Abstract
Background.
Human basal-like breast cancer (BLBC) has a poor prognosis and is often identified by expression of the epidermal growth factor receptor (EGFR). BLBC remains a major clinical challenge because its pathogenesis is not well understood, thus hindering efforts to develop targeted therapies. Recent data implicate the forkhead box C1 (FOXC1) transcription factor as an important prognostic biomarker and functional regulator of BLBC, but its regulatory mechanism and impact on BLBC tumorigenesis remain unclear.
Methods.
The association between FOXC1 and EGFR expression in human breast cancer was examined by immunohistochemistry in formalin-fixed tissues and analysis of the TCGA database. The regulation of FOXC1 by EGFR activation was investigated in MDA-MB-468 cells using immunoblotting, qRT-PCR, and luciferase activity assays. This EGFR effect on FOXC1 expression was confirmed using the MDA-MB-468 xenograft model.
Results.
Both FOXC1 mRNA and protein levels significantly correlated with EGFR expression in human breast tumors. EGFR activation induced FOXC1 transcription through the ERK and Akt pathways in BLBC. EGFR inhibition in vivo reduced FOXC1 expression in xenograft tumors. We also found that FOXC1 knockdown impaired the effects of EGF on BLBC cell proliferation, migration, and invasion.
Conclusions.
Our findings uncover a novel EGFR-FOXC1 signaling axis critical for BLBC cell functions, supporting the notion that intervention in the FOXC1 pathway may provide potential modalities for BLBC treatment.
Keywords: Akt, basal-like breast cancer, EGFR, ERK, FOXC1
INTRODUCTION
Basal-like breast cancers (BLBCs), which express genes characteristic of basal/myoepithelial cells in the normal mammary gland, comprise up to 25% of all breast cancers.1 They under-express estrogen receptor (ER), progesterone receptor,2 and HER-2, encompassing 60-90% of ER-/PR-/HER-2-(triple-negative breast cancers). BLBC usually presents with high histologic grade, aggressive clinical features, poor prognosis, and a propensity to metastasize to the brain and lung.3 Chemotherapy is the only modality of systemic therapy for BLBC and shows limited success against this subtype of breast cancer. A better understanding of the molecular mechanisms of BLBC and the development of effective targeted therapy are urgently needed.
Forkhead box (FOX) proteins constitute a family of evolutionarily conserved transcription factors characterized by a common 110-amino acid DNA-binding FOX domain. FOX proteins have been shown to regulate a wide variety of biological processes, including development, differentiation, proliferation, apoptosis, migration, invasion, and tumorigenesis.4 FOXC1, a FOX family member, plays important roles in the development of the brain, heart, and eye during the embryonic stage.5 In addition to its important role in development, FOXC1 has been recently found to be overexpressed in different types of cancer including breast cancer,6-8 hepatocellular carcinoma,9 prostate cancer,10 pancreatic adenocarcinoma,11 and non-small cell lung cancer.12 FOXC1 expression is associated with poor clinical outcome in these cancers. Notably, FOXC1 is specifically overexpressed in BLBC,13 and its overexpression induces cellular traits commonly associated with this breast cancer subtype, such as epithelial-to-mesenchymal transition (EMT) and increased cell proliferation, and migration.6,7 Recently, it has been reported that FOXC1 expression is a hallmark for circulating breast tumor cells with mesenchymal phenotypes.14 Thus, FOXC1 may serve as a functional regulator of BLBC. A recent study showed that BRCA1 in cooperation with GATA3 represses FOXC1 expression to inhibit the pathogenesis of BLBC.15 Of note, analysis of an oligonucleotide comparative genomic hybridization array revealed that the FOXC1 gene is not amplified in basal-like tumors.16 The mechanism for the exclusive induction of FOXC1 in BLBC is poorly understood.
A commonly accepted surrogate biomarker for BLBC is epidermal growth factor receptor (EGFR), which is abnormally activated by overexpression or constitutive mutation in many epithelial tumors. EGFR is widely used together with several other proteins in immunohistochemical detection of BLBC tumors and its high expression is associated with poor prognosis. 17,18 Numerous lines of evidence have shown the critical role of EGFR in cancer cell functions. It is still not clear whether EGFR and other BLBC-related genes form signaling pathways or networks dictating BLBC traits. Because both FOXC1 and EGFR are critical markers and functional regulators for BLBC, we hypothesize that EGFR may crosstalk with FOXC1 and that EGFR/FOXC1 signaling may orchestrate BLBC cellular traits. Our studies corroborate the association of EGFR and FOXC1 in human breast cancers. We demonstrate that EGFR activation can potently increase FOXC1 expression through ERK and Akt pathways in BLBC cells. This mechanism integrates the function of several key molecules that have been implicated in the regulation of human BLBC cells. We also delineate the role of FOXC1 in EGF-elicited cell functions. Taken together, our findings provide insight into the role of a novel EGFR/FOXC1 axis in BLBC pathogenesis.
MATERIALS AND METHODS
Detailed methods for in vitro migration/invasion, in vivo experiments, immunoblotting, and reverse transcription-PCR, and transfection are provided in the supplement.
Cell culture and cell proliferation assays
All cell lines were purchased from American Type Culture Collection. Cell proliferation was assessed by CellTiter-Glo Luminescent cell viability assay (Promega, Madison, WI).The 2-kb FOXC1-promoter from the transcription start site was cloned into the pGL4-luc vector (Promega). Details about the reagents are provided in the supplement.
Immunohistochemistry (IHC)
IHC in formalin-fixed breast cancer tissues was performed as described previously using a generated mouse monoclonal FOXC1 antibody.13
In vivo experiments
Animal studies were conducted with the approval of the institutional animal care and use committee. Details are described in the supplement.
Statistical analysis
All experiments were performed 3 times with samples measured in triplicate. Results are expressed as mean ± standard deviation, unless otherwise stated. GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA) was used for statistical analysis. Correlation analysis between EGFR and FOXC1 expression in human cancer samples was analyzed for significance with Pearson r test, P < 0.05 was considered statistically significant.
RESULTS
FOXC1 expression correlates with EGFR expression in human BLBC
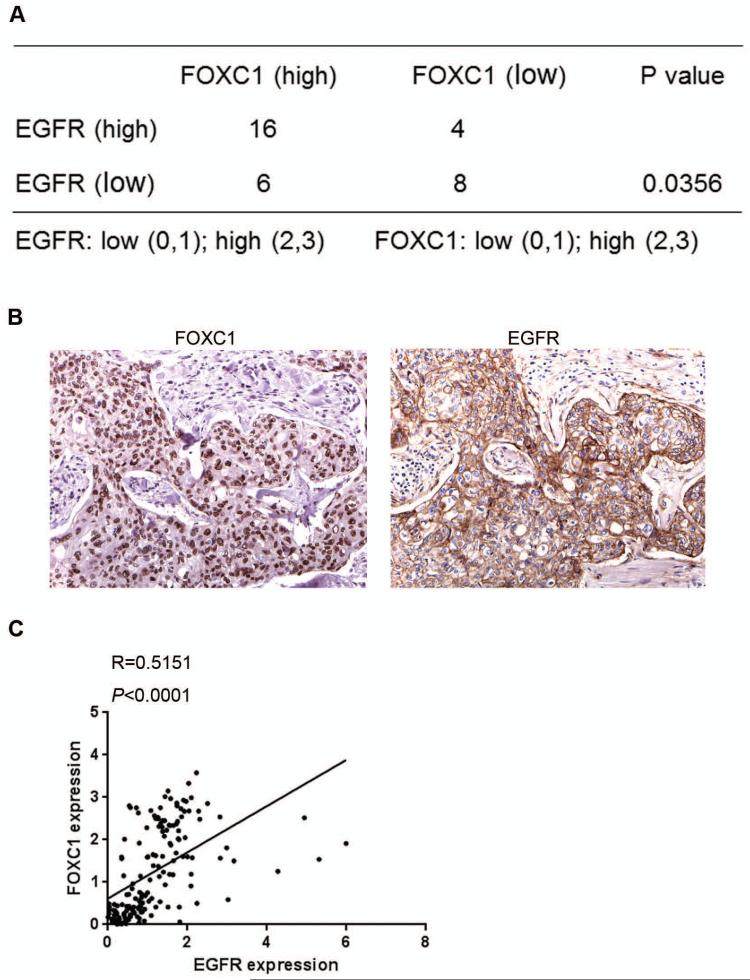
Because both FOXC1 and EGFR are critical markers and functional regulators of BLBC, we set out to assess the association between EGFR and FOXC1 expression. To this end, we performed IHC of EGFR and FOXC1 in 34 human triple-negative breast tumors. Quantitative IHC scoring showed that FOXC1 protein levels were significantly associated with EGFR protein levels (P=0.0356; Fig. 1A). The representative pictures for FOXC1 and EGFR IHC in breast cancer samples are shown in Fig. 1B. Consistent with this result, a positive correlation between FOXC1 and EGFR mRNA in invasive breast cancers was observed in The Cancer Genome Atlas database (Fig. 1C). 19
Figure 1. FOXC1 expression significantly correlates with EGFR expression in human BLBC tumors.
(A) Distribution of EGFR and FOXC1 by immunohistochemical staining scores in 34 human triple-negative samples (P=0.0356). Association was examined using the Fisher's Exact Test. (B) Representative images for FOXC1 and EGFR IHC in a human breast tumor. Magnification: × 400. (C) Correlation between EGFR and FOXC1 mRNA expression in invasive breast cancer samples from the TCGA database (R=0.5151, P<0.0001).
EGFR activation up-regulates FOXC1 expression in BLBC cells
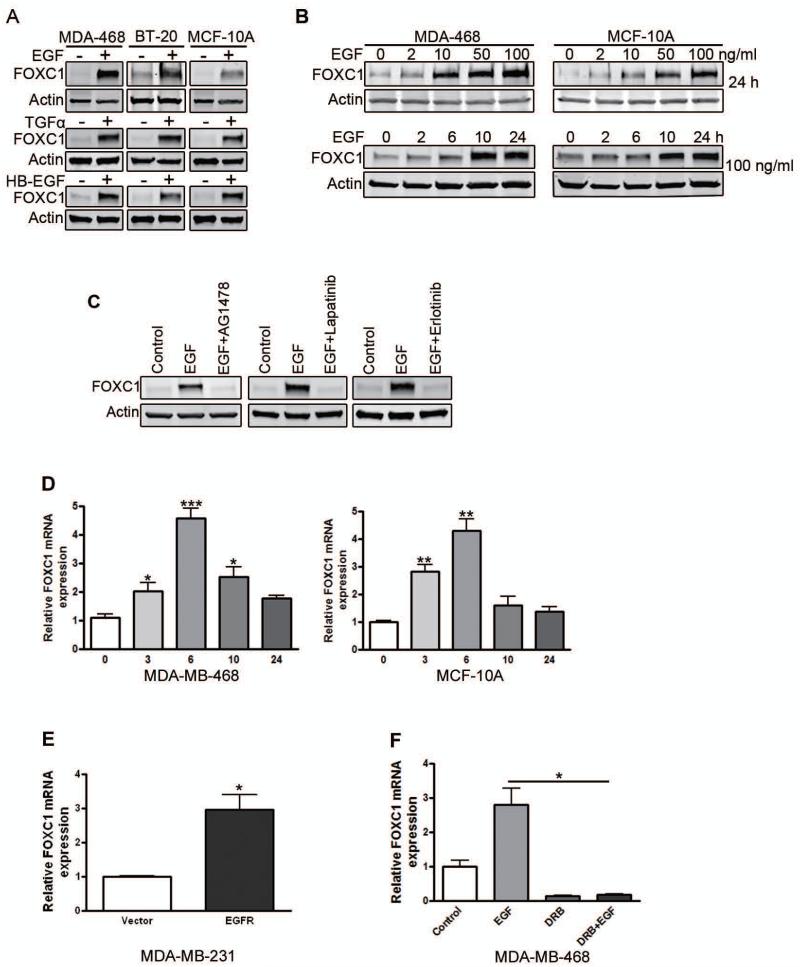
Next, we examined whether EGFR activation regulates FOXC1 expression. EGFR-positive MDA-MB-468 and BT-20 cells were treated with EGF for 24 h. Immunoblotting showed that EGF treatment induced FOXC1 protein expression (Fig. 2A). Similar results were found in MCF-10A mammary epithelial cells (Fig. 2A). Transforming growth factor (TGF)-α and heparin-binding EGF-like growth factor (HB-EGF), ligands of the EGF family, also upregulated FOXC1 expression (Fig. 2A). Dose-response and time-course studies using MDA-MB-468 and MCF-10A cells confirmed that EGF is a potent inducer of FOXC1 expression (Fig. 2B). To examine whether EGFR activation is required for EGF-elicited FOXC1 up-regulation, we pretreated MDA-MB-468 cells with the small-molecule EGFR inhibitors AG1478, Lapatinib, or Erlotinib, followed by EGF treatment. Immunoblotting showed that EGFR inhibitors blocked the EGF induction of FOXC1 expression (Fig. 2C).
Figure 2. EGFR activation up-regulates FOXC1 expression in BLBC cells.
(A) MDA-MB-468 and BT-20 EGFR-positive human breast cancer cells and MCF-10A human mammary epithelial cells were serum-starved overnight and treated with EGF, TGFα, or HB-EGF (50 ng/ml) for 24 h. FOXC1 level was analyzed by immunoblotting. Actin was used as a loading control. (B) Serum-starved MDA-MB-468 cells were treated with increasing concentrations of EGF for 24 h (top) or with EGF (100 ng/ml) for the indicated time periods (bottom), followed by immunoblotting analysis. (C) MDA-MB-468 cells were serum starved overnight. Then cells were treated with EGF for 24 h in the presence or absence of the EGFR inhibitor AG1478 (1 μM), Lapatinib (1 μM) or Erlotinib (3 μM). FOXC1 protein levels were measured by immunoblotting. (D) MDA-MB-468 cells (left) and MCF-10A cells (right) were serum-starved overnight and treated with EGF for the indicated time periods. FOXC1 mRNA levels were analyzed by qRT-PCR. *, P<0.05; ***, P<0.0001; **, P<0.001. (E) MDA-MB-231 cells were transfected with pBABE-EGFR construct for 48 h. FOXC1 mRNA levels were detected by qRT-PCR. *, P<0.05. (F) MDA-MB-468 cells were serum-starved overnight. Then cells were treated with EGF for 6 h after DRB pretreated for 1 h. FOXC1 mRNA levels were detected by qRT-PCR. *, P<0.05. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. Data represent mean ± SD from 3 independent experiments.
We then proceeded to investigate the mechanism for the EGF-induced up-regulation of FOXC1 expression. Using qRT-PCR, we found that EGF increased FOXC1 mRNA levels in MDA-MB-468 and MCF-10A cells (Fig. 2D). We then transiently transfected MDA-MB-231 cells, which normally express low levels of EGFR, with an EGFR construct. qRT-PCR analysis showed that EGFR overexpression also increased FOXC1 mRNA levels (Fig. 2E). To test whether EGFR activation induces FOXC1 at the transcriptional level, we pretreated MDA-MB-468 cells with the transcription inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), followed by 6 h of EGF treatment. As shown in Fig. 2F, DRB blocked the EGF induction of FOXC1 mRNA levels, suggesting that EGF activates FOXC1 transcription.
To further explore the mechanism whereby EGF induces FOXC1 transcription, we generated a 2-kb FOXC1 promoter reporter construct. Luciferase assays with a panel of human breast cancer cell lines revealed that EGF potentiated FOXC1 promoter activity specifically in BLBC cells (Supplementary Fig. 1A), consistent with the notion that EGFR is preferentially expressed in BLBC cells. Using MDA-MB-468 and MCF-10A cells, we further showed that EGF increased FOXC1 promoter activity, but not pGL4-luc control vector activity (Supplementary Fig. 1B). In agreement with these results, EGFR overexpression in MDA-MB-231 cells also dramatically increased FOXC1 promoter activity (Supplementary Fig. 1C). As expected, the EGFR inhibitor AG1478 suppressed the EGF effect on the FOXC1 promoter (Supplementary Fig. 1D). Taken together, these results establish EGFR as a positive regulator of FOXC1 expression in breast cancer cells.
EGFR inhibition abrogates the growth of MDA-MB-468 xenograft mammary tumors
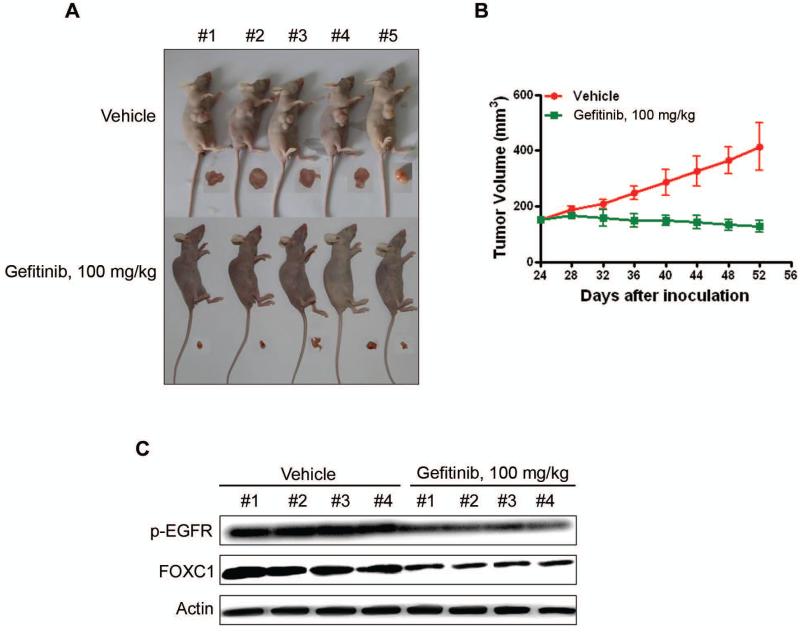
The antineoplastic and FOXC1-inhibitory activities of the EGFR inhibitor Gefitinib were evaluated in an in vivo xenograft model. The mammary glands of twenty nu/nu BALB/c mice were subcutaneously injected with MDA-MB-468 cells. When tumors grew to ~150 mm3, the mice were randomized to receive treatment with vehicle or Gefitinib (100 mg/kg/d) for 20 days (n=10). Tumors were harvested 8 days after treatment cessation. As expected, Gefitinib abrogated the growth of the xenograft tumors (Fig. 3A and B). Immunoblotting of tumor cell lysates showed pronounced decreases in phosphorylated EGFR and FOXC1 levels in the treatment group compared with the control group (Fig. 3C). The body weight, motor activity, and feeding behavior of the mice showed no significant differences between the drug-treated and control groups (data not shown). These results indicate that EGFR inhibition represses FOXC1 expression in vivo.
Figure 3. EGFR inhibitor Gefitinib abrogates the growth of xenograft mammary tumor in nude mice.
BALB/c nu/nu nude mice were subcutaneously inoculated with MDA-MB-468 cells. When tumors reached ~150 mm3, these mice were randomized into 2 groups (n=10) for Iressa treatment. (A) Comparisons of mice and tumors of between the two groups are shown. (B) The tumor growth curves are plotted. Error bars represent 95% confidence intervals, P<0.05. (C) The levels of p-EGFR and FOXC1 in the tumor tissues were detected by immunoblotting with the indicated antibodies.
EGFR activation induces FOXC1 through the Ras/ERK and phosphatidylinositol 3-kinase (PI3K)/Akt pathways
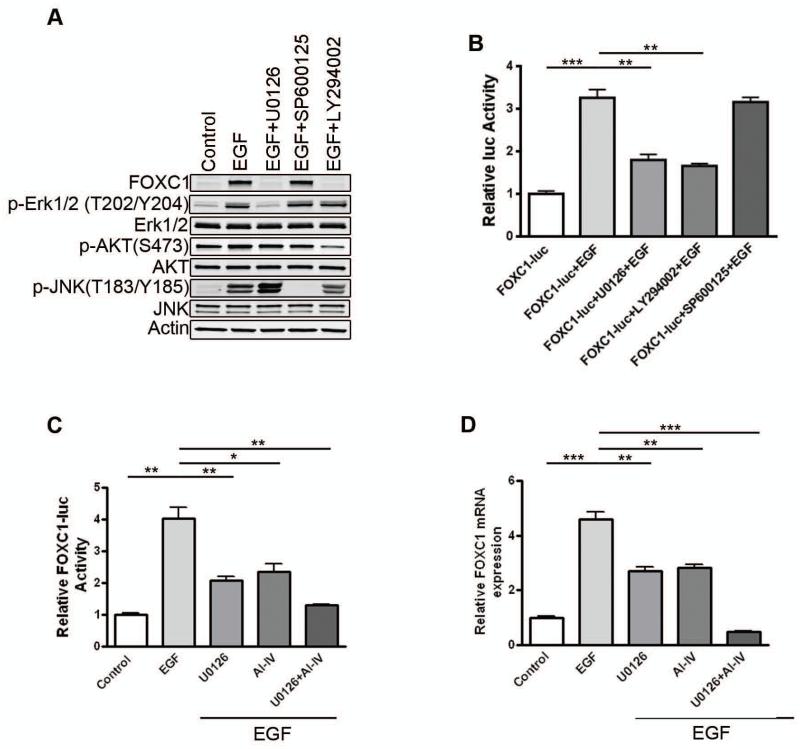
It is well-established that Ras/ERK and PI3K/Akt are two major signaling pathways downstream of EGFR.20 Indeed, EGF treatment of MDA-MB-468 cells activated ERK and Akt (Supplementary Fig. 2A), while the EGFR inhibitor AG1478 abolished the effect of EGF (Supplementary Fig. 2A). Immunoblotting showed that inhibition of ERK and PI3K by the respective small-molecule inhibitors U0126 and LY294002 blocked the EGF induction of FOXC1, while JNK inhibition by SP600125 did not (Fig. 4A). Likewise, luciferase reporter assays indicated that ERK and PI3K inhibition, not JNK inhibition, suppressed the EGF-elicited FOXC1 promoter activity (Fig. 4B). A specific Akt inhibitor AI-IV yielded a similar effect (Fig. 4C). Notably, simultaneous blockade of ERK and Akt exerted a more pronounced inhibitory effect on the EGF-mediated increase of FOXC1 promoter activity (Fig. 4C) and mRNA levels (Fig. 4D). The ERK signaling pathway is known to play a critical role in tumor cell function.21 BLBC has been shown to harbor high ERK activity and was sensitive to small-molecule MEK inhibitors in vitro and in vivo. 22,23 To further elucidate the involvement of ERK in FOXC1 expression, we transfected ERK2 into MDA-MB-231 cells. It was found that ERK2 overexpression markedly increased FOXC1 promoter activity (Supplementary Fig. 2B), mRNA levels (Supplementary Fig. 2C), and protein levels (Supplementary Fig. 2D). In agreement with these results, overexpression of the ERK activator H-Ras induced FOXC1 promoter activation in MDA-MB-231 cells (Supplementary Fig. 2E).
Figure 4. EGFR activation up-regulates FOXC1 expression through ERK and PI3K/Akt pathways.
(A) MDA-MB-468 cells were-serum starved overnight. Cells were treated with EGF for 24 h in the presence or absence of 5 μM U0126 (MEK inhibitor), 20 μM LY294002 (PI3K inhibitor), or 10 μM SP600125 (JNKII inhibitor) after preincubation with these inhibitors for 30 minutes. p-ERK (T202/Y204), ERK, p-Akt (S473), Akt, p-JNK (T183/Y185), JNK, and FOXC1 levels were analyzed by immunoblotting. (B) MDA-MB-468 cells were transiently transfected with the FOXC1-luc and then pretreated with the inhibitors for 30 minutes followed by EGF treatment for 24 h in the presence or absence of these inhibitors, followed by luciferase assays. **, P<0.001; ***, P<0.0001. (C) MDA-MB-468 cells were transiently transfected with the FOXC1 promoter-luc and then pretreated with 5 μM U0126, 1 μM AI-IV (Akt inhibitor) or their combination for 30 minutes followed by EGF treatment for 24 h in the presence or absence of these inhibitors, followed by luciferase assays. *, P<0.05; **, P<0.001. (D) MDA-MB-468 cells were serum-starved overnight, then pretreated with 5 μM U0126, 1 μM AI-IV or their combination for 30 minutes followed by EGF treatment for 6 h in the presence or absence of these inhibitors. FOXC1 mRNA levels were detected by qRT-PCR. **, P<0.001; ***, P<0.0001. Data represent mean ± SD of 3 independent experiments.
The PI3K/Akt pathway is highly activated in BLBC due to a loss of PTEN and inositol polyphosphate-4-phosphatase-II (INPP4B) or amplification of PIK3CA.19 Akt3 is reportedly upregulated and hyperactive in ER-negative breast cancers compared with other breast cancers.24,25 To further ascertain the role of PI3K/Akt signaling in FOXC1 expression, we overexpressed a constitutively active myristoylated (Myr) PI3K catalytic subunit p110α in MDA-MB-231 cells. The mutant p110α dramatically enhanced FOXC1 promoter activity (Supplementary Fig. 2F). Similarly, overexpression of a hyperactive Akt1 or Akt3 (Myr-Akt) containing a myristolation membrane-targeting sequence also induced FOXC1 promoter activity (Supplementary Fig. 2G), mRNA levels (Supplementary Fig. 2H), and protein expression (Supplementary Fig. 2I). Taken together, these data indicate that ERK and Akt activation is sufficient to elicit FOXC1 transcription.
FOXC1 plays an essential role in EGF-mediated cell proliferation, migration, and invasion
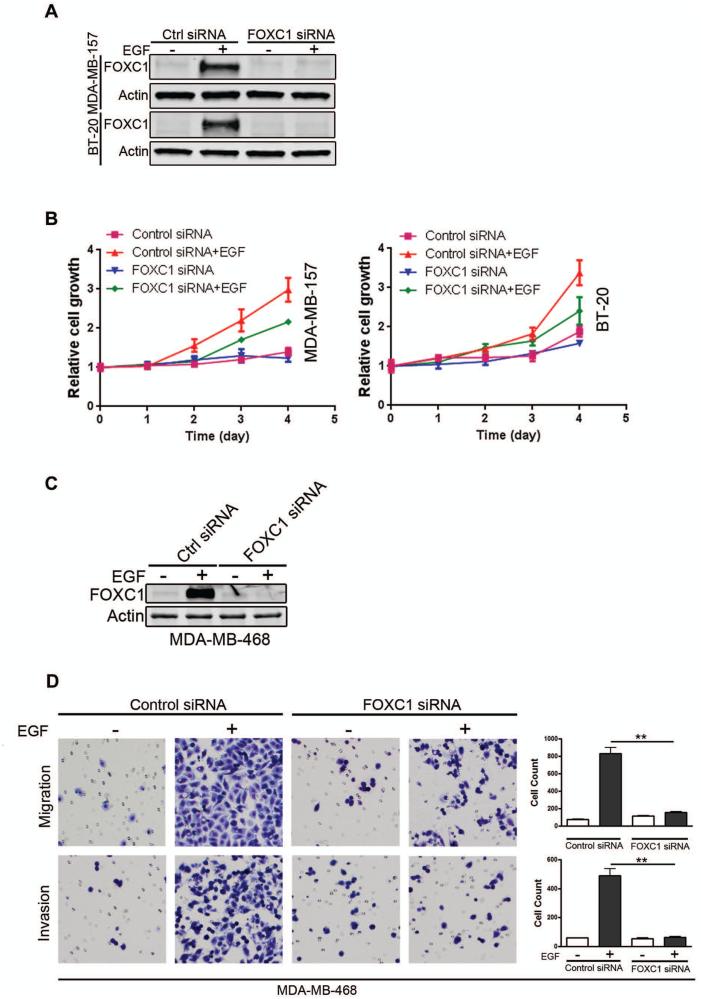
To address the question whether FOXC1 plays an important role in EGF-regulated cell function, we used control or FOXC1 siRNA to inhibit endogenous and EGF-induced FOXC1 levels in basal-like MDA-MB-157 and BT-20 breast cancer cells. Immunoblotting confirmed the knockdown of FOXC1 (Fig. 5A). Control or FOXC1 siRNA-transfected cells were then stimulated with EGF. Cell viability assays showed that FOXC1 knockdown impeded EGF-elicited cell proliferation (Fig. 5B). Given these results, we examined whether FOXC1 is essential for EGF-regulated, cell cycle-associated gene expression. Immunoblotting showed that overexpression of FOXC1 increased c-Myc and cyclin D1 levels (Supplementary Fig. 3A) while knockdown of FOXC1 impaired the induction of c-Myc, cyclin D1, E2F1, and p-Rb by EGF (Supplementary Fig. 3B). These genes are well-documented to be critically involved in cell cycle progression.26-28 Collectively, these data suggest that FOXC1 is essential for EGF-induced cell proliferation.
Figure 5. FOXC1 is essential for EGF-mediated cell proliferation, migration, and invasion.
(A) MDA-MB-157 or BT-20 cells were transiently transfected with control or FOXC1 siRNA for 24 h, and then treated with EGF for another 24 h after starvation overnight. FOXC1 knockdown was confirmed by immunoblotting. (B) Control or FOXC1-knockdown cells were stimulated with EGF and cell proliferation was evaluated by CellTiter-Glo assays at the indicated time points. Data represent 3 independent experiments. (C) MDA-MB-468 cells were transfected with control or FOXC1 siRNA for 24 h and then treated with EGF for 24 h. FOXC1 expression was assessed using immunoblotting. (D) Migration and invasion of control and FOXC1-knockdown cells were analyzed using transwell chamber assays. Migrating or invasive cells were counted in 3 randomly selected fields. Data represent mean ± SD. **, P<0.001.
To study the role of FOXC1 in EGF-elicited cell migration and invasion, we performed FOXC1 knockdown in MDA-MB-468 cells. In these cells, EGF treatment induces EMT and migration.29 As expected, FOXC1 siRNA transfection prevented the EGF induction of FOXC1 in MDA-MB-468 cells (Fig. 5C). Boyden chamber assays showed that FOXC1 knockdown abolished the increase of cell migration and invasion by EGF (Fig. 5D). We next examined the expression of matrix metalloproteinases (MMPs), as they are major proteases involved in cancer cell invasion.30 As shown in Supplementary Fig. 3C, overexpression of FOXC1 increased MMP-2 and MMP-9 levels. Conversely, knockdown of FOXC1 abrogated EGF-induced MMP-2 and MMP-9 expression and activity (Supplementary Fig. 3D, E). Of note, MMP-7 has also been shown to be regulated by FOXC1.31 Together, these results indicate that FOXC1 orchestrates EGF-elicited cell migration and invasion.
DISCUSSION
In this study, we show that EGFR activation up-regulates FOXC1 expression via ERK- and PI3K/Akt-mediated activation in BLBC. We demonstrate that FOXC1 is required for EGF-elicited cell proliferation, migration, and invasion. These findings uncover a regulatory mechanism of FOXC1 expression and an important role of the EGFR-FOXC1 signaling axis in BLBC cell function. Although EGFR has been widely accepted as a biomarker for BLBC, it is not well understood how EGFR modulates the basal-like phenotype. The induction of FOXC1 by hyperactive EGFR provides new insight into the aggressive behaviors of these cancers.
A critical issue arising from this study is the specificity of the Ras/ERK and PI3K/Akt pathways in the EGF/EGFR regulation of FOXC1. Many receptor tyrosine kinases (RTKs) such as HER-2 act through these pathways. Interestingly, FOXC1 levels are low in HER-2 (+) and luminal breast cancers, but high in BLBC. This unique expression pattern of FOXC1 may be attributed to the combined effect of several regulatory mechanisms. First, the FOXC1 promoter region is hypermethylated in ER+ breast cancer,32 which may explain why FOXC1 levels are suppressed in non-basal tumors despite ERK and Akt activation. In contrast, basal-like tumors possess lower frequency of aberrant promoter methylation compared to other subtypes, and FOXC1 gene methylation is absent in basal-like tumors. 6,33 This may lead to chromatin configuration permissive of transcription factor binding elicited by growth factor signaling. Notably, recent studies have shown that a small subset of breast cancer cells, called luminobasal and basal-HER2 cells, 34,35 exhibit the basal phenotype and express high levels of FOXC1 (X. Cui, unpublished data).
Second, PI3K/Akt and Ras/ERK activation is more prevalent in ER− /PR− and basal-like tumors.14,22,24,27 FOXC1 may serve as an indicator for high PI3K/Akt and Ras/ERK activities in BLBC. Of note, FOXC1 levels were low in some EGFR-positive BLBC cells. This might be due to the inactivation of EGFR in these cells or interrupted signal transduction downstream of EGFR, preventing transmission of signals from EGF/EGFR to the FOXC1 promoter. In addition, activation of ERK and Akt by other RTKs in BLBC may explain why FOXC1 is highly expressed in some EGFR-negative cases. Lastly, EGFR induces NF-κB and other transcription factors, some of which are preferentially activated in BLBC.36,37 Whether these transcription factors dictate the effect of Akt and ERK on FOXC1 in BLBC remains to be determined.
Recent studies show that the PKC and CARMA3 pathways are involved in EGFR-induced effects in cancer cells.38,39 It is unknown whether their levels correlate with BLBC. PKC can act downstream or upstream of Ras, inducing ERK and Akt activation. We found that PKC is not involved in the EGF regulation of FOXC1 (Y. Jin, unpublished data).
It is known that cyclin D1 and c-Myc are instrumental in EGF-elicited tumor cell proliferation. Numerous studies suggest that MMPs are critical for EGF-elicited cancer cell invasion and metastasis. Our results implicate FOXC1 as an important mediator for the EGF effect on these genes and BLBC cell functions. EGFR-directed drugs have been evaluated in breast cancer clinical trials; however, the results were disappointing.40 There is a need to exploit new predictive biomarkers for EGFR-targeted therapy to identify those patients with “EGFR-addicted” breast cancers dependent on this signaling for growth and progression. Our results might implicate FOXC1 as a read-out of EGFR activity and a useful marker in this regard. We propose that intervention of the EGFR/FOXC1 pathway may provide effective modalities for the treatment of BLBC.
Supplementary Material
Synopsis.
FOXC1 expression is regulated by EGFR activation via Akt and ERK signaling, and FOXC1 mediates the effect of EGF/EGFR in human basal-like breast cancer cells.
ACKNOWLEDGEMENTS
We thank Guifa Li for the FOXC1 promoter construct. This work was supported by National Institutes of Health (CA151610), the Avon Foundation (02-2010-068), David Salomon Translational Breast Cancer Research Fund, and the Fashion Footwear Charitable Foundation of New York, Inc. to XC and the State Key Development Program for Basic Research of China (2011CB707705) and the Science and Technology Program of Guangdong (2010B031600133, 2011B031800323) to YC.
Footnotes
Competing interest: XC is a named inventor on patent applications filed relevant to the role of FOXC1 in cancer. The other authors declare no conflict of interest.
REFERENCES
- 1.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008 May 20;26(15):2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 2.Batra SK, Castelino-Prabhu S, Wikstrand CJ, et al. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1995 Oct;6(10):1251–1259. [PubMed] [Google Scholar]
- 3.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer research. 2008 May 1;68(9):3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 4.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nature reviews. Cancer. 2007 Nov;7(11):847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 5.Kume T, Deng KY, Winfrey V, Gould DB, Walter MA, Hogan BL. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998 Jun 12;93(6):985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 6.Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proceedings of the National Academy of Sciences of the United States of America. 2008 Sep 16;105(37):14076–14081. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray PS, Wang J, Qu Y, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer research. 2010 May 15;70(10):3870–3876. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- 8.Taube JH, Herschkowitz JI, Komurov K, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2010 Aug 31;107(35):15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia L, Huang W, Tian D, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2012 Aug 22; doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]
- 10.Peraldo-Neia C, Migliardi G, Mello-Grand M, et al. Epidermal Growth Factor Receptor (EGFR) mutation analysis, gene expression profiling and EGFR protein expression in primary prostate cancer. BMC cancer. 2011;11:31. doi: 10.1186/1471-2407-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Gu F, Liu CY, Wang RJ, Li J, Xu JY. High level of FOXC1 expression is associated with poor prognosis in pancreatic ductal adenocarcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012 Dec 16; doi: 10.1007/s13277-012-0617-7. [DOI] [PubMed] [Google Scholar]
- 12.Wei LX, Zhou RS, Xu HF, Wang JY, Yuan MH. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012 Dec 22; doi: 10.1007/s13277-012-0629-3. [DOI] [PubMed] [Google Scholar]
- 13.Ray PS, Bagaria SP, Wang J, et al. Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value: a retrospective immunohistochemical study. Annals of surgical oncology. 2011 Dec;18(13):3839–3847. doi: 10.1245/s10434-011-1657-8. [DOI] [PubMed] [Google Scholar]
- 14.Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013 Feb 1;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkocz D, Crawford NT, Buckley NE, et al. BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene. 2012 Aug 9;31(32):3667–3678. doi: 10.1038/onc.2011.531. [DOI] [PubMed] [Google Scholar]
- 16.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009 Jan 15;15(2):441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 17.Heideman MR, Hynes NE. AXL/epidermal growth factor receptor (EGFR) complexes in breast cancer - culprits for resistance to EGFR inhibitors? Breast cancer research : BCR. 2013 Oct 29;15(5):315. doi: 10.1186/bcr3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004 Aug 15;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 19.TCGA Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nature reviews. Molecular cell biology. 2006 Jul;7(7):505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 21.Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert opinion on therapeutic targets. 2012 Jan;16(1):103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balko JM, Cook RS, Vaught DB, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012 Jul;18(7):1052–1059. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeflich KP, O'Brien C, Boyd Z, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009 Jul 15;15(14):4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 24.Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012 Jun 21;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatani K, Thompson DA, Barthel A, et al. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. The Journal of biological chemistry. 1999 Jul 30;274(31):21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- 26.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature reviews. Cancer. 2009 Nov;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nature reviews. Drug discovery. 2009 Aug;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mears AJ, Jordan T, Mirzayans F, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. American journal of human genetics. 1998 Nov;63(5):1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui X, Kim HJ, Kuiatse I, Kim H, Brown PH, Lee AV. Epidermal growth factor induces insulin receptor substrate-2 in breast cancer cells via c-Jun NH(2)-terminal kinase/activator protein-1 signaling to regulate cell migration. Cancer research. 2006 May 15;66(10):5304–5313. doi: 10.1158/0008-5472.CAN-05-2858. [DOI] [PubMed] [Google Scholar]
- 30.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature reviews. Molecular cell biology. 2007 Mar;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. The Journal of biological chemistry. 2012 Jul 13;287(29):24631–24640. doi: 10.1074/jbc.M112.375865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muggerud AA, Ronneberg JA, Warnberg F, et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast cancer research : BCR. 2010;12(1):R3. doi: 10.1186/bcr2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dejeux E, Ronneberg JA, Solvang H, et al. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Molecular cancer. 2010;9:68. doi: 10.1186/1476-4598-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagaria SP, Ray PS, Wang J, et al. Prognostic value of basal phenotype in HER2-overexpressing breast cancer. Annals of surgical oncology. 2012 Mar;19(3):935–940. doi: 10.1245/s10434-011-2032-5. [DOI] [PubMed] [Google Scholar]
- 35.Haughian JM, Pinto MP, Harrell JC, et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proceedings of the National Academy of Sciences of the United States of America. 2012 Feb 21;109(8):2742–2747. doi: 10.1073/pnas.1106509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr., Sledge GW., Jr. Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Molecular and cellular biology. 1997 Jul;17(7):3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi N, Ito T, Azuma S, et al. Constitutive activation of nuclear factor-kappaB is preferentially involved in the proliferation of basal-like subtype breast cancer cell lines. Cancer science. 2009 Sep;100(9):1668–1674. doi: 10.1111/j.1349-7006.2009.01228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang T, Grabiner B, Zhu Y, et al. CARMA3 is crucial for EGFR-Induced activation of NF-kappaB and tumor progression. Cancer research. 2011 Mar 15;71(6):2183–2192. doi: 10.1158/0008-5472.CAN-10-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W, Xia Y, Cao Y, et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Molecular cell. 2012 Dec 14;48(5):771–784. doi: 10.1016/j.molcel.2012.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burness ML, Grushko TA, Olopade OI. Epidermal growth factor receptor in triple-negative and basal-like breast cancer: promising clinical target or only a marker? Cancer J. 2010 Jan-Feb;16(1):23–32. doi: 10.1097/PPO.0b013e3181d24fc1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.