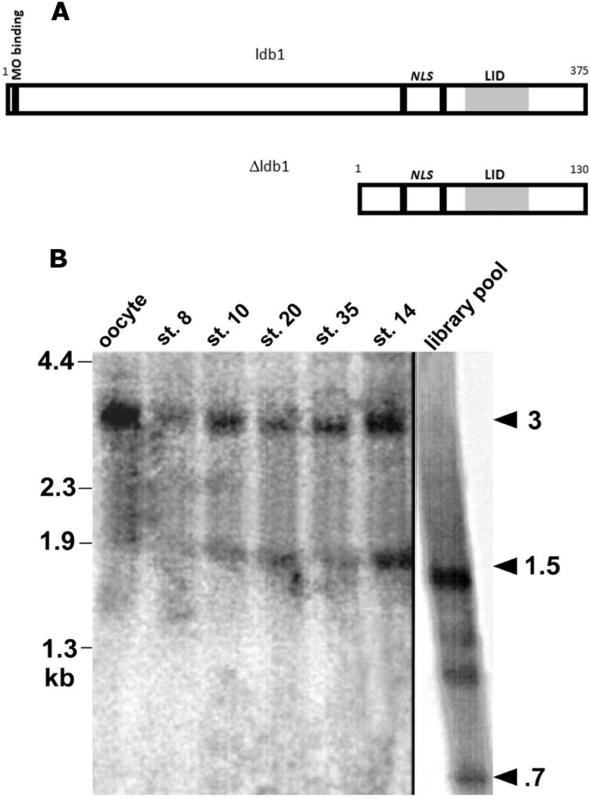
Fig. 1.
A: Comparisons of sizes of Δldb1 fragment and full-length ldb1; nuclear localization signals (NLS) and MO binding site indicated. The functional LIM-binding domain of ldb1/ldb2 (LIM interaction domain, LID) is shaded. B: Northern analysis of endogenous and synthetic transcripts for ldb1 expression (Δldb1 probe). Lanes 1-5: Five embryo equivalents total RNA each from St. VI oocyte, St. 8, St. 10, St. 20, and St. 35 embryos. Lane 6: 1.5 μg St. 14 poly(A)+ RNA. Lane 7: 1 μg pooled synthetic RNA from 10,000 member library fraction. Library pool contains bands corresponding to Δldb1 (700-750 bp) and ldb1 (1.5 kb). Oocyte/embryos contain endogenous 3kb and 1.5-1.8 kb ldb1 transcripts; not Δldb1.
Identity of Δldb1 and ldb1
Δldb1 is apparently not a naturally occurring splice variant since it appeared in library but not embryo transcripts when analyzed by Northern blot (Fig. 1B). Additionally, Δldb1 has no leader sequence or untranslated region 5’ to the putative start site, as one would expect of an alternative transcript. Rather, our data are consistent with the suggestion that Δldb1 was a fortuitously active fragment generated in library construction. Analysis of stage 14 poly(A)+ RNA and total RNA from oocyte through stage 35 reveals endogenous 3kb and ~1.5kb bands (Fig. 1B). A larger (1.1 kb) ldb1 fragment was cloned and used to prepare an in situ hybridization probe and RNA for expression in functional assays.
A test of ldb1 activity in our oocyte-animal cap assay was performed by injecting ldb1 RNA in place of Δldb1. foxe3 expression was detected in 6/31 cases (19%) in comparison to 28% for Δldb1. This and other experiments suggest that Δldb1 activity is representative of the activity of the whole gene ldb1, eliminating Δldb1 as a dominant inhibitory form of ldb1. Δldb1 does appear to have higher activity in functional assays than the full-length construct, even after molarity was normalized by injection of two times the amount of full-length ldb1. This increased activity may be due to higher rates of translation or differential RNA stability, or to the presence of negative regulatory regions present in the amino terminus of ldb1 protein. We investigated these issues by analysis of Δldb1 and ldb1 translation products.
Nuclear dependence of ldb1 activity
To address the higher activity of Δldb1 in the functional assay relative to full-length ldb1, we observed the translation of each in the oocyte. Oocytes were metabolically labeled with 35S-methionine following injection with 20 ng Δldb1 or 40 ng ldb1 RNA (Fig. 2A). High levels of translation were observed for Δldb1 but not for ldb1. Injected oocytes used for metabolic labeling were also used in functional assays, and consistent results were obtained for Δldb1 and ldb1 as reported above. Thus, the injection of ldb1 was able to activate induction of animal caps despite being translated at a much lower rate than Δldb1.
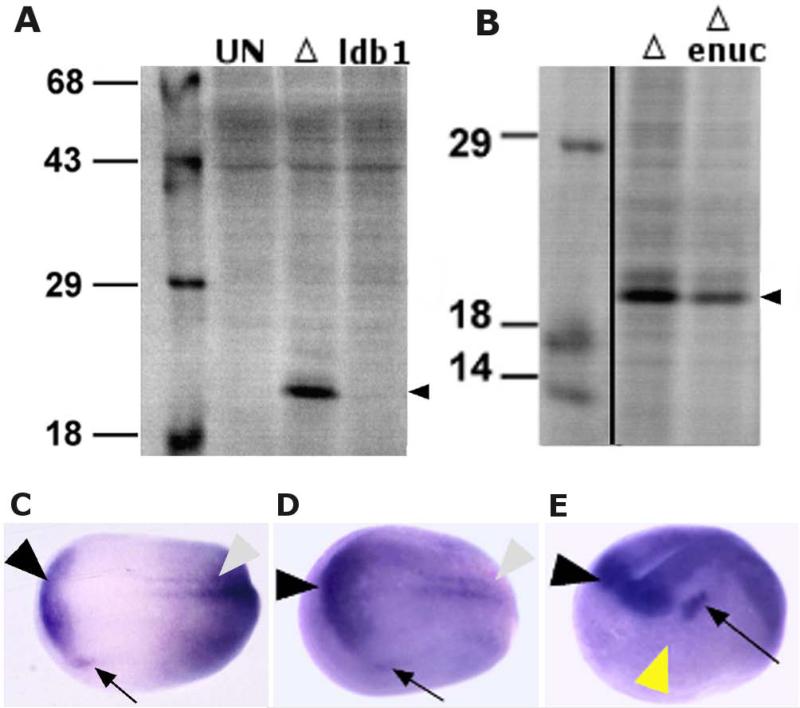
Fig. 2.
Δldb1 RNA is highly enriched in neural tissue at neural tube stages, and is translated efficiently in oocytes with or without a nucleus. A-B: 35S- labeled protein products from metabolic labeling of oocytes expressing ldb1 or Δldb1; Δldb1 is abundant. Uninjected oocytes (UN), oocytes injected with 20 ng Δldb1 RNA (Δ), and oocytes injected with 40 ng ldb1 (ldb1) were cultured in 35S-Met and visualized by SDS-PAGE (A); arrowhead indicates Δldb1. 20 ng Δldb1 RNA was injected into normal and enucleated oocytes for translation (B); arrowhead indicates Δldb1. C-E: Whole-mount in situ hybridization of ldb1 at Stage 15 (C), anterior neural plate (black arrowhead), lateral expression (arrow), and posterior neural expression (gray arrowhead) indicated; Stage 17 (D); anterior neural plate (black arrowhead), lateral expression (presumptive ganglia; arrow), and posterior domain (gray arrowhead) indicated; and Stage 18 (E), PLE region (yellow arrowhead) flanked on dorsal side by anterior neural expression (black arrowhead) and posterior side by presumptive cranial ganglion expression (arrow) of ldb1.
The oocyte-animal cap screen was expected to yield a secreted rather than a nuclear factor. The nuclear localization and LIM-HD-binding data for ldb1 (Agulnick et al., 1996) led to the suggestion that Δldb1 may act on the oocyte nucleus to activate a signaling molecule target that is responsible for the response in animal caps. The nuclear dependence of Δldb1 activity was tested by enucleating oocytes to be used in conjugates with animal caps. Ectoderm placed on manually enucleated, Δldb1-injected oocytes was positive in only 2/73 cases (<3%). We also tested full-length ldb1 in enucleated oocytes and found no cases to be positive. These results indicate that the ability of ldb1 to mediate induction is dependent on the nucleus.
To eliminate the possibility that enucleation of the oocytes was injurious or inhibitory to Δldb1 translation, we monitored translation by 35S metabolic labeling of injected oocytes. In normal and enucleated oocytes, Δldb1 was translated efficiently (Fig. 2B), even though activity was not detectable in the oocyte-animal cap assay. These data indicate that the positive response seen in the functional assay results described earlier is dependent upon a nuclear-mediated pathway for the action of Δldb1.
Pattern of ldb1 expression
The expression of ldb1 was initially reported as ubiquitous (Agulnick et al., 1996), but we found it is highly enriched in neural tissues of the embryo through early development; this is consistent with the expression reported by Hiratani et al. (2003). By stage 15, expression becomes enriched in the anterior neural plate (Fig. 2C), and sections reveal that expression is limited to the neural tissue and not in the mesoderm of the neural plate. ldb1 is expressed in the ectoderm anterior and ventral to the olfactory placodal region on the neural fold in addition to higher levels of expression in neural tissue. By stage 17, the pattern becomes more enriched in the anterior neural plate, and the lateral domains (cranial ganglia) become clearer (Fig. 2D). By stage 18/19 the expression is prominent in anterior neural tissue, posterior spinal cord, and anterior lateral domains, and the PLE (yellow arrowhead, Fig. 2E) is flanked on dorsal and posterior sides by ldb1 expression.
The expression of ldb1 appears to be localized to regions of the embryo consistent with a role in activation of foxe3 in oocyte-animal cap assays: ldb1 is expressed in domains adjacent to and overlapping foxe3 expression domains. Notably, it is strongly expressed in the tissue known to provide positive lens-inducing signals, the anterior neural plate.
Demonstration of ldb1 placode-inducing activity in animal caps
Because foxe3 is expressed in both lens and nonlens tissues, the specificity of ectodermal response to ldb1 observed in oocyte-animal cap assays was tested using various ectodermal markers induced by ldb1-expressing tissue. Lens-competent animal cap tissue will not express early lens markers either in isolation or in combination with another animal cap; however, we hypothesized injection of RNA involved in the signaling pathway could enable an animal cap to act as a lens-inducing tissue upon a lens-competent animal cap and activate the early phases of lens specification. We combined animal caps from Δldb1-injected embryos with lineage-labeled animal caps and assayed recombinants for the expression of genes expressed in head ectoderm or neural tissue in addition to foxe3 (Fig. 3). Other specific marker genes included nrl-maf (Ogino and Yasuda, 1996; PLE from stage 24); dlx5 (Papalopulu and Kintner, 1993; presumptive nasal ectoderm, cement gland, and neural crest, but not PLE, from mid-neural plate stages); ag1 (Sive et al., 1989; cement gland primordium from late gastrula stages); snai2 (Mayor et al., 1995; cranial and trunk neural crest from late gastrula stages); sox2 (Mizuseki et al., 1998; Schlosser et al., 2008; broadly in early ectoderm, exclusively in neural tissue from stage 13 and in cranial placodes); ncam1 (Kintner and Melton, 1987; all neural tissue from early neural plate stages).
Fig. 3.
Δldb1-induced activation of early lens, olfactory, and cement gland markers in ectodermal recombinants. A: Schematic of recombinant procedure; Stage 10-11 Δldb1 RNA-injected ectoderm combined with Stage 11-11.5 lineage-labeled ectoderm, cultured to the equivalent of Stages 18-26, processed for in situ hybridization with foxe3, nrl-maf, dlx5, and ag1, and sectioned for analysis. B-C: Expression of foxe3 and lineage labeling of ectoderm, Stage 24. D-E: Expression of nrl-maf and lineage labeling of ectoderm, Stage 26. F-G: Expression of dlx5 and lineage labeling of ectoderm, Stage 18. H-I: Expression of ag1 and lineage labeling of ectoderm, Stage 18. Arrowheads indicate identical locations on each pair of images. Table: Expression of genes in recombinants; markers of the lens, nose, and cement gland are detected in Δldb1-injected recombinants, markers of neural and neural crest tissue are not.
Recombinants of Δldb1-injected (250 pg) ectoderm (stage 10-11) with responding lineage-labeled animal cap ectoderm (stage 10-10.5 for sox2 and ncam1; stage 11-11.5 for nrl-maf, dlx5, snai2, ag1, and foxe3) were assayed for expression of these transcripts; early lens, olfactory epithelium, and cement gland genes were activated in the responding animal caps. foxe3 (Fig. 3B-C) activation was tested in recombinants and found to be expressed in 53% of cases (31/58); parallel recombinants were also made between full length ldb1-injected (500 pg) animal cap ectoderm and lineage-labeled ectoderm, and foxe3 expression was detected in 37% of those cases (10/27). nrl-maf (Fig. 3D-E) was activated in 30% of cases (7/23), dlx5 (Fig. 3F-G) in 80% of cases (17/21), and ag1 (Fig. 3H-I) in 100% of cases (18/18). All of these RNAs are expressed in ectodermal structures of the head region. By contrast, RNAs expressed in neural tissue and neural crest were not activated by Δldb1. sox2 (0/8), ncam1 (0/18), and snai2 (0/17) showed no positive expression in responding animal caps.
In some cases, staining was observed in the Δldb1-injected animal cap as well as in responding ectoderm. This was observed for dlx5, ag1, and in a small number of cases (5/58) for foxe3. This suggests that ldb1 could act in a cell-autonomous or tissue-autonomous manner under some conditions or in certain tissues. When analyzed with regard to whether expression appeared in the RNA-injected cap, lineage-labeled (responding) cap, or both, we find expression in injected tissue is most prevalent for the genes expressed in cement gland and olfactory epithelium. ag1 and dlx5, both expressed in the cement gland, are more frequently activated in the injected animal cap (33% and 57%, respectively). nrl-maf expression is confined to the responding animal cap.
To further address the range of presumptive tissues induced by Δldb1 expression, foxe3 positive caps induced by Δldb1 were cryosectioned to assess whether expression was localized to the inner or outer ectoderm. Expression in the inner (sensorial) layer would be characteristic of a lens response, since lens is formed exclusively from the inner layer. Localization of expression in the outer layer or both layers suggests an olfactory response, since the olfactory placode forms from both layers of ectoderm. Our results indicate both lens and olfactory responses may be occurring since responding ectoderm showed expression in the inner layer, outer layer, and both in the oocyte-animal cap assay and ectodermal recombinants (Fig. 3). Cryosections of recombinants of Δldb1-expressing tissue and responding tissue cultured to later tailbud stages (26-35) were analyzed for histologically identifiable tissues (placodal thickenings or cement gland formation); no evidence for these cell types was observed (data not shown). Additionally, these recombinants were tested for the expression of crystallin mRNA by in situ hybridization but were negative for crystallin expression (0/8).
The observation that early lens, olfactory, and cement gland responses are all activated by ldb1 suggests that ldb1 is involved in the early (possibly shared) steps in the determination of all these structures; however, genes indicative of early stage neural and neural crest tissue are not activated. These data also imply that ldb1 may play a cell-or tissue-autonomous role in anterior ectodermal induction in addition to its role in the adjacent anterior neural plate.
ldb1 enhances endogenous placode-inducing activity of anterior neural plate tissue
The ability of ldb1 to mediate induction of early head ectodermal structures raised the possibility that endogenous inducing signals operate by an ldb1-mediated pathway, and that signaling might be increased by Δldb1 overexpression. Since the early signal for lens induction comes predominantly from the anterior neural plate, recombinants between this tissue and lens-competent animal cap ectoderm activate expression of early lens markers (Zygar et al., 1998) as well as crystallin protein (Grainger, unpublished) in the ectoderm. In the present study, we injected 1 ng Δldb1 RNA into embryos at the one cell stage, cultured these to stage 14, explanted the neural plate and combined it with stage 11-11.5 lineage-labeled animal cap ectoderm (Fig. 4A). The recombinants were cultured to stage 23 and analyzed for expression of foxe3.
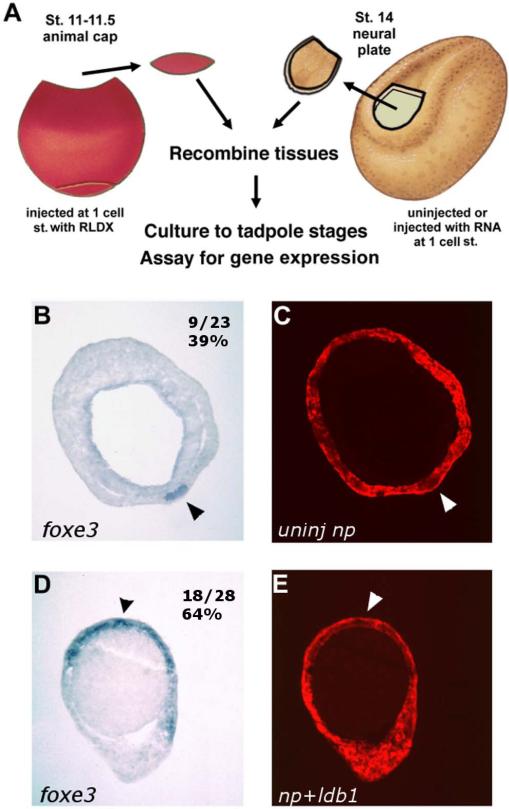
Fig. 4.
The ability of anterior neural tissue to induce foxe3 expression is enhanced by ldb1. A: Schematic diagram of recombinant procedure; Stage 11-11.5 lineage-labeled animal cap ectoderm combined with control anterior neural plates or anterior neural plates from embryos injected with Δldb1 RNA into the animal pole at the 1-cell stage, cultured to Stage 23, processed for in situ hybridization, and sectioned for analysis. B-C: Expression of foxe3 (B) and lineage labeling (C) of responding ectoderm in recombinant with control neural plate (39% expressing foxe3). D-E: Expression of foxe3 (D) and lineage labeling (E) of responding ectoderm in recombinant with Δldb1-injected neural plate (64% expressing foxe3).
Injection of Δldb1 augments the placode-inducing capability of the anterior neural plate in these recombinants. Control anterior neural plates induced ectodermal foxe3 expression in 9/23 cases (39%), as shown in Fig. 4B-C. Δldb1-injected neural plates, however, induced ectodermal foxe3 expression in 18/28 cases (64%; Fig. 4D-E). These differences are quantitatively significant (p < .05, chi-square analysis); there is also a qualitative difference in the level of expression of foxe3. foxe3 staining was in all cases broader by 2- to 10-fold in the responding ectoderm in Δldb1-overexpressing recombinants (Fig. 4D) than in control recombinants (Fig. 4B). In preliminary dose-response experiments, positive response was seen with 400 pg to 1 ng Δldb1 (or 1 to 2 ng ldb1).
ldb1 is able to complement the endogenous ability of anterior neural plate tissue to induce foxe3 expression in ectoderm. Whether this enhancement of foxe3 expression is due exclusively to early lens expression domains of this gene is not known; however, foxe3 is expressed in the inner (sensorial) layer of all positively responding ectoderm in recombinants. This enhancement of neural plate activity was observed over a range of approximately 100- to 500-fold over the endogenous ldb1 mRNA level. The results suggest that ldb1 is involved in the endogenous regulatory mechanisms upstream of early anterior placode-inductive signaling.
ldb1 is essential for normal eye development
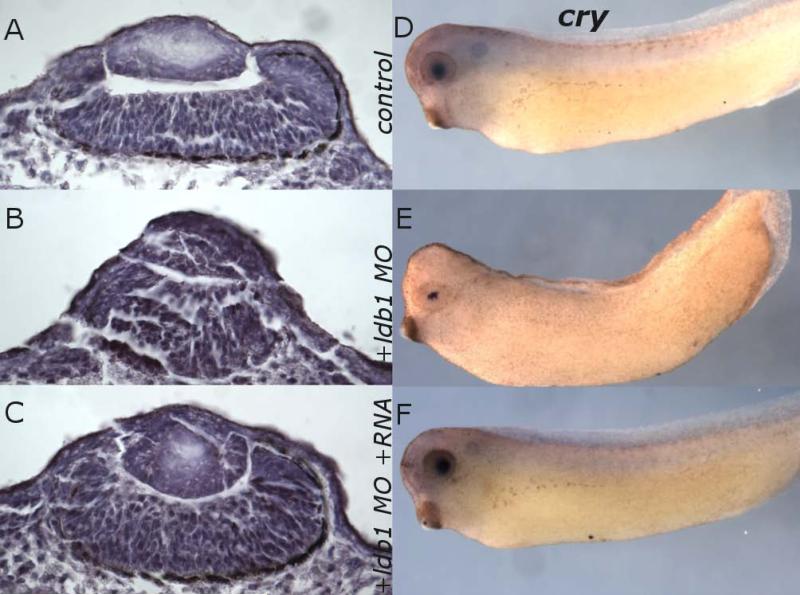
To analyze the phenotypic consequences of ldb1 deficiency in development, we injected 1-cell stage embryos with MO designed to block ldb1 translation (but not affect Δldb1 translation; see Fig. 1). Specific morphological defects appeared by tailbud stages, with a range of severity of phenotypes observed: reduced eye size and enlarged cement gland were prominent at stages 21-26 (Fig. 5C, D). In many cases development proceeded only to stage 26. 83% of MO-injected embryos died by stage 34, with only 1% of control embryos dying during that time. The remaining 17% exhibited somewhat reduced lens size, reduced γ-crystallin expression domain, abnormal lens organization, and abnormal development of retina including loss of retinal pigment epithelium when analyzed in histological sections or processed for in situ hybridization for γ-crystallin at stage 34. (Fig. 6B, E). Although the results were variable, the eye defects were clear. In younger embryos, MO-mediated depletion of ldb1 also led to decreased foxe3 and rax expression domains, as demonstrated by injection at the 1 cell stage with 65ng ldb1 MO followed by culture to stages 22-26 and in situ hybridization for foxe3 and rax. These MO-injected embryos exhibited markedly decreased or eliminated foxe3 and rax expression in all cases (foxe3 n=17; rax n=16; Fig. 5C, D).
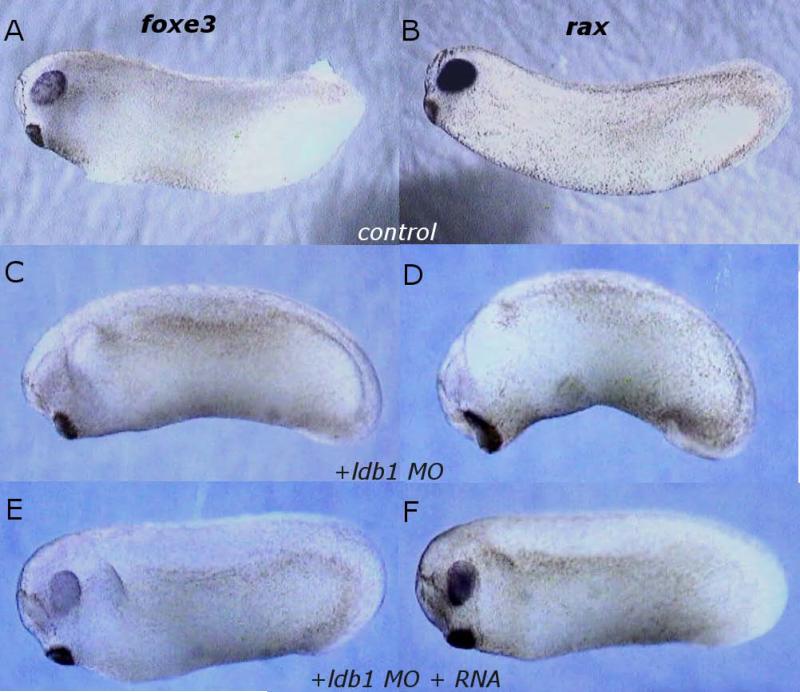
Fig. 5.
Expression of foxe3 and rax decreased in ldb1 MO-injected embryos. A-B: Control in situ hybridization expression pattern of foxe3 (A) and rax (B) at Stage 26. C-D: ldb MO-injected embryos, expression of foxe3 (C) and rax (D) at Stage 25-26. E-F: ldb1 MO- and Δldb1 RNA-coinjected embryos, expression of foxe3 (E) and rax (F) at Stage 25-26.
Fig. 6.
Eye morphology and retinal pigmentation are disrupted, and lens γ-crystallin expression is reduced in ldb1 MO-injected embryos. A-C: Sections of control (A), ldb1 MO-injected (B), and ldb1 MO + Δldb1 RNA-injected (C) embryos at Stage 34. D-F: In situ hybridization for γ-crystallin in control (D), ldb1 MO-injected (E), and ldb1 MO + Δldb1 RNA-injected (F) embryos at Stage 34.
To test the specificity of the knockdown, Δldb1, which lacks the sequence targeted by the MO, was coinjected to rescue the MO effects. When embryos were coinjected with 65ng ldb1 MO and 0.11ng Δldb1 RNA and cultured to stages 22-26, then analyzed for foxe3 and rax expression by in situ hybridization, expression was restored in all samples (foxe3 n=18; rax n=23; Fig. 5E, F): foxe3 to normal levels in 37% and rax expression to normal levels in 30% of cases, with reduced expression in the other cases. Normal development ensued in 76% of these embryos cultured to stage 34 and beyond. In rescued embryos the eyes appear indistinguishable from controls (Fig. 6A,C; D,F). Taken together, these data suggest a relationship between ldb1 and normal expression of eye and lens markers, that generalized MO toxicity was not responsible for phenotypes seen in MO injections, and that normal eye and lens development require ldb1.
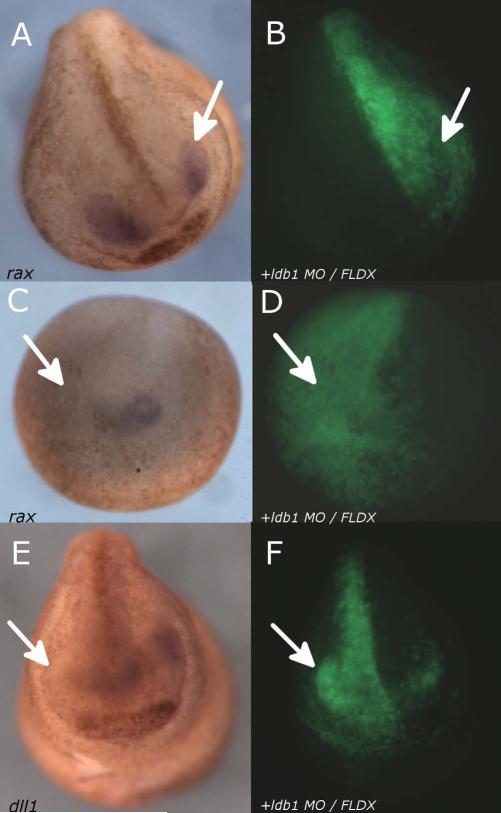
Since it has been shown that dll1 plays a role in foxe3 activation (Ogino et al., 2008), we examined the link between ldb1 depletion and dll1 expression. We also examined the link between ldb1 depletion and rax expression, since it is a definitive marker of the retina (Bailey et al., 2004). We coinjected 35ng ldb1 MO and 45ng FLDX into one of two dorsal blastomeres at the 4-cell stage and cultured to stages 15 or 21 before processing for in situ hybridization for rax or dll1 (Fig. 7). The reduction of endogenous ldb1 on the injected side results in the reduction or elimination of rax in 72% of cases (n=18; Fig. 7A,C) and reduction or elimination of dll1 in 90% of cases (n=10; Fig. 7E), with the uninjected side remaining essentially normal.
Fig. 7.
Expression of rax and dll1 reduced in embryos injected unilaterally with ldb1 MO. A-D: Embryos injected into one of two dorsal blastomeres at the 4-cell stage with 35ng ldb1 MO and 45ng FLDX, then cultured to Stage 21 (A-B) or Stage 15 (C-D) and processed for in situ hybridization for rax (A,C); lineage label in B, D. E-F: Embryo injected into one of two dorsal blastomeres at the 4-cell stage with 35ng ldb1 MO and 45ng FLDX, then cultured to stage 21 and processed for in situ hybridization for dll1(E); lineage label in F. Arrows indicate reduction in expression on the injected side compared to the uninjected side.
DISCUSSION
We have isolated a truncated form of the previously identified transcriptional co-regulator ldb1 in an assay designed to identify inducers of early-stage lens induction, expanding upon ldb1's known role in development. Our data indicate that its role in induction is not as a direct signaling molecule; rather, its actions are indirect and carried out through the nucleus. Expression of this gene in oocytes or animal cap tissue, or enriched expression in the anterior neural plate, appears to mediate early inductive responses linked to several anterior ectodermal head structures. Presumably by acting with endogenous transcription factor partners, ldb1 activates early signals from the neural plate to the adjacent non-neural ectoderm and activates genes expressed in the presumptive lens, olfactory epithelium, and cement gland. This activation as well as later eye development can be inhibited by depletion of ldb1, and restored using ldb1 RNA which is not a target of the depletion.
Oocyte-animal cap assay: cloning of directly and indirectly acting genes
Our oocyte-animal cap assay was anticipated to select for genes that were acting as direct signaling molecules to mediate ectodermal induction following translation of their mRNA. Δldb1's role in mediating induction via the nucleus, however, was demonstrated using enucleated oocytes: while injected Δldb1 RNA is translated efficiently in the absence of the nucleus, its function is eliminated. Our assay system, using the active transcriptional machinery of the Stage VI oocyte, yielded a clone which functioned through a multistep process. Thus, the system is capable of identifying a far wider range of molecules than had previously been thought. As to why a direct-acting molecule was not identified, it is possible that more than one factor secreted at the same time is necessary for a positive response; alternatively, the amount of RNA encoding a directly-acting molecule added at the largest pool size of 105 clones, or even at 104 clones, may have been insufficient to yield a positive result. Therefore, this assay appears useful for the selection of molecules which can be amplified by a nuclear-mediated mechanism.
The results of analysis at the sequence, expression pattern, and functional levels indicate that Δldb1 is a truncation of ldb1 and that the functions of the longer clone (mediated by the NLS and LID domains; see Fig. 1A) are conserved in the truncation. One possible reason for the selection of Δldb1 in our screen was its efficient translation in oocytes. The high rate of translation of Δldb1, not observed for ldb1, as well as the potent effect of injected Δldb1 in oocytes which already contain ldb1 RNA suggest that ldb1 is subject to translational control, being either inefficiently translated or non-translated in the oocyte. The idea that ldb1 is under translational repression in the oocyte is consistent with the observation that very small amounts (one-quarter to one-half of endogenous message levels) of Δldb1 were able to mediate head ectodermal inductive activity in early stages of the screen. Additionally, differential stability of Δldb1 and ldb1 products may account for observed differences in activity, since the N-terminal domain of ldb1 known to be sufficient for rnf12-mediated ubiquitination (Hiratani et al., 2003) and subsequent degradation is lacking in Δldb1.
ldb1 activates anterior ectodermal genes in naive tissue
To determine the extent of the inducing ability of ldb1 with cofactors present in early embryogenesis, we overexpressed Δldb1 in two tissues: animal cap ectoderm and the anterior neural plate. Only genes of the anterior non-neural ectoderm – lens, olfactory epithelium, and cement gland – were activated in response.
The expression of foxe3 at the anterior neural fold during neurula stages encompasses a region which includes the olfactory placodal field but also includes midline tissue thought to give rise to forebrain structures (Eagleson and Harris, 1990). This observation underscores the difficulty that has complicated the analysis of early olfactory induction. In particular, the olfactory placode is formed from inner and outer layers of ectoderm and neural and non-neural ectoderm, so interactions between these tissues are difficult to separate. For example, dlx5 was expressed in the outer layer of ectoderm in all positive recombinants (Fig. 3F and data not shown), suggesting that the dlx5 is associated with an early cement gland response, but an early olfactory response cannot be ruled out. The common induction of early genes of the head we observe to be mediated by ldb1 not only strengthens the link between the early phases of lens and olfactory induction (Pandit et al., 2011), but also suggests an early broad activation of anterior ectodermal properties shared by many presumptive anterior structures in this preplacodal region (Bhattacharyya and Bronner, 2013).
ldb1 may be acting in a cell autonomous or non-cell autonomous manner. Cement gland and olfactory epithelium markers are activated in the Δldb1-injected animal cap in ectodermal recombinants, and this raises the possibility that the response in these tissues is mediated by a cell or tissue autonomous mechanism. It is also possible that in addition to activating autocrine, juxtacrine, or paracrine signaling molecules (as is suggested by the activation of foxe3 in the oocyte-animal cap assay), ldb1 can act directly on transcription factors in some anterior ectodermal regions. The ability of a truncated ldb1 (containing the NLS and LID domains, nearly identical to Δldb1) fusion construct to induce β-globin transcription in murine erythroid cells (Deng et al., 2012) demonstrates that these domains of ldb1 can efficiently engage binding partners to effect transcription.
The increase in foxe3 expression observed in ldb1-overexpressing neural plate recombinants was distributed over a much broader region than control recombinants (Fig. 4). This broad continuous domain may represent the activation of a contiguous region of presumptive olfactory and lens ectoderm as well as other domains, as discussed above. The ability of the neural tissue to increase signaling suggests that ldb1-family cofactors may be limiting in at least some aspects of anterior ectodermal induction. There are clearly upper limits to the amount of exogenous ldb1 which can effect a positive response. In addition, the endogenous signal may be attenuated by limiting concentrations of ldb1. However, as suggested by the work of Hiratani et al. (2003), the balance of LIM domain-binding proteins, LIM-only proteins, and LIM-homeodomain transcription factors may be easily disrupted by a shift in levels of any of these partners.
Reduction of endogenous ldb1 disrupts normal eye development
While the assays discussed above all suggest that ldb1 is sufficient to activate a pathway leading to foxe3 expression, the MO depletion of ldb1 further demonstrates the necessity of ldb1 to normal gene expression and patterning in and around the anterior neural plate. Our MO-mediated knockdown of endogenous ldb1 shows that reduction of ldb1 affects subsequent expression of foxe3, rax, and dll1. These findings support the idea that ldb1 plays an important role in shifting fates during anterior neural and placodal development. Although we tested three markers, the mouse ldb1 knockout (Mukhopadhyay et al., 2003) suggests the effects may be much more widespread, since severe neural, head, and heart defects were observed. Additionally, the morphological abnormalities we observe with altered ldb1 expression (such as cement gland enlargement, eye defects, and later disintegration) are reminiscent of the severe anterior deficiencies and later apoptotic cell death noted in null mutant mice (Mukhopadhyay et al., 2003).
foxe3 was clearly activated in our ldb1 overexpression experiments (Figs. 3 and 4) and knocked down in our MO studies (Fig. 5); these phenomena may be linked to one another by a direct mechanism in which ldb1 is acting as part of a regulatory system that activates a lens-inducing factor. Alternatively, the knockdown may be due to a separate phenomenon whereby ldb1 depletion is acting to inhibit retinal development and thus affecting foxe3 expression in an indirect manner by a more generic effect on retina formation. Thus, the MO could be altering the activity of a different binding partner or target of ldb1 or the same one as that altered by overexpression. The greatest level of reduction in expression in response to MO knockdown was exhibited by dll1, and this suggests that the dll1 may be a direct target of ldb1 and its relevant partner, and that notch-dll1 signaling may be affected by misexpression of ldb1. It was previously demonstrated that notch-dll1 signaling plays a role in the activation of foxe3 transcription (Ogino et al., 2008), establishing a link between dll1 ligand expression in the presumptive optic vesicle and foxe3 expression in the PLE. The question then arises whether this signaling pathway was activated and exogenous ldb1-induced dll1 expression provided the source of signaling to induce expression of foxe3 in the responding animal cap in our expression cloning assay. It is a distinct possibility, since the notch receptor is expressed in the animal cap by stage 10.5 (Miazga and McLaughlin, 2009). This also provides a possible explanation for why we did not observe a secreted protein band in the supernatant of our metabolically-labeled Δldb1-injected oocytes (data not shown), since dll1 is a membrane-tethered ligand (Chitnis et al., 1995).
Interactions of ldb1 with other transcription factors
Identification of other members of the LIM domain-binding protein family such as CLIM/ldb2 (Bach et al., 1997) has provided key insights on the functional domains of this group of proteins. Deletion analysis of ldb1 and ldb2 revealed that while the N-terminal self-association domain mediates formation of trimers and higher order oligomers (Cross et al., 2010) and may be sufficient for chromatin looping and transcriptional activation of β-globin in erythroid cells (Deng et al., 2012; Krivega et al., 2014), the C-terminal LIM interaction domain (LID) mediates interactions with transcription factors including lhx1 and other lhx family members and otx family members such as pitx1. A tested region of only 63 amino acids was found to be necessary for efficient interaction and synergy with transcriptional binding partners in the pituitary (Bach et al., 1997), and shows the required region to be from 21 amino acids upstream of the nuclear localization signal (NLS) to the C-terminus. By comparison, Δldb1 begins 15 amino acids upstream of the first NLS and 40 amino acids upstream of the second NLS and contains the LID (see Fig. 1). A detailed deletion analysis of ldb1 found a 38 amino acid region to be sufficient for LID function (Jurata and Gill, 1997), and this region as well is contained within the boundaries of Δldb1. Thus, it is clear that Δldb1 contains the functional domains necessary to act as a transcriptional co-regulator.
ldb proteins have been identified as key factors in the assembly of transcriptional complexes in a wide variety of developing tissues, including roles in hematopoiesis (Meier et al., 2006; Song et al., 2010; Li et al., 2011), limb patterning (Tzchori et al., 2009), neural patterning (Ostendorff et al., 2006; Zhao et al., 2007), and other processes including cancer development (Teufel et al., 2010). Since targeted ablation of the mouse ldb1 gene results in such a severe pleiotropic phenotype, including truncation of head structures anterior to the hindbrain, it is necessary to use a more subtle approach to address the role of ldb in the development of eye and placodal structures. ldb1 null ES cells have also been generated (Hwang et al., 2008), and were demonstrated to be incapable of neuronal differentiation when treated so as to mimic in vivo neural induction.
ldb proteins have been demonstrated to act positively to assemble LIM-homeodomain (lhx) proteins and other transcription factors (reviewed by Matthews and Visvader, 2003), participate in multiprotein complexes (Gungor et al., 2007) and to play a key role in activation of downstream targets in a dose-responsive manner. Since over- or underexpression of ldb1 has been demonstrated to disturb lhx-ldb stoichiometry and lead to misexpression of its binding partners in the eye (lhx2, lhx3, lhx9) and their downstream targets (Hiratani et al., 2003), evidence is accumulating to suggest that disruption of ldb levels in early development may have multiple effects on target tissues. Further, established binding partners of ldb such as lhx2 have been demonstrated to not only be expressed in the eye but also to be necessary for eye formation (Viczian et al., 2006). In our hands, overexpression of ldb1 in whole embryos, both alone and in combination with otx2, caused a reduction in eye size (data not shown). ldb1 acts as a cofactor in a number of transcriptional complexes and may act in a competitive or dominant negative manner when overexpressed in some contexts. Positive transcriptional regulation of gsc, a homeobox gene expressed in the mesoderm signaling the anterior neural plate, was demonstrated to be exerted by lhx1 and ldb1 with further synergistic activation exerted by otx2, suggesting a model of lhx1 and ldb1 acting upstream of otx2, and then together with otx2 on downstream targets; the same study demonstrated gsc is capable of inhibiting the activity of its own promoter (Mochizuki et al., 2000). When we overexpress ldb1 in whole embryos, gsc expression is reduced or eliminated (data not shown). otx2 is also present in the oocyte (Pannese et al., 1995), suggesting that at least part of the signal activated by Δldb1 may be mediated by interactions with otx2 in the oocyte. This, together with evidence that the severe head phenotype of the mouse ldb1 ablation is due to disruption of an otx2-mediated pathway (Mukhopadhyay et al., 2003), lends support to the idea that otx2 is involved in regulation of the endogenous early placode-inducing signal. Further, since spatio-temporal modulation of otx2 activity limits cement gland formation to the anterior end of the embryo (Gammill and Sive, 1997), up- or downregulation of a key binding partner would be expected to alter the size of structures forming in the placodal region, as we have observed.
Downregulation of ldb proteins by LIM-domain-only (LMO) proteins have been well-documented (Calle-Mustienes et al., 2003; Matthews et al., 2008), and evidence has been presented that ldb1 may be negatively regulated by transcriptional intermediary factors through an ubiquitin-mediated degradation process (Howard et al., 2010 a,b) or stabilized by ubiquitination at specific ldb1 residues (Howard et al., 2010 b). Germane to its role in establishing lens-forming bias and other anterior ectodermal properties, it will be illuminating in the future to focus directly on the activation of ldb1 and to confirm the specific targets activated by ldb1 and its binding partners.
Positive and negative signals for anterior ectodermal patterning
A complex series of positive and negative instructions are necessary to form the ectodermal structures of the vertebrate head. These consist of both early signals as well as later interactions which result in the specific determination and positioning of sense organs. The ldb1-mediated inductive activation of lens, olfactory, and cement gland genes we have observed leads to the development of a model for the role of ldb1 in early anterior ectodermal induction. An ldb1-activated signal may pattern or bias the head ectoderm and allow later specific determination by other sources. This idea of a common early mechanism is consistent with the evolutionary and embryological ideas of a common placodal stage put forth by Gans and Northcutt (1983), Jacobson (1966), and Schlosser and Ahrens (2004). The proximity of presumptive tissues in and around the anterior neural plate is also consistent with this idea.
Eye field transcription factors (otx2, rax, six3, pax6, lhx2, and others) work synergistically to specify the eye field (Zuber at al., 2003) and to activate transcription of secreted molecules and other ligands that exert a lens-forming bias, lens specification, and ultimately lens differentiation. This delicate balance of levels and timing of such signaling molecules is subject to disruption by over- or underexpression of ldb1, and as we have shown, affects the expression of molecules in the eye field (rax, dll1) as well as in the responding lens ectoderm (otx2, foxe3). We propose that ldb1 may be acting synergistically in multiple converging pathways in the development of the retina and the specification of the lens. The reduction in rax expression (Figs. 5 and 7) and eye size, the shifting of fates in the non-neural ectoderm (enlargement of cement gland), and the later disorganization of the retina and lens (Fig. 6) resulting from ldb1 depletion may all represent a reduction in the size and normal development of the eye field. While the reduction in dll1 expression may be also due to an indirect mechanism, our data, as well as the established link between notch-dll1 and foxe3 in the specification of the lens (Ogino et al., 2008) and the activation of foxe3 in our oocyte-animal cap assays, all lead us to suggest that dll1 may be a direct target of ldb1 and its transcriptional cofactors.
EXPERIMENTAL PROCEDURES
Tissue recombinants
Embryos were cultured in 1/10X Normal Amphibian Medium (NAM; Slack, 1984); all dissections were performed in 3/4X NAM. All embryo stages are according to Nieuwkoop and Faber (1994). Tissues were lineage-labeled by injection at the 1-cell stage with RLDX or FLDX (Molecular Probes). To remove neural plates from underlying tissues, embryos were dissected in 0.01% trypsin (Sigma T-8253) and rinsed in 0.02% soybean trypsin inhibitor (Sigma T-9003). Tissues were dissected in clay-lined dishes and the pieces held together by clay. Recombinants were cultured to stages 18 to 24, then subjected to in situ hybridization.
cDNA library synthesis
Embryos were dorsalized with 0.3M LiCl for 5 minutes at the 32-cell stage to a dorso-anterior index (DAI) of 8 (Kao and Elinson, 1988). Embryos were collected at stage 14 and RNA isolated by the acid guanidinium/phenol method (Chomzcynski and Sacchi, 1987). A Poly(A)+ RNA-enriched fraction was isolated using an oligo d(T)-cellulose Type 3 (Collaborative Biomedical Products) column (Sive et al., 2000). 5 μg of this RNA was used to make a directional plasmid cDNA library using the SuperScript Plasmid System for cDNA Synthesis and Plasmid Cloning (Gibco BRL). 20 ng cDNA was ligated into 50 ng SalI-NotI cut pCS105 vector and one-quarter of this ligation was transformed into Epicurean Coli XL2-Blue ultracompetent cells (Stratagene). Inserts were released with SalI-NotI; the average insert size was 1.75kb with a range from .5-3kb. Complexity was estimated at 2 × 106. Aliquots of the cDNA library were linearized with NotI and transcribed with SP6 RNA polymerase according to Sambrook et al. (1989). These transcripts were used in the oocyte-animal cap assay.
Northern analysis
RNA ran on 1-1.2% agarose-formaldehyde gels was blotted onto GeneScreen (DuPont/New England Nuclear), transferred (Sambrook et al., 1989), and UV-crosslinked at 120 J/cm2 in a Fisher Biotech UV Crosslinker (Fisher Scientific). 32P-labeled random-primed cDNA probes were made with the SalI-NotI fragment of Δldb1 purified with Geneclean (Bio101). Hybridizations were 12-18 hours at 42°C in hybridization buffer containing 50% formamide (Sambrook et al., 1989). Filters were washed in 0.2X SSC, 0.1% SDS at 37°C for 2 hours, exposed to a phosphorimager screen for 8-16 hours (Molecular Dynamics), and analyzed using ImageQuant software (Molecular Dynamics).
PCR Cloning of ldb1
Full-length ldb1 was cloned using primers made from the ends of the coding region reported in GenBank (accession number U74360; Agulnick et al., 1996). Following isolation of a 1.1 kb fragment from a neurula stage cDNA library (Kintner and Melton, 1987), it was cloned into the BamHI-EcoRI site of pCSII. Sequencing (UVa Biomolecular Research Facility, Charlottesville, VA) confirmed sequence identity to ldb1.
Oocyte-animal cap assay
Oocytes were obtained by surgical isolation of Stage VI ovarian fragments and defolliculation by 1 hour treatment with 1 mg/ml collagenase A (Boehringer-Mannheim) in Ca++/Mg++-free OR2 (Sive et al., 2000). Oocytes were washed in OR2 containing Ca++/Mg++ and transferred to Oocyte Culture Medium (OCM; Wylie et al., 1996). Following overnight culture, oocytes were injected with 20 nl (1 ng/nl) RNA and incubated for 8-24 hours at 20°C. Animal cap ectoderm was prepared for co-culture with oocytes by dissection from mid-gastrula (stage 11-11.5) embryos. Oocytes were immobilized in impressions made in clay-lined dishes. Animal caps were placed on the oocytes and held together by curved glass coverslip fragments. Oocyte-cap recombinants were cultured until control embryos reached stage 23-25 at 20°C, then separated. Animal caps were fixed 1 hour in MEMFA (3.8% formaldehyde, 0.1M MOPS pH 7.4, 2 mM EGTA, 1 mM MgSO4).
To test oocyte translation and secretion in functional assays, 20 pg inhbb (Sokol et al., 1991) RNA was injected into oocytes and incubated as above. Blastula (stage 8-9) animal cap ectoderm was co-cultured with oocytes and assayed for presence of muscle protein with the 12/101 antibody (Kintner and Brockes, 1984). Muscle-inducing activity was observed with 2-200 pg inhbb mRNA, even in the presence of 500-fold excess co-injected stage 14 poly(A)+ RNA.
Enucleated oocyte-animal cap assays were performed as above, modified according to the procedure of Dabauvalle et al. (1988) by 5 minute treatment in 0.5X Modified Barth's Solution (MBS) and extrusion of the nucleus through a small incision at the animal pole. Following enucleation, oocytes were healed in potassium phosphate solution (90mM phosphate pH 7.2, 10mM NaCl, 1mM MgSO4) and recovered in full-strength MBS.
Sib selection of positive clones
Sib selection of a clone with inducing activity was carried out according to the procedure of Smith and Harland (1991, 1992). Six pools of 100,000 cDNAs from the stage 14 plasmid cDNA library described above were initially tested under varying RNA concentrations, age of responding tissue, and culture conditions. Since 4/6 pools showed positive results for otx2 expression we moved to analyze 10 pools of 10,000 and begin splitting pools 10-fold. At each step, colonies were grown on 10 LB-ampicillin plates and collected in 7 ml LB. A glycerol stock was prepared from 0.5 ml, and the remaining 6.5 ml was used to prepare DNA by alkaline lysis method. Pooled DNA was linearized with NotI and transcripts were synthesized with SP6 polymerase. The RNA pool with the highest activity was selected; the glycerol stock was titrated and used to plate out 10 plates with one-tenth the colonies from the previous step. Activity was traced to a single clone, which was identified as a truncated form of the gene ldb1. Sequence was read from the SP6 and T3 promoter/primers of pCS105 and the ldb1 downstream primer.
Metabolic labeling and SDS-PAGE
To observe protein synthesis in oocytes, RNA-injected and control oocytes were incubated in 5 μl per oocyte 1mCi/ml 35S-Methionine in Oocyte Culture Medium (OCM) for 24-36 hours at 20°C. Leupeptin, chymostatin, and pepstatin (Boehringer-Mannheim) were added at 0.25 mg/ml. Following incubation, all supernatant was separated from the oocytes, and 1 μl 0.3M PMSF added to oocytes. The oocytes were rinsed in OCM then homogenization buffer (HB; 100mM NaCl, 20mM Tris pH 7.6, 1% Triton X-100, 1mM PMSF). Oocytes were homogenized in 5 μl HB per oocyte. Following a 10 minute 4°C 10,000 × g spin, the cytoplasmic portion was analyzed by SDS-PAGE. The supernatant incubated with 2.5 oocytes or the homogenate from 1 oocyte was analyzed in each lane. Following fixation in 25% methanol-7% acetic acid destain, the gel was dried, exposed to a phosphorimager screen (Molecular Dynamics), and analyzed using ImageQuant software (Molecular Dynamics).
In situ hybridization, immunohistochemistry, and histology
In situ hybridization probes were prepared for foxe3 (probe courtesy of Milan Jamrich), otx2 (probe courtesy of Richard Harland), nrl-maf (probe courtesy of Kunio Yasuda), γ-crystallin (Smolich et al., 1993), dlx5 (probe courtesy of Nancy Papalopulu), ag1 (probe courtesy of Hazel Sive), ncam1 (probe courtesy of Nancy Papalopulu), snai2 (probe courtesy of Roberto Mayor), sox2 (Grainger, unpublished), rax (Andreazzoli et al., 1999), and dll1 (Chitnis et al., 1995). Probes were prepared for Δldb1 and ldb1. In situ hybridization was carried out according to Sive et al. (2000). Whole-mount immunocytochemistry with 12/101 muscle antibody (Kintner and Brockes, 1984; Developmental Studies Hybridoma Bank) was carried out as described by Sive et al. (2000).
Morpholino oligonucleotides
ldb1 MO (GTCCCACATCTCGATCCAGCATGGT) from Gene Tools LLC. For bilateral injections, ldb1 MO (65 ng) or ldb1 MO (65 ng) plus 0.11 ng Δldb1 RNA was injected into each zygote, in approximately 8nl volume. For unilateral injections, 35ng ldb1 MO and FLDX were coinjected into one of two dorsal blastomeres at the 4-cell stage.
Key Findings.
Transcriptional cofactor ldb1 isolated in a functional screen for early lens-inducing activity
Overexpression and recombinant assays suggest ldb1 is sufficient to activate a pathway leading to activation of lens and other anterior head ectodermal properties
Depletion of ldb1 results in a reduction of the eye field and disruption of lens development
This study lays the groundwork for linking an ldb1-mediated regulatory event to activation of lens-inducing signals, possibly dll1
ACKNOWLEDGEMENTS
The authors thank Drs. Marilyn Fisher and Takuya Nakayama for helpful comments and discussions, and Dr. Carol Hurney for generous assistance.
Grant Sponsor: NIH
Grant Numbers: EY05542, EY06675, EY10283 and EY17400 to RMG
REFERENCES
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of Xrx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Baker CVH, Bronner-Fraser M. Vertebrate Cranial Placodes I. Embryonic Induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner ME. Clonal analyses in the anterior pre-placodal region: implications for the early lineage bias of placodal progenitors. Int J Dev Biol. 2013;57:753–757. doi: 10.1387/ijdb.130155mb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle-Mustienes E, Lu Z, Cortes M, Andersen B, Modolell J, Gomez-Skarmetaa JL. Xenopus Xlmo4 is a GATA cofactor during ventral mesoderm formation and regulates Ldb1 availability at the dorsal mesoderm and the neural plate. Dev Biol. 2003;264:564–581. doi: 10.1016/j.ydbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene dll1. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Chomzcynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analyt Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cross AJ, Jeffries CM, Trewhella J, Matthews JM. LIM domain binding proteins 1 and 2 have different oligomeric states. J Mol Biol. 2010;399:133–144. doi: 10.1016/j.jmb.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Dabauvalle MC, Doree M, Bravo R, Karsenti E. Role of nuclear material in the early cell cycle of Xenopus embryos. Cell. 1988;52:525–533. doi: 10.1016/0092-8674(88)90465-5. [DOI] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale TA, Elinson RP. Inductive events in the patterning of the Xenopus laevis hatching and cement glands, two cell types which delimit head boundaries. Dev Biol. 1993;158:245–253. doi: 10.1006/dbio.1993.1183. [DOI] [PubMed] [Google Scholar]
- Eagleson GW, Harris WA. Mapping of the presumptive brain regions in the neural plate of Xenopus laevis. J Neurobiol. 1990;21:427–440. doi: 10.1002/neu.480210305. [DOI] [PubMed] [Google Scholar]
- Fini ME, Strissel KJ, West-Mays JA. Perspectives on eye development. Dev Genet. 1997;20:175–185. doi: 10.1002/(SICI)1520-6408(1997)20:3<175::AID-DVG1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Gallagher BC, Henry JJ, Grainger RM. Inductive processes leading to inner ear formation during Xenopus development. Dev Biol. 1996;175:95–107. doi: 10.1006/dbio.1996.0098. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Sive H. Identification of otx2 target genes and restrictions in ectodermal competence during Xenopus cement gland formation. Development. 1997;124:471–481. doi: 10.1242/dev.124.2.471. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–274. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Grainger RM. Embryonic lens induction: shedding light on vertebrate tissue determination. Trends Genet. 1992;8:349–355. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- Grainger RM, Mannion JE, Cook TL, Jr, Zygar CA. Defining intermediate stages in cell determination: acquisition of a lens-forming bias in head ectoderm during lens determination. Dev Genet. 1997;20:246–257. doi: 10.1002/(SICI)1520-6408(1997)20:3<246::AID-DVG7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. PNAS. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JJ, Grainger RM. Early tissue interactions leading to embryonic lens formation in Xenopus laevis. Dev Biol. 1990;141:149–163. doi: 10.1016/0012-1606(90)90110-5. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Yamamoto N, Mochizuki T, Ohmori S, Taira M. Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development. 2003;130:4161–4175. doi: 10.1242/dev.00621. [DOI] [PubMed] [Google Scholar]
- Howard PW, Ransom DG, Maurer RA. Transcription intermediary factor 1gamma decreases protein expression of the transcriptional cofactor, LIM-domain-binding 1. Biochem Biophys Res Commun. 2010;396:674–678. doi: 10.1016/j.bbrc.2010.04.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PW, Jue SF, Ransom DG, Maurer RA. Regulation of LIM-domain-binding 1 protein expression by ubiquitination of Lys134. Biochem J. 2010;429:127–136. doi: 10.1042/BJ20091461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang M, Gorivodsky M, Kim M, Westphal H, Geum D. The neuronal differentiation potential of Ldb1-null mutant embryonic stem cells is dependent on extrinsic influences. Stem Cells. 2008;26:1490–1495. doi: 10.1634/stemcells.2007-1099. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. The determination and positioning of the nose lens and ear. I. Interaction within the ectoderm and between the ectoderm and underlying tissues. J Exp Zool. 1963a;154:273–284. doi: 10.1002/jez.1401540303. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. The determination and positioning of the nose lens and ear. II. The role of the endoderm. J Exp Zool. 1963b;154:285–292. doi: 10.1002/jez.1401540304. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. Inductive processes in embryonic development. Science. 1966;152:25–34. doi: 10.1126/science.152.3718.25. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KR, Elinson RP. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol. 1988;127:64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Moody SA, Jamrich M. A novel fork head gene mediates early steps during Xenopus lens formation. Development. 1999;126:5107–5116. doi: 10.1242/dev.126.22.5107. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastemal cells derived from dedifferentiating muscle in newt limb regeneration. Nature. 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Melton DA. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- Krivega I, Dale RK, Dean A. Role of LDB1 in the transition from chromatin looping to transcription activation. Genes Dev. 2014;28:1278–1290. doi: 10.1101/gad.239749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, Bresnick EH, Zhao K, Westphal H, Love PE. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig KD, Kirschner MW. Use of an oocyte expression assay to reconstitute inductive signaling. PNAS. 1995;92:6234–6238. doi: 10.1073/pnas.92.14.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Reports. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Bhati M, Craig VJ, Deane JE, Jeffries C, Lee C, Nancarrow AL, Ryan DP, Sunde M. Competition between LIM-binding domains. Biochem Soc Trans. 2008;36:1393–1397. doi: 10.1042/BST0361393. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Jamrich M. Foxe view of lens development and disease. Development. 2007;134:1455–1463. doi: 10.1242/dev.000117. [DOI] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- Miazga CM, McLaughlin KA. Coordinating the timing of cardiac precursor development during gastrulation: A new role for notch signaling. Dev Biol. 2009;333:285–296. doi: 10.1016/j.ydbio.2009.06.040. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and sox2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–87. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Karavanov AA, Curtiss PE, Ault KT, Sugimoto N, Watabe T, Shiokawa K, Jamrich M, Cho KWY, Dawid IB, Taira M. Xlim-1 and LIM Domain Binding Protein 1 cooperate with various transcription factors in the regulation of the goosecoid promoter. Dev Biol. 2000;224:470–485. doi: 10.1006/dbio.2000.9778. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J. Normal Table of Xenopus laevis (Daudin) Garland Publishing, Inc.; NY: 1994. [Google Scholar]
- Ogino H, Fisher M, Grainger RM. Convergence of a head-field selector otx2 and notch signaling: a mechanism for lens specification. Development. 2008;135:249–258. doi: 10.1242/dev.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Involvement of maf gene family in crystallin gene regulation. Tanpakushitsu Kakusan Koso - Protein, Nucleic Acid, Enzyme. 1996;41:1050–1057. [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Sequential activation of transcription factors in lens induction. Develop Growth Differ. 2000;42:437–448. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, Bach I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev Dyn. 2006;235:786–791. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- Pandit T, Jidigam VK, Gunhaga L. BMP-induced L-Maf regulates subsequent BMP-independent differentiation of primary lens fibre cells. Dev Dyn. 2011;240:1917–1928. doi: 10.1002/dvdy.22692. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. Xenopus Distal-less related homeobox genes are expressed in the developing forebrain and are induced by planar signals. Development. 1993;117:961–975. doi: 10.1242/dev.117.3.961. [DOI] [PubMed] [Google Scholar]
- Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. The Xenopus homologue of otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development. 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; N.Y.: 1989. [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servetnick M, Grainger RM. Changes in neural and lens competence in Xenopus ectoderm: evidence for an autonomous developmental timer. Development. 1991;112:177–188. doi: 10.1242/dev.112.1.177. [DOI] [PubMed] [Google Scholar]
- Sive HL, Hattori K, Weintraub H. Progressive determination during formation of the anteroposterior axis in Xenopus laevis. Cell. 1989;58:171–180. doi: 10.1016/0092-8674(89)90413-3. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; NY.: 2000. [Google Scholar]
- Slack JM. Regional biosynthetic markers in the early amphibian embryo. J Emb Exp Morph. 1984;80:289–319. [PubMed] [Google Scholar]
- Smith WC, Harland RM. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- Smolich BD, Tarkington SK, Saha MS, Stathakis DG, Grainger RM. Characterization of Xenopus laevis gamma-crystallin-encoding genes. Gene. 1993;128:189–195. doi: 10.1016/0378-1119(93)90562-h. [DOI] [PubMed] [Google Scholar]
- Sokol S, Christian JL, Moon RT, Melton DA. Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell. 1991;67:741–752. doi: 10.1016/0092-8674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel A, Maass T, Strand S, Kanzler S, Galante T, Becker K, Strand D, Biesterfeld S, Westphal H, Galle PR. Liver-specific Ldb1 deletion results in enhanced liver cancer development. J Hepatol. 2010;53:1078–1084. doi: 10.1016/j.jhep.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love PE, Porter FD, Westphal H, Yang Y. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development. 2009;136:1375–1385. doi: 10.1242/dev.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczian AS, Bang AG, Harris WA, Zuber ME. Expression of Xenopus laevis Lhx2 during eye development and evidence for divergent expression among vertebrates. Dev Dyn. 2006;235:1133–1141. doi: 10.1002/dvdy.20708. [DOI] [PubMed] [Google Scholar]
- Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Maternal β-catenin establishes a dorsal signal in early Xenopus embryos. Development. 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. PNAS. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- Zygar CA, Cook TL, Jr, Grainger RM. Gene activation during early stages of lens induction in Xenopus. Development. 1998;125:3509–3519. doi: 10.1242/dev.125.17.3509. [DOI] [PubMed] [Google Scholar]