Abstract
Microsatellite-expansion diseases are a class of neurological and neuromuscular disorders caused by the expansion of short stretches of repetitive DNA (e.g. GGGGCC, CAG, CTG …) within the human genome. Since their discovery 20 years ago, research into how microsatellites expansions cause disease has been examined using the model that these genes are expressed in one direction and that expansion mutations only encode proteins when located in an ATG-initiated open reading frame. The fact that these mutations are often bidirectionally transcribed combined with the recent discovery of repeat associated non-ATG (RAN) translation provides new perspectives on how these expansion mutations are expressed and impact disease. Two expansion transcripts and a set of unexpected RAN proteins must now be considered for both coding and “non-coding” expansion disorders. RAN proteins have been reported in a growing number of diseases, including spinocerebellar ataxia type 8 (SCA8), myotonic dystrophy type 1 (DM1), Fragile-X tremor ataxia syndrome (FXTAS), and C9ORF72 amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD).
Keywords: RAN translation, SCA8, DM1, FXTAS, ALS, FTD, microsatellite expansions, dipeptide repeat
Overview of microsatellite expansion disorders
Microsatellite expansion disorders are a growing family of neurological and neuromuscular diseases caused by the expansion of short (3–6 nucleotides) repetitive sequences in the human genome [1]. The position of the expansion mutation, within or outside an ATG-initiated open reading frame (ORF), has provided the framework for research into the molecular consequences of these mutations [2]. For example, research into CAG expansions within ATG-initiated ORFs (e.g. Huntington’s disease (HD) and several ataxias), has focused almost exclusively on understanding the pathogenic effects of the resulting ATG-initiated polyGln proteins [3, 4]. In contrast dominant diseases caused by expansion mutations located outside ATG-initiated ORFs (e.g. myotonic dystrophy) have focused on the toxic effects of expanded RNA transcripts and the dysregulation of RNA binding proteins [5, 6]. Cell culture and animal models to study these diseases have been built with the expectation that expansions in coding regions encode a single mutant protein and non-coding expansions do not encode proteins [7]. While substantial data support polyGln toxicity and RNA gain of function mechanisms [8], recent discoveries that fundamentally change our understanding of how genes are expressed must now be considered. First, a growing number of expansion mutations are known to be bidirectionally transcribed producing expansion RNAs in both directions [2, 9–11]. Second, in 2011, Zu et al., [12] demonstrated that expansion mutations can express proteins in all three reading frames without an AUG initiation codon. This novel type of translation is called repeat associated non-ATG (RAN) translation [12]. These discoveries have uncovered previously unappreciated expansion RNAs and novel sets of disease-specific expansion proteins. Our current understanding of the molecular biology of RAN translation and progress towards understanding its role in disease will be discussed.
The discovery of RAN translation in SCA8
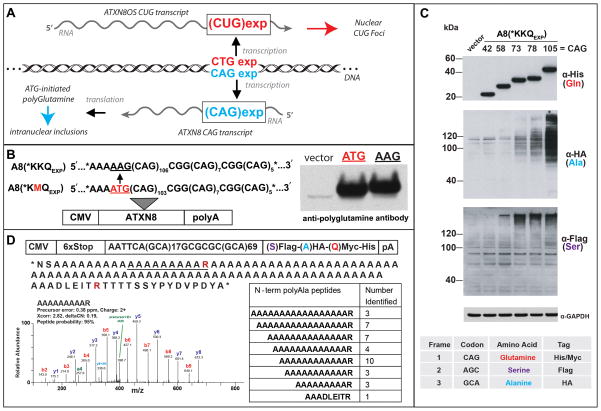
Spinocerebellar ataxia type 8 (SCA8) is a dominantly inherited neurodegenerative disorder caused by an expanded CTG•CAG repeat in the ATXN8 gene [13]. This expansion mutation is bidirectionally transcribed producing both CUG and CAG expansion transcripts and an ATG-initiated polyGln expansion protein (Fig. 1A) [10]. RAN translation was first discovered in SCA8 by Zu et al. [12] when a control experiment to block ATG-initiated ATXN8 polyGln translation did not prevent expression of the protein (Fig. 1B). The discovery that a polyGln protein was produced without an ATG-initiation codon, thought to be required to set the reading frame, raised the possibility that proteins might also be made in the other two frames (e.g. GCA and AGC). Surprisingly, experiments with epitope-tagged constructs demonstrated that expanded repeat tracts produce homopolymeric proteins in all three reading frames, polyGln, polyAla and polySer, without an ATG initiation codon (Fig. 1C) [12]. Because these results were completely unexpected, Zu et al. [12] performed a series of control experiments to detect these proteins and to characterize the RNA transcripts. Characterization of this repeat-associated non-ATG (RAN) translation showed no evidence of RNA editing or frame shifting. Mass spectrometry of the polyAla RAN protein showed a series of N-terminal peptides with varying numbers of alanines with no peptides containing an N-terminal methionine suggesting translational initiation occurs without incorporating an N-terminal methionine initiation codon (Fig. 1D) [12]. Immunofluorescence of transfected cells frequently showed RAN proteins from one or two reading frames, and occasionally all three frames, accumulate in individual cells. Additional studies in HEK293T cells showed: 1) hairpin forming CAG but not non-hairpin forming CAA repeats express polyGln RAN proteins; 2) CUG expansion transcripts also express proteins in three reading frames; 3) longer CAG repeats are associated with simultaneous expression of RAN proteins from all three reading frames; 4) the resulting homopolymeric proteins are toxic [12]. In summary, these results demonstrated for the first time that CAG and CUG expansion mutations can express proteins in three reading frames without the canonical ATG initiation codon thought to be required for translational initiation (see Figure 2 and Table 1 for summary).
Figure 1. The discovery of RAN translation in SCA8.
(A) Bidirectional transcription at the SCA8 locus produces CUG expansion transcripts that form RNA foci and CAG expansion transcripts that produce a short ATG-initiated poly-glutamine expansion protein [10]. (B) Surprisingly, mutating the only ATG initiation-codon upstream of the CAG repeat did not prevent the expression of the poly-glutamine protein [12]. (C) Protein blot showing repeat expansion proteins detected by epitope tags are expressed from all three reading frames (poly-glutamine, poly-alanine and poly-serine) without an ATG-initiation codon. Expression of these repeat-associated non-ATG (RAN) proteins is repeat-length dependent, with simultaneous expression from multiple reading frames observed from longer repeat tracts [12]. (D) Mass-spectrometry of the poly-Alanine protein was performed on cell lysates transfected with a modified epitope-tagged CAGEXP construct that encoded an arginine interruption within the polyAla protein to allow trypsin digestion. MS and RNA analysis confirmed that polyAla proteins are expressed without an AUG initiation codon and identified a series of peptides that suggest translation initiation may occur in the polyAla frame at sites throughout the repeat tract [12].
Figure 2. One repeat - multiple RNA and protein products.
Schematic diagram showing potentially toxic RNA and protein products expressed from a repeat expansion mutation through a combination of bidirectional transcription, ATG-initiated and repeat associated non-ATG (RAN) translation. In vitro studies predict ATG-initiated and RAN translation can both occur when the repeat is located in an open reading frame (ORF) [12]. While a single ATG-initiated protein is illustrated, multiple ATG-initiated proteins may be produced if there are multiple ORFs. Additionally, RAN translation of the expanded repeat results in the expression of up to six distinct RAN proteins. For example, a CTG•CAG expansion can produce poly-Gln, poly-Ala and poly-Ser RAN proteins from the CAG transcript and poly-Leu, poly-Ala and poly-Cys RAN proteins from the CUG transcript. Each RAN protein, depending upon flanking sequences, may contain distinct C-terminal regions and an ATG-initiated protein in the same reading frame may also have a distinct N-terminal region.
Table 1.
In vitro and in vivo evidence for RAN translation
| Repeat | In Vitro evidence of RAN proteins | In Vivo evidence of RAN proteins | Reference | |
|---|---|---|---|---|
| SCA8 | CAG•CTG | GlnSa,b,g, AlaSa,b,c,d,f,g, SerSa,b,g LeuASa, AlaASa, CysASa |
AlaSj,m | Zu et al., 2011 [12] |
| DM1 | CAG•CTG | GlnASa,e,f, AlaASa, SerASa | GlnASi,j,l,m | Zu et al., 2011 [12] |
| FXTAS | CGG•CCG | GlySa, AlaSa,d,g | GlySh,j,m | Todd et al., 2013 [24] |
| C9ORF72 ALS FTD | G4C2•G2C4 | GlyProSe, GlyAlaSe | GlyProS/ASl, GlyAlaSl, GlyArgSl | Mori et al., 2013a [47] |
| GlyProS/ASl | Ash et al., 2013 [44] | |||
| GlyProS/ASk | Almeida et al., 2013 [36] | |||
| GlyAlaSl | Mackenzie et al., 2013 [45] | |||
| GlyProS/ASe, ProArgASe | GlyProS/ASl, ProArgASl, ProAlaASl | Gendron et al., 2013 [38] | ||
| GlyProS/ASk | Donnelly et al., 2013 [37] | |||
| GlyArgSl, GlyAlam ProArgASl, ProAlaASl |
Mori et al., 2013b [46] | |||
| GlyProSa,f, GlyArgSa,e,f GlyAlaa,f GlyProS/ASa,e, ProArgASa,e,f, ProAlaASa,e,f |
GlyProSl,m, GlyArgSl,m, GlyAlam GlyProASl, ProArgASl,m, ProAlaASl,m |
Zu et al., 2013 [48] | ||
| GlyArgSl, GlyAlaSl GlyProS/ASl, ProArgASl, ProAlaASl |
Mann et al., 2013 [49] |
NS = sense protein
NAS = antisense protein
NS/AS = sense or antisense protein (antibodies against the repeat cannot distinquish sense GlyPro from antisense GlyPro)
epitope tagged detection of RAN proteins
tritium labelling of RAN proteins
mass spectrometry of repeat
mass spectrometry of flanking sequences
detection of RAN proteins with antibody against repeat
detection of RAN proteins with antibody against C-terminal region
toxicity of RAN protein
detection of RAN proteins with antibody against C-terminal tag (drosophila)
detection of RAN proteins with antibody against repeat (mouse)
detection of RAN proteins with antibody against C-terminal region (mouse)
detection of RAN proteins with antibody against repeat (IPSC)
detection of RAN proteins with antibody against repeat (human tissue)
detection of RAN proteins with antibody against C-terminal region (human tissue)
In vivo evidence for RAN translation in SCA8 and DM1
Zu et al., [12] extended these results by testing the hypothesis that CAG•CTG expansion mutations express RAN proteins in vivo. They developed a peptide antibody that recognizes the C-terminal region of an SCA8 polyAla expansion protein predicted by RAN translation. Immunohistochemistry (IHC) and immunofluorescence (IF) experiments showed that a novel SCA8 polyAla RAN protein accumulates in cerebellar Purkinje cells in both an SCA8 BAC mouse model of the disease and SCA8 human autopsy tissue [12]. The detection of RAN proteins in Purkinje cells, a cell type which shows prominent degeneration in SCA8 patients, is consistent with a role for RAN translation in disease (Table 1).
Zu et al., [12] also provided evidence that RAN translation occurs in myotonic dystrophy type 1. Myotonic dystrophy type 1 is a multisystemic neuromuscular disorder caused by a CTG•CAG expansion mutation in the 3′ UTR of the DMPK gene [14–16]. Research into the pathogenic mechanisms of DM1 has focused on an RNA gain of function mechanism in which expanded CUG transcripts sequester MBNL proteins leading to alternative splicing dysregulation [17–20]. Although there is strong evidence supporting an RNA gain of function mechanism in DM1, it is not yet clear which aspects of the disease are explained by this mechanism. Because DM1 is bidirectionally transcribed [9, 12] and CAG expansion mutations can express proteins without an ATG-initiation codon, Zu et al. [12], tested the hypothesis that RAN translation across DM1-CAG expansion transcripts produces a DM1-polyGln expansion protein. A monoclonal antibodies against the polyGln expansion itself [21] and the predicted C-terminal flanking sequence [12], showed the accumulation of a polyGln RAN protein in DM1 patients and mice [12]. DM1 polyGln-RAN proteins were found at low frequency in patient myoblasts, skeletal muscle and heart and were more common in blood. The co-localization of DM1-polyGln aggregates with caspase-8 [12], an early indicator of polyGln-induced apoptosis [22], is consistent with a role for polyGln toxicity in DM1 [23] (Table 1).
RAN proteins in FXTAS
Todd et al. [24] recently showed evidence that RAN translation contributes to Fragile X-associated tremor ataxia syndrome (FXTAS). FXTAS, a late onset cerebellar disorder characterized by gait incoordination, dementia and tremors [25], is caused by a premutation expansion (50–200 copies) of a CGG•CCG repeat in the 5′ UTR the FMR1 gene [26]. In contrast to Fragile X full mutations (>200 copies) that shut down FMR1 RNA expression [27], premutation expansions result in increased levels of FMR1 CGGEXP transcripts [28]. While several studies of FXTAS support an RNA gain-of-function mechanism [29, 30], the large ubiquitinated aggregates found in FXTAS patient brains appear more similar to aggregates found in protein-mediated neurological disorders [24].
Using a fly model of FXTAS, Todd et al. [24] noticed the puzzling accumulation of GFP aggregates in flies containing an upstream CGG expansion mutation. This observation suggested the possibility that RAN translation might occur across FXTAS CGG expansion mutations. Todd et al. [24] went on to demonstrate that a polyGly expansion protein is expressed and accumulates in FXTAS fly and mouse models as well as human autopsy tissue. Mass spectrometry detected fragments upstream of the CGG repeat suggesting that translation in the polyGly frame can initiate 5′ of the repeat. This polyGly RAN protein accumulates in neuronal inclusions in the hippocampus, frontal cortex and cerebellum in FXTAS but not control autopsy tissue. Todd et al [24] also demonstrated that 5′ sequence differences between Dutch and NIH FXTAS mouse models affect polyGly RAN protein expression in transfected cells, a result which demonstrates 5′ flanking sequences are important for polyGly expression. Mutations which block polyGly protein expression were used to show polyGly RAN proteins contribute to toxicity in cell culture and fly models independent of RNA gain of function effects. Additionally, these sequence differences explain why ubiquitin-positive, polyGly positive inclusions are found the Dutch but not the NIH mutant mice [31, 32]. This group also showed that a polyAla RAN protein is expressed from a second reading frame in transfected cells [24] but it is not yet clear if polyAla RAN proteins are expressed in vivo. Given that the CGG repeats of FXTAS are bidirectionally transcribed [11], it is possible that antisense RAN proteins may also be expressed. Taken together these results show polyGly RAN proteins accumulate in patient brains and suggest a role for RAN translation in FXTAS (see Table 1 for summary).
RAN translation in C9ORF72 ALS/FTD
C9ORF72 amyotrophic lateral sclerosis (ALS)/frontotemporal degeneration (FTD) is caused by a GGGGCC•GGCCCC repeat expansion in intron 1 of the C9ORF72 gene [33, 34]. The discovery of the C9ORF72 expansion mutation has generated substantial excitement because it connects a large body of research on microsatellite expansion mutations to the most common known cause of ALS and dementia – two diseases with a high impact on society. Several diseases mechanisms have been proposed for C9ORF72 ALS/FTD in which the expansion causes: a) decreased levels of C9ORF72 transcripts and protein [33, 35]; b) RNA gain of function effects [36–43]; c) and most recently, the expression and accumulation of toxic RAN-proteins [36, 38, 44–49].
C9 Sense RAN Proteins
RAN translation of the sense GGGGCC expansion is predicted to result in the expression of three dipeptide proteins: GlyPro (GP), GlyArg (GR) and GlyAla (GA). Support for the accumulation of RAN-proteins in C9ORF72 ALS/FTD autopsy brains was first reported using antibodies against the predicted dipeptide repeat motifs (GP, GR and GA) [44, 47] and more recently using antibodies to both the repeats and unique C-terminal regions [48]. Immunostaining shows evidence that RAN proteins accumulate in neuronal inclusions in the cerebellum, hippocampus and other brain regions of C9ORF72 ALS/FTD but not in control autopsy tissue [44, 47, 48]. The inclusions are similar in shape and abundance to previously characterized p62-positive/phospho-TDP-43 negative ALS/FTD inclusions [44, 47] suggesting that C9-RAN proteins play a key role in the neuropathology of this disease (Table 1).
C9 Antisense Foci
Following previous discoveries of bidirectional transcription in DM1 [9], SCA8 [10] and other expansion disorders [2] several groups have recently showed that the G4C2 expansion mutation is also bidirectionally expressed, and that antisense RNA foci accumulate in patient autopsy brains [46, 47], patient derived cell lines [40], and peripheral blood [48]. The Zu et al. study [48] showed C9ORF72 antisense transcript levels are dramatically elevated in C9(+) but not C9(−) brains; but no similar upregulation of the AS transcript was found in blood of ALS/FTD patients [48]. Additionally, double labeling of sense and antisense foci showed that in the majority of cells in the brain [41, 48] and the blood [48] express either sense or antisense foci with only a minority of cells positive for both (Table 1).
C9 Antisense RAN proteins
Additionally, several groups have recently reported that antisense RAN protein aggregates accumulate in C9ORF72-positive autopsy brains [38, 46, 48, 49]. The repeat motifs for these antisense RAN proteins are polyProArg, (PR), polyProAla (PA) and polyGlyPro (GP). Although GP motifs are expressed in both sense and antisense directions, the sense GP protein has a unique C-terminal end not found in the antisense GP protein (GPAS) [48]. Similarly, it is important to note that five of the six C9-RAN proteins are predicted to contain completely different C-terminal flanking sequences that may also affect their function and pathogenicity [48]. C9 RAN proteins were initially detected using antibodies raised against individual dipeptide repeat motifs [44, 47]. Because these repeat motifs are also found in a number of other proteins, Zu et al [48] developed a panel of antibodies that recognize either individual repeat motifs or the unique C-terminal regions of the sense and antisense C9-RAN proteins (Table 1). Similar to the observation for RNA foci, sense and antisense RAN proteins have been infrequently detected in the same cell [46, 48], although in one study all six proteins were shown to be expressed in the same brain region [48]. Histological studies of affected brain regions showed a striking pattern of clustered RAN protein aggregates that differs within and between patients[48]. Understanding why these differences occur and if they are driven by variability in RNA or triggers of RAN protein expression may provide insight into why patients with expansion mutations are often healthy for decades followed by a rapid decline in health.
Common themes and new directions
RAN in expansion mutations
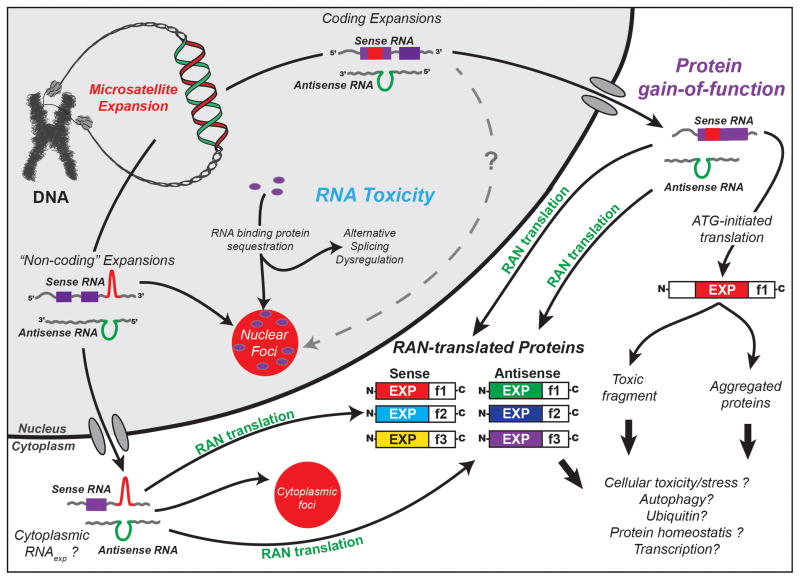
Historically, protein gain of function (e.g. HD, SCA1, SBMA) and RNA gain of function diseases (e.g. DM1, FXTAS, SCA8) have been considered to have separate molecular mechanisms. In recent years, however, these lines have begun to blur. For example, in Huntington’s disease, nuclear aggregates with expanded CAG HTT mRNA have been observed [4, 50, 51] and SCA3 Drosophila studies demonstrate a toxic RNA component for this polyGln disorder [52]. Additionally four diseases with expansion mutations located in “non-coding” regions have been shown to express proteins in one or more reading frames [53]. While these diseases are characterized by expansion RNAs that accumulate in nuclear foci, the recent observation of cytoplasmic RNA foci C9ORF72 ALS/FTD [37, 40, 42] and the growing list of RAN proteins [12, 24, 38, 44–48] suggest at least some expansion transcripts make it out to the cytoplasm (Fig. 3).
Figure 3. Triple threat - three disease mechanisms for microsatellite expansion disorders.
Expanded microsatellite repeats have been traditionally classified as either coding disorders or non-coding disorders that give rise to protein gain- or loss-of-function or RNA toxicity mechanisms. For traditional “coding” disorders, the repeat expansion is translated as part of a larger open-reading frame (ORF) and results in the expression of a mutant protein that disrupts normal cellular function and induces toxicity. For example Huntington’s disease (HD), a late-onset neurodegenerative disorder, is caused by a CAG expansion within the first exon of huntingtin gene that is translated as a polyglutamine tract in the huntingtin protein, HTT [78]. For traditional “non-coding” disorders (blue), the repeat expansion remains in the RNA transcript, accumulates as RNA foci that sequester RNA-binding proteins and lead to a loss of their normal function. For example, in myotonic dystrophy, CUG(G) expanded RNA transcripts sequester MBNL proteins from their normal splicing targets leading to a MBNL loss-of-function and alternative splicing dysregulation [19, 79–81]. The recent discovery of repeat associated non-ATG (RAN) translation [12] adds a third pathway for disease. RNA transcripts from both “non-coding” and “coding” disorders may undergo RAN translation. Once in the cytoplasm, these transcripts are capable of producing proteins in all three reading frames, which may contribute to cellular toxicity/stress. Depending upon the flanking sequences, each of these RAN proteins will have a distinct expanded peptide repeats (colored boxes) and unique different C-terminal regions (f1, f2 and f3). If the repeat is also within an ATG-initiated open-reading frame, this ATG-initiated protein will share the expanded peptide repeat and C-terminal region with one of the RAN proteins but will have an additional N-terminal region. Further complexity is added by fact that many expansion mutations are bidirectionally transcribed [2], which doubles the number of distinct RAN proteins that may be produced. While individual RAN proteins have been observed in SCA8[12],DM1[12] and FXTAS[24] patients, sense and antisense RNA foci and RAN proteins in all six reading frames been shown to accumulate in C9ORF72 ALS/FTD patient cells [36–38, 44–48].
Conversely, the discovery of RAN translation raises the possibility that RAN proteins are also expressed from expansion mutations located within ATG-initiated open reading frames (e.g. HD, SBMA and SCA1) (Fig. 3). While RAN proteins have not yet been demonstrated for any of these diseases, in vitro experiments show that both ATG-initiated and RAN proteins can be expressed from the same minigene [48]. While disease-causing coding expansions tend to be much smaller than non-coding expansions, the typical size range for coding expansions (40 to 100 repeats) is well within the size range shown to be required for RAN translation. Based on in vitro studies, relatively short CAG expansions (e.g. 40–50) undergo RAN translation in the polyGln frame [12] raising the possibility that ATG-initiated polyGln and RAN polyGln proteins are both expressed in individuals with relatively short expansion mutations. These experiments also predict that individuals with longer expansions associated with more severe phenotypes (50–80 repeats), may express a cocktail of mutant proteins: ATG-initiated polyGln, plus RAN polyGln, RAN polyAla and RAN polySer proteins (Fig. 3). The simultaneous expression of RAN proteins in multiple reading frames from larger expansions in vitro [12], suggests the possibility that RAN translation may play a role in anticipation, or the decreased age of onset and increased severity, seen in individuals with longer repeat expansions.
Do RAN proteins contribute to disease?
To determine the potential contribution of RAN proteins to microsatellite expansion disorders, it will be important to understand multiple aspects of these proteins: (1) Toxicity. RAN proteins are found in disease-relevant tissues (e.g. polyAla in SCA8 Purkinje cells) and several of the RAN proteins have been shown to be toxic in transfected cells [12, 24, 48] and model systems [24] suggesting these protein are toxic in vivo. While the contribution of these proteins to disease remains to be demonstrated, this process is complicated by the need to tease apart the underlying toxicity of the RNA transcripts required to make the RAN proteins and the proteins themselves. (2) Regulation. RAN proteins have been reported in a variety of tissues including brain, muscle and blood [12, 24, 38, 44–48]. There is also considerable variability in RAN protein accumulation within a particular tissue (e.g. RAN protein aggregates cluster at variable sites in C9ORF72 brains [48]). This variability suggests that RAN protein aggregation may be triggered focally and then spread to neighboring cells in a prion-like manner [48, 54–62]. Alternatively, RAN-protein expression may be triggered under conditions of stress [63]. Understanding how, when and where RAN proteins are translated may help explain the variability in age of onset, penetrance and phenotypes of C9ORF72 ALS/FTD and other disorders. (3) Function. Expanded RAN proteins are likely to have aberrant cellular functions, similar to the ATG-initiated polyGln expansion proteins (e.g. mutant Huntingtin and ataxin-1) (Fig. 3). RAN translation can also occur, in vitro, across repeat lengths [12, 24, 48] that occur normally within the human genome, such as within the SCA1 [64, 65] and TATA-binding protein genes [66]. While in vitro observations do not always predict what occurs in vivo, it is possible that RAN proteins may be expressed across relatively short repeats throughout the genome and that these putative proteins have a normal cellular function. Understanding and characterizing RAN proteins will be important for understanding their potential role in disease and normal biology.
Requirements for RAN translation
Sequence
Although there is strong evidence that RAN proteins accumulate in four different microsatellite expansion diseases, how expanded repeats can express proteins in multiple reading frames without an ATG initiation codon is just beginning to be explored. Experiments from Zu et al., [12] show that hairpin-forming CAG but not non-hairpin forming CAA repeats express polyGln RAN proteins in HEK293T cells, suggesting RNA structure is important for RAN translation. Structured RNAs appear to be a common theme as expanded CUG [12], CGG [24], G4C2 [44, 47, 48] and G2C4 [38, 46, 48] expansions, all known to express RAN proteins, form hairpin [67, 68] or G-quadruplex structures [69, 70]. The increased propensity of longer repeat tracts to adopt RNA structures may explain why longer CAG, CGG and G4C2 repeat tracts are typically associated with higher levels of RAN protein accumulation and expression of RAN proteins in multiple reading frames [12, 24, 47, 48].
Translation initiation
Typically, cell free in vitro translation systems are used to understand the biochemical requirements for translation initiation. Information on the molecular requirements for RAN translation is limited because non-ATG translation is less permissive in rabbit reticulocyte lysates (RRL). Expression of repeat proteins across CAG expansions in cell free lysates does not occur in the polyAla frame. Expression in the polyGln and polySer frames is limited and strongly favored by the presence of close cognate initiation codons (e.g. ATT, ATC) upstream of the repeat [12]. Labeling experiments in RRL lysates using S35 show that translation in this cell free system initiates with a Met-tRNAiMet [12]. The incorporation of an N-terminal methionine in RRLs is not surprising because a previously documented alternative initiation codon (ATT) was available. RAN translation of CAGEXP transcripts in cells is likely to use a different initiation mechanism as: 1) initiation in cells does not require close cognate initiation codons; 2) mass spectrometry of the polyAla showed no evidence for an N-terminal methionine; 3) translation occurs robustly in all three reading frames [12]. Additionally, initiation-start sites appear to be different between reading frames and repeat motifs. For CAG repeats, initiation in the polyAla frame appears to occur at multiple GCA codons throughout the repeat tract, while initiation in the polyGln frame occurs close to or at the beginning of the repeat tract [12]. In FXTAS, MS analysis of the polyGly protein suggests initiation can occur upstream of the CGG repeat tract [24]. Because the polyGly tracts in these experiments were too large for MS analysis, it is possible that initiation of polyGly proteins also occurs within the repeat tract. Little is known regarding the initiation of RAN translation for the G4C2 and G2C4 repeats, however immunoblots using 5′ and 3′ epitope tags show that translation proceeds through as many as 120 repeats in transfected cells [45]. A better understanding of how and where RAN translation initiates and terminates will be important for determining the molecular mechanisms of disease and for developing future therapeutic strategies to block the expression of RAN proteins.
RAN translation and microsatellite expansion disorder therapies
Recent therapeutic efforts for microsatellite expansion disorders include targeting the RNA for degradation or disrupting interactions with RNA binding proteins [37, 39, 40, 71–76]. While these strategies, which employ antisense oligonucleotides (ASO) or small molecules, have met with a variety of successes [37, 39, 40, 71–76], bidirectional transcription and RAN translation have brought to light new questions. Will knocking down the sense transcript be sufficient to treat these disorders? Will sense transcript knockdown have unintended consequences on the antisense transcript or RAN translation? In SCA7, decreasing the antisense transcript in mutant transgenic mice leads to epigenetic changes and an increase in sense transcript expression [77], suggesting that sense and antisense transcripts regulation may be linked. Two studies using ASOs targeting the sense strand in C9ORF72 ALS/FTD iPS cells rescued several phenotypes including RNA foci and dysregulated gene expression [37, 42]. One study did not observe RAN proteins in their cells [42] and the other study did not detect a reduction in GP RAN proteins [37]. For the latter study, because GP proteins are expressed from both sense and antisense transcripts [48], the ASO knockdown of sense transcripts would not be expected to knockdown antisense GP [38]. A third study saw reduction of sense RNA foci but did not see rescue of gene expression phenotypes in patient fibroblasts after ASO knockdown of G4C2 sense transcripts. This group found antisense G2C4 RNA foci in these cells and showed they were unaffected by ASO treatment. These authors suggested that the presence of antisense G2C4 foci in the treated cells may explain the lack of rescue in these cells [40]. These studies highlight the importance of considering both sense and antisense transcripts as well as ATG- and RAN-proteins in therapeutic treatment strategies for microsatellite expansion diseases.
Conclusions
The discovery of RAN translation has important implications for understanding fundamental mechanisms of gene expression and disease. For microsatellite expansion disorders, bidirectional transcription and RAN translation raises the possibility that a cocktail of mutant transcripts and proteins contributes to many of these diseases (Fig. 2). For example, polyAla, polySer, polyLeu, polyCys and CUGEXP transcripts may contribute to the pathogenesis of some of the CAG polyGln diseases. Additionally, novel RAN proteins may contribute to diseases currently thought to be caused by RNA gain-of-function effects. Because >50% of the human genome consists of repetitive DNA, RAN translation could reveal an abundant, yet previously unrecognized, category of proteins that may shift current views of proteome complexity and fundamental aspects of cell biology.
Acknowledgments
The authors wish to thank Dr. Tao Zu and Dr. Maurice Swanson for helpful comments and suggestions. This work was supported by the National Institutes of Health [PO1NS058901 and RO1 NS040389], Muscular Dystrophy Association, Keck Foundation Grant, and the Myotonic Dystrophy Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447(7147):932–40. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19(R1):R77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6(10):743–55. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Orr HT. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22(3):251–5. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr HT. Toxic RNA as a driver of disease in a common form of ALS and dementia. Proc Natl Acad Sci U S A. 2013;110(19):7533–4. doi: 10.1073/pnas.1305239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 7.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77(5):825–43. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20(3):483–9. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38(7):758–69. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 11.Ladd PD, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16(24):3174–87. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- **12.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108(1):260–5. doi: 10.1073/pnas.1013343108. Discovery of repeat-associated non-ATG translation and demonstration of RAN proteins in SCA8 and DM1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koob MD, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21(4):379–84. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 14.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 15.Buxton J, et al. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355(6360):547–8. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- 16.Fu YH, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255(5049):1256–8. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 17.Shin J, Charizanis K, Swanson MS. Pathogenic RNAs in microsatellite expansion disease. Neurosci Lett. 2009;466(2):99–102. doi: 10.1016/j.neulet.2009.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulos MG, et al. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3(1):a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charizanis K, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75(3):437–50. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KY, et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol Med. 2013;5(12):1887–900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trottier Y, et al. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10(1):104–10. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- 22.UM, et al. Extended polyglutamine selectively interacts with caspase-8 and -10 in nuclear aggregates. Cell Death Differ. 2001;8(4):377–86. doi: 10.1038/sj.cdd.4400819. [DOI] [PubMed] [Google Scholar]
- 23.Gomes-Pereira M, et al. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 2007;3(4):e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Todd PK, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78(3):440–55. doi: 10.1016/j.neuron.2013.03.026. Discovery that CGG expansion mutations express polyGly RAN proteins which accumulate FXTAS flies, mice and autopsy brains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126(1):1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagerman RJ, et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57(1):127–30. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 27.Pieretti M, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66(4):817–22. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 28.Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1(2):103–5. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 29.Sofola OA, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55(4):565–71. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin P, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55(4):556–64. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willemsen R, et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet. 2003;12(9):949–59. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 32.Entezam A, et al. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395(1–2):125–34. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 36.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. This group showed RAN proteins are expressed in iPS cells and the ASO knockdown of sense transcripts reverses some of the molecular phenotypes in these cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1192-8. These authors provide in vivo evidence that antisense foci and antisense RAN protein aggregates accumulate in patient brains and in vitro evidence for G2C4 antisense RAN translation in transfected cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gendron TF, Cosio DM, Petrucelli L. c9RAN translation: a potential therapeutic target for the treatment of amyotrophic lateral sclerosis and frontotemporal dementia. Expert Opin Ther Targets. 2013 doi: 10.1517/14728222.2013.818659. [DOI] [PubMed] [Google Scholar]
- **40.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1318835110. These authors show CNS delivery of ASOs that knockdown the sense C9ORF72 transcript is well tolerated in wildtype mice. Additionally, these authors identified an RNA signature in patient fibroblasts which was not corrected with ASO sense knockdown. These authors suggest that the presense of unexpected antisense RNA foci may explain these results and that both sense and antisense transcripts will need to be considered for successful therapeutic strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Mizielinska S, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1200-z. These authors show sense and antisense foci are a consistently found in C9ORF72(+) human autopsy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–46. doi: 10.1016/j.neuron.2013.02.004. This paper was one of the first to detect aggregated RAN proteins in C9ORF72 ALS/FTD autopsy tissue using an antibody that recognized the GP dipeptide repeat motif. This study and the study by Mori et al. provided independent confirmation of the Zu et al. 2011 study that RAN proteins can be expressed across microsatellite expansion mutations and may be important in disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackenzie IR, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- *46.Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013 doi: 10.1007/s00401-013-1189-3. These authors provided evidence that RAN translation in C9ORF72 ALS/FTD patients also occurs in the antisense direction and confirmed antibodies directed against the GA repeat motif itself and the C-terminus recognize the same aggregates in patient cells. [DOI] [PubMed] [Google Scholar]
- **47.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–8. doi: 10.1126/science.1232927. This paper was one of the first to detect aggregated RAN proteins in C9ORF72 ALS/FTD autopsy tissue using an antibody that recognized the GP dipeptide repeat motif. Additionally, this study provided evidence that RAN translation across G4C2 expansions can occur in tranfected cells. This study and the study by Ash et al. 2013 provided independent confirmation of the Zu et al. 2011 study that RAN proteins can be expressed across microsatellite expansion mutations and may be important in disease. [DOI] [PubMed] [Google Scholar]
- **48.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1315438110. These authors demonstrated C9ORF72 antisense transcripts are elevated in patient brains and that antisense RNA foci accumulate in patient brains and blood. This group was the first to develop a panel of antibodies that recognize all six sense and antisense repeat motifs and the unique C-terminal ends of these RAN proteins. In vitro data show RAN proteins are toxic to cells. These authors suggest that variations in and clustering of RAN protein aggregates may underlie the variability in phenotypes and the reduced penetrance of C9ORF72 ALS/FTD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mann DM, et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun. 2013;1(1):68. doi: 10.1186/2051-5960-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Mezer M, et al. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39(9):3852–63. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20(19):3811–21. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li LB, et al. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453(7198):1107–11. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22(R1):R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costanzo M, Zurzolo C. The cell biology of prion-like spread of protein aggregates: mechanisms and implication in neurodegeneration. Biochem J. 2013;452(1):1–17. doi: 10.1042/BJ20121898. [DOI] [PubMed] [Google Scholar]
- 55.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106(31):13010–5. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kordower JH, et al. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14(5):504–6. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- 57.Li JY, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14(5):501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 58.de Calignon A, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73(4):685–97. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, et al. Trans-synaptic spread of tau pathology in vivo. PLoS One. 2012;7(2):e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polymenidou M, Cleveland DW. Prion-like spread of protein aggregates in neurodegeneration. J Exp Med. 2012;209(5):889–93. doi: 10.1084/jem.20120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–53. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen C, et al. alpha-Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. J Clin Invest. 2011;121(2):715–25. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orr HT, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4(3):221–6. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 65.Ranum LP, et al. Molecular and clinical correlations in spinocerebellar ataxia type I: evidence for familial effects on the age at onset. Am J Hum Genet. 1994;55(2):244–52. [PMC free article] [PubMed] [Google Scholar]
- 66.Koide R, et al. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8(11):2047–53. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 67.Kiliszek A, et al. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39(16):7308–15. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kiliszek A, et al. Crystallographic characterization of CCG repeats. Nucleic Acids Res. 2012;40(16):8155–62. doi: 10.1093/nar/gks557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fratta P, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reddy K, et al. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288(14):9860–6. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sicot G, Gomes-Pereira M. RNA toxicity in human disease and animal models: from the uncovering of a new mechanism to the development of promising therapies. Biochim Biophys Acta. 2013;1832(9):1390–409. doi: 10.1016/j.bbadis.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325(5938):336–9. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamori M, Gourdon G, Thornton CA. Stabilization of expanded (CTG) (CAG) repeats by antisense oligonucleotides. Mol Ther. 2011;19(12):2222–7. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Warf MB, et al. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106(44):18551–6. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106(33):13915–20. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70(6):1071–84. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Labbadia J, Morimoto RI. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–85. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302(5652):1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 80.Mankodi A, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10(19):2165–70. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 81.Liquori CL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293(5531):864–7. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]