Abstract
Molecular transduction of biological signals is understood primarily in terms of the cooperative structural transitions of protein macromolecules, providing a mechanism through which discrete local structure perturbations affect global macromolecular properties. The recognition that proteins lacking tertiary stability, commonly referred to as intrinsically disordered proteins, mediate key signaling pathways suggests that protein structures without cooperative intramolecular interactions may also have the ability to couple local and global structure changes. Presented here are results from experiments that measured and tested the ability of disordered proteins to couple local changes in structure to global changes in structure. Using the intrinsically disordered N-terminal region of the p53 protein as an experimental model, a set of proline and alanine to glycine substitution variants were designed to modulate backbone conformational propensities without introducing non-native intramolecular interactions. The hydrodynamic radius (Rh) was used to monitor changes in global structure. Circular dichroism spectroscopy showed that the glycine substitutions decreased polyproline II (PPII) propensities relative to the wild type, as expected, and fluorescence methods indicated that substitution-induced changes in Rh were not associated with folding. The experiments showed that changes in local PPII structure cause changes in Rh that are variable and that depend on the intrinsic chain propensities of proline and alanine residues, demonstrating a mechanism for coupling local and global structure changes. Molecular simulations that model our results were used to extend the analysis to other proteins and illustrate the generality of the observed proline and alanine effects on the structures of intrinsically disordered proteins.
Keywords: intrinsically disordered protein, proline, alanine, polyproline II, hydrodynamic radius
Introduction
Intrinsic disorder is a common structural motif observed in many eukaryotic proteins most-widely found in transcription factors and other regulatory proteins that transduce molecular signals (1–4). Proteins, or protein domains, that have been classified as intrinsically disordered exhibit wide degrees of conformational freedom and flexibility (5), an absence of correlated dynamics between residue positions (6,7), and, overall, a persistent lack of tertiary stability (8,9). In addition to signal transduction, protein phosphorylation usually occurs within intrinsically disordered regions (10) and, accordingly, many critical biological processes are mediated by intrinsically disordered proteins (IDPs), such as transcriptional regulation, chromatin remodeling, and cell cycle control (11–13).
Induced folding studies (14–16) have led to quantitative descriptions of IDP signaling activity (17–19) in terms of allostery arising from the statistical thermodynamics of disorder-to-order transitions (i.e., cooperative folding). These allostery models have been useful for explaining distinctive signaling-related phenomena, such as conditional cooperativity (20) and agonism/antagonism switching (21). A key feature of these models is the induced folding of disordered structure from binding that results in the thermodynamic coupling of distant protein regions. Binding-induced folding has been observed for many IDPs (16,20–25) and characterized in detail, in terms of structures (23), binding kinetics (24), and interaction energies (25). Global organization of IDP structure in the absence of binding or folding has also been reported (26,27). The physicochemical mechanisms that organize disordered protein structures on a global scale without cooperative folding are not yet fully understood.
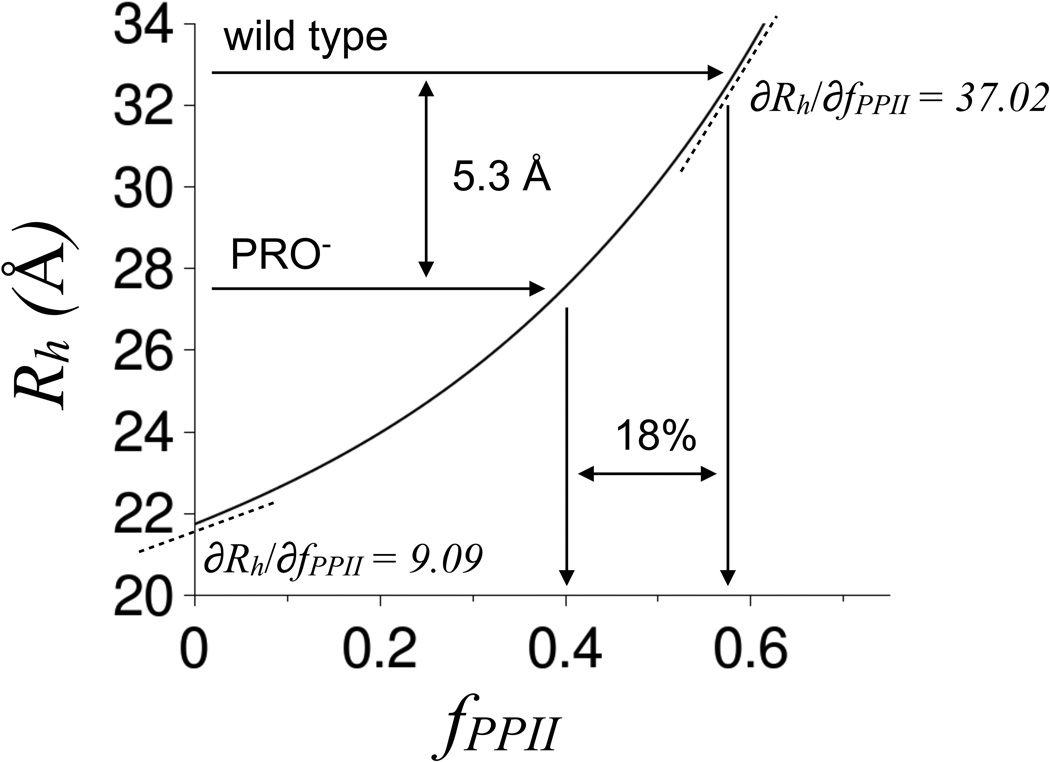
The study presented here quantitatively investigates coupled structural properties in an IDP in the absence of detectable folding. The intrinsically disordered N-terminal region of the p53 protein, p53(1–93), was used as the model IDP since this protein fragment is structurally dynamic (22,28,29), exhibits regulatory activity in the context of the full-length protein (30,31), and is proline-rich with high net charge; all common IDP properties (2–4). Motivation for the present work was the observation that substitution of all proline residues for glycine decreased the hydrodynamic radius (Rh) of p53(1–93) by 5.3 Å, though only a 2 Å reduction was expected from lower polyproline II (PPII) propensities in the substitution variant (32). The experimental data showed that changes in intrinsic backbone conformational propensities could be coupled to changes in the global structure and, specifically, that changes in local PPII propensities could cause changes in Rh greater than the predictions of structure-based models.
To assess the relationships between global structure, as reported by Rh, and position-specific conformational propensities, the structural characteristics of a set of proline (PRO) and alanine (ALA) to glycine (GLY) substitution variants of p53(1–93) were measured. The amino acid substitutions were designed to modulate intrinsic backbone conformational propensities without introducing non-native or long-range intramolecular interactions. Circular dichroism (CD) spectroscopy was used to estimate secondary structure content in each p53(1–93) variant while dynamic light scattering (DLS) and size exclusion chromatography (SEC) techniques were used to measure Rh. Analysis of the results estimated that per-residue PPII propensities for ALA and PRO in the wild type were 48% and 76%, respectively. Rh dependence on intrinsic PPII propensities followed an exponential trend, consistent with simulation data reported previously (32), showing an increased dependence on the amount of PPII structure when Rh had expanded values and a reduced dependence with compacted Rh. To test if Rh compaction caused by the PRO and ALA to GLY substitutions buried hydrophobic surface, fluorescence methods were used to measure solvent accessibility of tryptophan groups. Overall, the results showed that disordered structures can fine-tune Rh sensitivity to local structure perturbations through intrinsic backbone conformational propensities and thus regulate the linkage between local and global structure changes in the absence of cooperative folding. Extrapolation of the p53(1–93) results to other IDPs using Rh values from published reports suggested that most IDPs have similar structural characteristics in their disordered states.
Materials and Methods
Expression and purification of recombinant protein
Recombinant human p53(1–93) and staphylococcal nuclease were expressed in bacterial cells and isolated to >95% purity using the protocols described elsewhere (33,34). Purified bovine carbonic anhydrase, chicken albumin, and horse myoglobin were purchased from Sigma-Aldrich (St. Louis, MO) and further processed by ion exchange chromatography to remove residual contaminants.
Circular dicroism spectroscopy
CD spectra were recorded using a Jasco J-710 spectropolarimeter equipped with a PFD-425S peltier unit (Jasco, Easton, MD) and employed a 1-mm path-length quartz cuvette. Samples were equilibrated at each temperature for 10 min. Spectra were collected with a resolution of 0.5 nm, a scan rate of 20 nm/min, and were the average of eight scans. Reported spectra were baseline corrected for solvent and buffer contributions.
Dynamic light scattering
DLS readings used noninvasive backscatter optics and were measured using a Zetasizer Nano ZS with peltier temperature control from Malvern Instruments (Worcestershire, UK). Samples were filtered using 0.2 mm PVDF syringe-driven filters from EMD Millipore Corporation (Billerica, MA). All measurements used 1-cm path-length quartz cuvettes. Solvent viscosity was calculated using the solvent builder software program provided by Malvern, which calculates the viscosity of dilute solutions from the solution contents (e.g., percentage weight of solutes and protein) based on the program Sednterp (35).
Size exclusion chromatography
SEC experiments used Sephadex G-75 (GE Healthcare, Piscataway, NJ) equilibrated in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7. Elution volumes were determined from chromatograms measured using a Bio-Rad BioLogic LP System equipped with a UV absorbance monitor (Hercules, CA). Each sample contained 0.2–0.5 mg/mL protein in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7 with 0.3 mg/mL blue dextran and 0.03 mg/mL 2,4-dinitrophenyl-L-aspartate added as indicator dyes to determine the void and total column volumes, respectively.
Flourescence spectroscopy
Fluorescence measurements were performed using a Horiba Jobin-Yvon Fluorolog 3 spectrofluorometer equipped with a Wavelength Electronics Model LF1–3751 temperature controller. Spectra of 0.5 µM p53(1–93) and of the corresponding buffer were recorded at 20°C and averaged three times from 305–440 nm using excitation wavelengths of 280 and 295 nm. The step width was 1 nm, the integration time 1 s. Isothermal acrylamide quenching of the tryptophan fluorescence for 0.5 µM p53(1–93) used an excitation wavelength of 295 nm. Following addition of buffered acrylamide solution, the protein sample was equilibrated for 5 min under slight stirring and the fluorescence intensity was recorded for 20 s at 359 nm and averaged. Thermal scans with 0.5 µM p53(1–93) were performed using a scan rate of 2°C min−1 from 20–75°C under slight stirring.
Results and Discussion
Computer simulated structural relationship between PPII content and Rh
Rh measured for p53(1–93) under normal solution conditions was 32.8 Å at 25°C, approximately 11 Å larger than statistical coil estimates for a 93-residue polypeptide (32). A statistical coil is defined as a polypeptide chain with no strongly preferred conformations (36). p53(1–93) shows a local maximum in its CD spectrum at 221 nm, indicating PPII propensities that were estimated at 12–17% by spectral deconvolution (33). To determine if 12–17% PPII propensities could explain the relatively large Rh that was measured for p53(1–93), simulations of randomly configured polypeptide chains using a hard sphere collision model (37) were performed to quantify the relationship between chain propensities for PPII and Rh in disordered structures (32). The results of those computer simulations were condensed to a set of simple equations that are reproduced here. The mathematical relationships outlined by these equations frame the experimental strategy of the current study and accordingly were used for data analysis.
The first equation in this set relates Rh (in Å) to the number of residues, N, using the standard power-law scaling relationship,
| (1) |
where Ro was found to be 2.16. The effects of N on Rh were insensitive to amino acid sequence in the simulations. The exponential term, v, was dependent on the applied sampling bias for PPII, SPPII, by,
| (2) |
where vo was the value of the exponential with no applied chain bias (i.e., for SPPII = 0), found to be 0.509. Elimination of randomly generated structures containing van der Waals contact violations (i.e., removal of steric conflicts from the simulation) caused the fractional number of PPII folded residues, fPPII, to differ from SPPII by a Gaussian function that can be represented as,
| (3) |
Equations 1–3 provide an estimate of the dependence of Rh on PPII content in disordered proteins of any size N.
From equations 1–3, PPII propensities of 12–17% increase Rh for a 93-residue disordered protein to 23.0–23.6 Å from the statistical coil value of 21.7 Å (see Figure 1). Since Rh for p53(1–93) was measured to be 32.8 Å, these data indicated that either the simulation results were in gross error, the Rh measurements were wrong, PPII propensities in p53(1–93) were much higher than 17%, or molecular properties other than PPII propensities were dominant in determining Rh for this proline-rich protein (22 of 93 residues are proline in the wild type sequence). The CD-estimated PPII propensities were supported by NMR analysis of the proline and pre-proline positions (38), whereas size exclusion chromatography and gel electrophoresis experiments indicated that the DLS-measured Rh values were reasonable (32). Surprisingly, the decrease in Rh, relative to the wild type, measured for a p53(1–93) variant that had all PRO residues substituted for GLY (referred to as PRO−) supported the simulation data in terms of the predicted change in fPPII. Figure 1 shows that an analysis of Rh measured for this PRO− variant, found to be 27.5 Å at 25°C, was consistent with an 18% decrease in fPPII relative to the wild type, in close agreement to the CD-estimated wild type PPII propensities (32).
Figure 1. Rh dependence on fPPII for a 93-residue disordered protein.
Rh measured for wild type and PRO− p53(1–93) are indicated in the figure. The curved line was calculated using equations 1–3 with N = 93. ∂Rh/∂fPPII for a statistical coil was estimated empirically by calculating the change in Rh and fPPII for the incremental change in SPPII of 0 to 0.01. ∂Rh/∂fPPII for wild type was estimated by a similar method. First, the wild type Rh (32.8 Å) was used to calculate a corresponding SPPII (SPPII,wt) using equations 1 and 2. Then ∂Rh/∂fPPII for the wild type was estimated by calculating the change in Rh and fPPII for the incremental change in SPPII from SPPII,wt to SPPII,wt + 0.01.
The simulation results thus seemed to capture some features of the relationship between intrinsic backbone conformational propensities and the global structural parameter Rh despite its simplicity. The simulation data also indicated that changes in Rh from incremental changes in fPPII (i.e., ∂Rh/∂fPPII) were strongly dependent on the degree of Rh compaction. This can be seen in Figure 1 by noting that ∂Rh/∂fPPII for wild type p53(1–93) was ~4X the statistical coil value. To test if Rh dependence on backbone conformational propensities could be affected by structural compaction in IDPs, two additional p53(1–93) variants were constructed. One had all 12 alanine residues in the wild type substituted for GLY (referred to as ALA−). The other used the same ALA-to-GLY substitution strategy applied to the PRO− variant (referred to as ALA−PRO−). ALA residues in disordered polypeptides show significant PPII propensities, with per-residue estimates as high as 80% (39,40). Thus, ALA should contribute to Rh in a manner similar to PRO. These two additional substitution variants also tested additivity of the effects of PPII structure on Rh in this system, by measuring the effects of ALA-to-GLY substitutions in the background of wild type and PRO−, and similarly by measuring the effects of PRO-to-GLY substitutions in the background of wild type and ALA−.
Proline and alanine contributions to PPII propensities
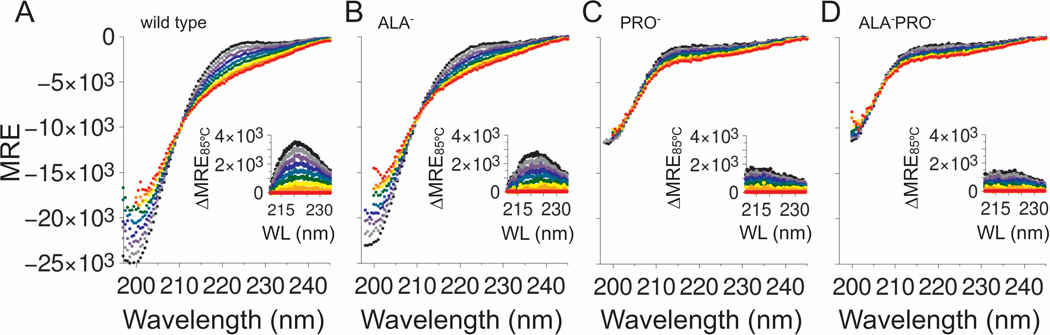
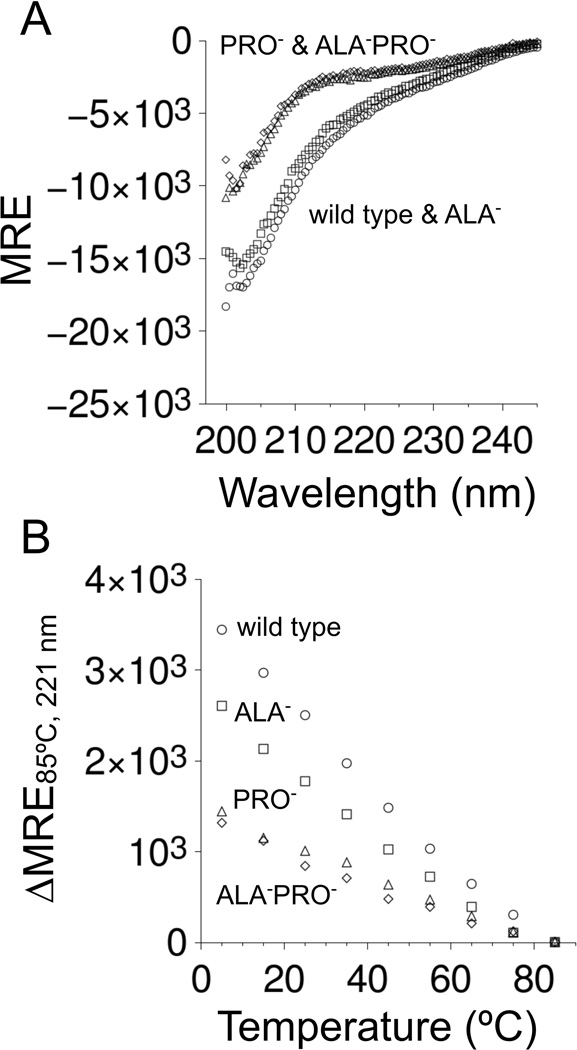
CD spectroscopy was used to estimate relative PPII propensities among the four p53(1–93) variants. The spectra measured under normal solution conditions for wild type, ALA−, PRO−, and ALA−PRO− from 5–85°C are provided in Figure 2. Spectra measured for ALA−PRO− were practically superimposable with the spectra measured for PRO−. A local maximum centered at 221 nm was observed in the variants that contained proline residues that was mostly absent in the variants that had no prolines (figure insets). A local CD maximum from 220–225 nm is often seen in proteins containing PPII (41–43). For p53(1–93), this local maximum disappeared with increased temperatures, consistent with known heat effects on PPII propensities (44). Overall, these data suggested that the proline-containing variants had significant PPII content at normal temperatures, whereas the variants lacking proline had negligible PPII content. Panel A of Figure 3 shows that the CD spectra at high temperatures differed noticeably between variants that contained proline residues (wild type and ALA−) in comparison to variants that had no prolines (PRO− and ALA−PRO−). Thus, at high temperatures where PPII content should be minor, structural differences were noted owing to the presence or absence of proline residues in the sequence.
Figure 2. CD spectrum of p53(1–93) from 5–85°C.
Panels show A) wild type, B) ALA− C) PRO−, and D) ALA−PRO−. All spectra were measured in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7 using 0.1–0.2 mg/mL of protein and reported in molar residue ellipticity (MRE) units of deg cm2 dmol−1 res−1. The data symbols (filled circles) were colored according to the temperature the spectrum was measured using: 5°C (black), 15°C (gray), 25°C (purple), 35°C (blue), 45°C (blue–green), 55°C (green), 65°C (yellow), 75°C (orange), and 85°C (red). The insets show the difference in MRE between the spectra at each temperature relative to the 85°C spectrum.
Figure 3. Effects of PRO-to-GLY and ALA-to-GLY substitutions on the CD spectrum of p53(1–93).
In both panels, wild type measurements were given with circles, ALA− with squares, PRO− with triangles, and ALA−PRO− with diamonds. Panel A shows the spectrum of each variant at 85°C. Panel B shows the temperature-dependence in the height of the local CD maximum that was observed at 221 nm (from insets of Figure 2).
Spectral deconvolution using the CDPro software package (45) was performed to quantify PPII content in the four p53(1–93) variants. Noted previously (32), CDPro predicted 12–17% PPII propensities for the wild type spectrum, depending on the algorithm (e.g., CONTIN/LL, CDSSTR) and basis set used. The results of spectral analysis performed in this manner were surprisingly insensitive to differences in the p53(1–93) spectrum owing to heat and PRO-to-GLY and ALA-to-GLY substitutions. For example, the CONTIN/LL algorithm and SP22X basis set (46) showed PPII content that ranged from 13.1–13.8% among the four variants at all temperatures (Figure S1). Since PRO-to-GLY substitutions (47) and increased temperatures (44) are known to decrease PPII content, and this was not detected by spectral deconvolution, only qualitative differences in PPII propensities among the p53(1–93) variants could be determined from CD readings. Using the local CD maximum at 221 nm as a metric for PPII content, Figure 3 panel B shows that in terms of the height of this local maximum, wild type > ALA− > PRO− ≳ ALA−PRO−. These CD data were thus interpreted qualitatively to indicate that at normal temperatures, wild type p53(1–93) had the highest levels of PPII content, with ALA− slightly less, and PRO− and ALA−PRO− significantly less.
Proline and alanine contibutions to Rh
To quantify the structural size of p53(1–93), its diffusion coefficient, D, was measured by DLS and converted to an apparent hydrodynamic radius (Rh) using the Stokes-Einstein equation,
| (4) |
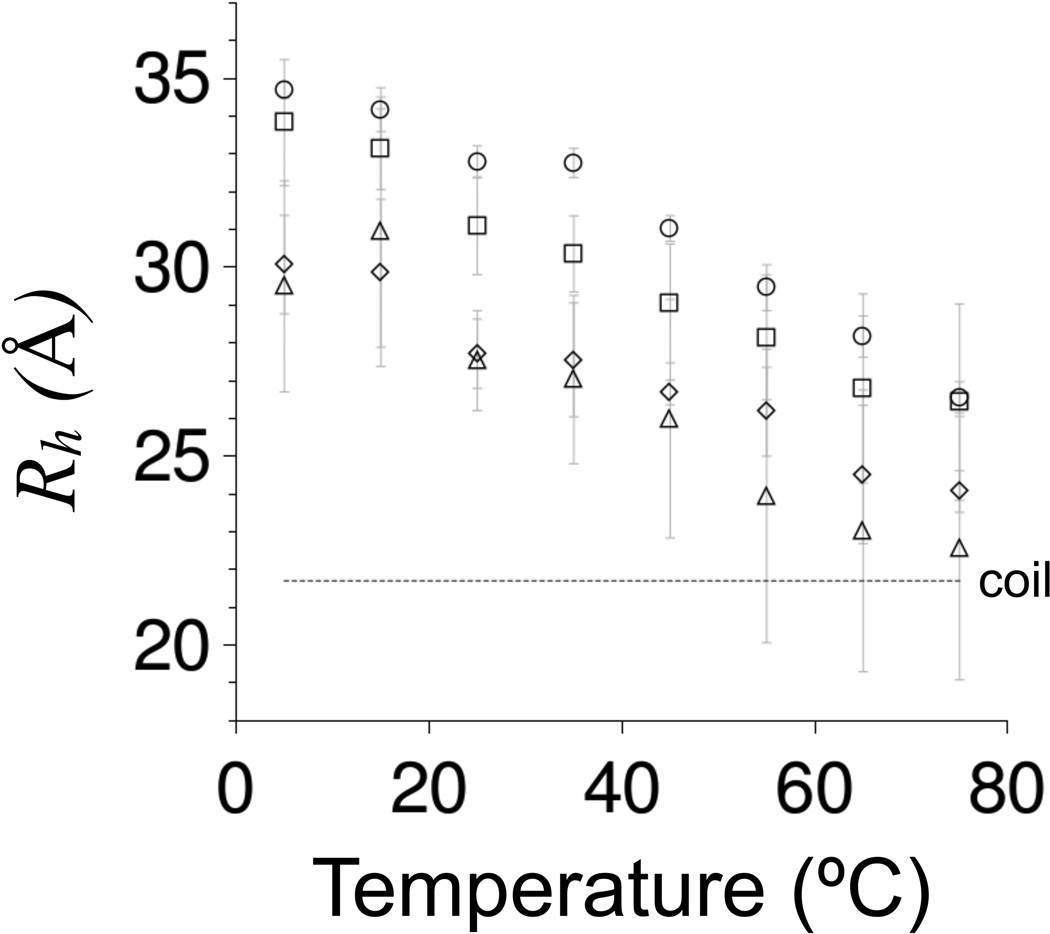
where k is the Boltzmann constant, T is absolute temperature, and η is solvent viscosity. The results from measuring Rh for each variant over a temperature range of 5–75°C using DLS are provided in Figure 4.
Figure 4. DLS-measured Rh for p53(1–93) from 5–75°C.
Rh values were shown using circles for wild type, squares for ALA−, triangles for PRO−, and diamonds for ALA−PRO−. Rh were measured using 0.25–0.75 mg/mL samples of protein in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7. The reported values are the average of at least 5 measurements performed at each temperature and error bars represent standard deviations. The stippled line labeled “coil” shows Rh calculated for a 93-residue disordered protein using equations 1 and 2 with SPPII = 0.
The three substitution variants each showed the same broad and gradual compaction in Rh caused by increased temperatures that was noted previously for the wild type (32), indicating heat-induced structural changes that were absent cooperative effects. At low temperatures, Rh for each variant was at least 7 Å larger than Rh calculated for a 93-residue statistical coil. At 75°C, Rh for PRO− and ALA−PRO− approached statistical coil dimensions, whereas Rh for wild type and ALA− remained significantly more expanded by ~5 Å. Similar to the spectroscopic observations at high temperatures using CD (Figure 3 panel A), these data indicated structural differences owing to proline residues even when proline-associated PPII content should be small. Also, compaction in Rh from glycine substitution followed the trend that was observed in the CD-estimated changes in PPII content. ALA−, which was estimated to have slightly less PPII content than the wild type, had a slightly smaller Rh at most temperatures. PRO− and ALA−PRO−, which were estimated to have significantly less PPII content than the wild type, had significantly smaller Rh.
To test if the DLS methods measured Rh accurately, size exclusion chromatography (SEC) experiments were performed at room temperature using the p53(1–93) variants and a set of protein standards. Correlating elution volumes (Ve) measured for the protein standards to molecular weight using the distribution coefficient,
| (5) |
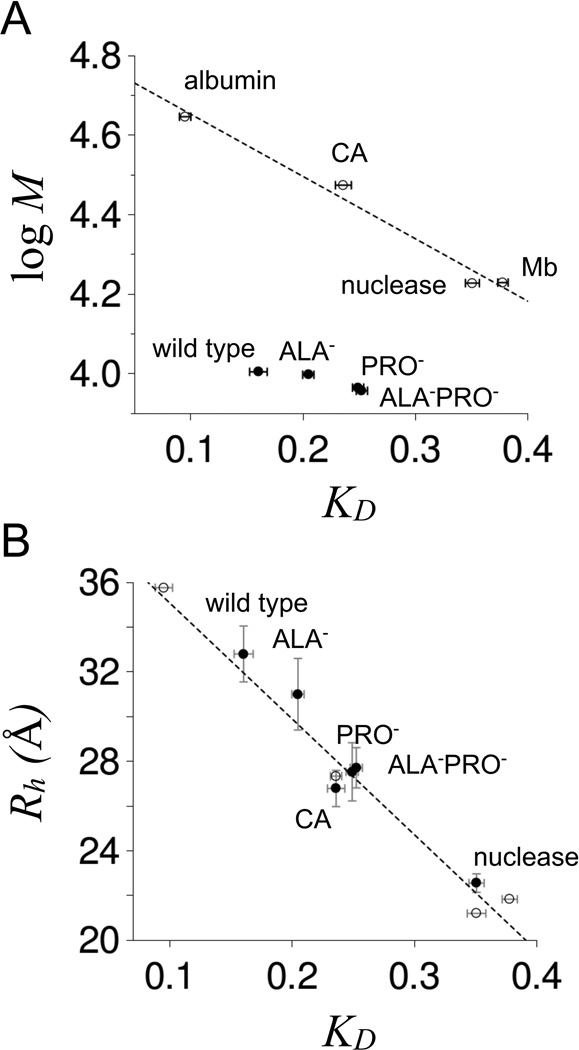
where Vt and Vo were the total and void volumes of the column, respectively, shows that the elution volume for each p53(1–93) variant was comparatively low, indicating Rh similar to folded proteins of much larger molecular weight, and Rh in a rank order of wild type > ALA− > PRO− ≳ ALA−PRO− (Figure 5 panel A). KD that were measured for the p53(1–93) variants are provided in Table I. Though KD for these variants did not trend with molecular weight when compared to folded proteins, KD for all tested proteins trended with Rh (Figure 5 panel B). Here, Rh was estimated as one-half the maximum distance between any two Cα atoms in the reported crystallographic structure for each protein standard (48–51) and then used to correlate linearly with KD. Plotting DLS-measured Rh against KD followed the same linear trend for the p53(1–93) variants, showing that SEC and DLS methods yield consistent Rh values that likewise compare favorably to Rh estimated from high resolution structures. Because of this strong agreement, KD values measured for the p53(1–93) variants were used to estimate Rh using the linear correlation that was observed with the protein standards. Rh determined in this manner yielded almost identical Rh values as the DLS measurements (Table I).
Figure 5. SEC-measuredKDfor p53(1–93) at room temperature.
Panel A shows a comparison of KD to molecular weight for wild type, ALA−, PRO−, and ALA−PRO− (filled circles). Open circles are KD measured for chicken albumin (44.3 kDa), bovine erythrocyte carbonic anhydrase (29.8 kDa, indicated as CA in the figure), staphylococcal nuclease (16.9 kDa), and horse myoglobin (16.95 kDa). Reported KD were the average of at least 3 measurements for each protein with error bars representing the standard deviations. Panel B shows a comparison of KD to Rh. Open circles are Rh estimated as one-half the maximum Cα-Cα distance in the crystallographic structures of albumin (48), carbonic anhydrase (49), nuclease (50), and myoglobin (51). The dashed line is a linear fit of Rh to KD applied to the open circles. Filled circles show Rh measured by DLS at 25°C for wild type, ALA−, PRO−, ALA−PRO−, carbonic anhydrase, and nuclease. KD were measured using 0.2 mg/mL samples of protein in 10 mM sodium phosphate, 100 mM sodium chloride, pH 7. DLS measurements used identical solution conditions except for 0.25–0.75 mg/mL protein concentrations.
Table I. Rh measured for p53(1–93) variants.
Rh values are given in Å. Rh,DSL refers to DLS measured values at 25°C (from Figure 4) and Rh,SEC is room temperature measured values using SEC (from Figure 5B). Rh,DSL and Rh,SEC averaged for each variant is given by Rh,avg. Rh,seq was calculated using the equation Rh = (1.24·fPRO + 0.904) (0.00759·Qnet + 0.963)·2.49·N0.509 determined by Marsh and Forman-Kay (52).
|
Rh,DSL |
KD |
Rh,SEC |
Rh,avg |
Rh,seq |
|
|---|---|---|---|---|---|
| wild type | 32.80 ± 1.25 | 0.160 ±0.008 | 31.95 | 32.4 | 32.2 |
| ALA− | 31.08 ± 1.28 | 0.205 ±0.005 | 29.65 | 30.4 | 32.2 |
| PRO− | 27.53 ± 1.32 | 0.249 ±0.005 | 27.36 | 27.4 | 24.3 |
| ALAPRO− | 27.71 ± 0.92 | 0.252 ±0.005 | 27.18 | 27.4 | 24.3 |
Forman-Kay and Marsh showed that Rh for IDPs scale with fractional proline content and net charge and can be predicted accurately from sequence (52). For p53(1–93), sequence predicted an Rh of 32.2 Å for the wild type and 24.3 Å for the variants lacking proline residues (Table I). For a 93-residue IDP with no proline residues and no net charge, the sequence-based equation predicted an Rh of 21.8 Å. These comparisons are important for a number of reasons. First, it shows that Rh measured for wild type p53(1–93) by DLS and SEC were consistent with general Rh trends observed in other IDPs – and thus the p53(1–93) values were not anomalous. Next, it demonstrates that estimates of statistical coil Rh from equations 1 and 2 (i.e., with SPPII = 0) are reasonable since they agree with IDP Rh values absent proline-induced or net charge-induced structural expansion. For the specific case of p53(1–93), our data indicated that proline effects on Rh depend on the presence of alanine residues at the wild type ALA positions. This observation was not predicted by the Forman-Kay and Marsh equation that calculates an identical proline contribution to Rh (7.9 Å) for both the wild type and ALA−.
Rh values measured for the p53(1–93) variants were also used to estimate per-residue PPII propensities for ALA and PRO. Using averaged Rh for wild type and PRO−, equations 1–3 predict that fPPII changed from 0.572 to 0.393 with the PRO-to-GLY substitutions. This represents a change in fractional PPII content of −0.179 and indicates a per-residue PPII propensity of 76% for each PRO in wild type. An identical calculation using the averaged Rh for ALA− gave a per-residue PPII propensity of 48% for each wild type ALA. Other research groups have shown that alanine residues favor PPII structure in disordered peptides, with propensities reported to be as low as 30% (53) and as high as 80% (39,40). Our measurements of ALA propensities for PPII were within these (admittedly large) extremes.
Figure 1 predicted that Rh dependence on fPPII should decrease with structural compaction. The experimental results clearly were in qualitative agreement with that prediction. The PRO-to-GLY substitutions compacted Rh by 5 Å when applied to the wild type, but caused only a 3 Å effect in ALA−. Similarly, the ALA-to-GLY substitutions applied to the wild type compacted Rh by 2 Å, but had a negligible effect on Rh in PRO−. Using the measured Rh values to compare changes in fPPII among the p53(1–93) variants, however, demonstrated quantitative differences between the simulation model and the experimental results. The PRO-to-GLY substitutions decreased fPPII by 0.179 when applied to the wild type and only by 0.117 when applied to ALA−. If the simulation and experimental results were in full quantitative agreement, these values should have matched. Similarly, fPPII decreased by 0.062 from wild type to ALA−, but had no change from PRO− to ALA−PRO−.
Contributions of hydrophobic burial to Rh compaction
The PRO-to-GLY and ALA-to-GLY substitutions compacted Rh for p53(1–93). Our analysis of the changes in Rh included only the effects of reduced PPII content. Since unfolding almost universally reports increased Rh values for stable proteins (54), Rh compaction is typically associated with protein folding rather than lowered chain propensities for PPII. To test for “folding” effects in Rh compaction of p53(1–93), experiments were performed to determine if hydrophobic surfaces were buried with the glycine substitutions. In this pursuit, intrinsic tryptophan fluorescence was measured in the presence of increasing acrylamide concentrations for wild type, ALA−, PRO−, and ALA−PRO−. Tryptophan residues are found at positions 23, 53, and 91 in p53(1–93). Acrylamide solutions quench tryptophan fluorescence by collisional contact and concomitant energy transfer, though this effect is more pronounced for exposed tryptophan groups relative to those that are structurally buried and protected from solvent (55–58).
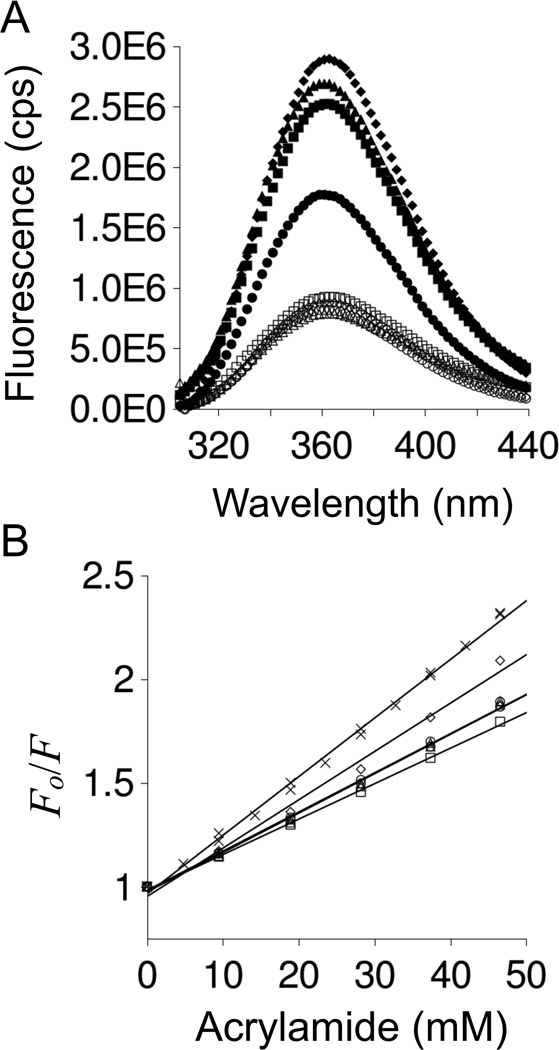
The intrinsic fluorescence of each variant at 20°C is provided in Figure 6 panel A. The emission maximum for each was ~360 nm from excitation at 280 and 295 nm, indicating tryptophan residues in polar environments consistent with fully exposed groups (59,60). Adding acrylamide decreased p53(1–93) fluorescence proportional to the amount that was added. This is shown in panel B, where fluorescence is given in terms of Fo/F. Fo refers to fluorescence in the absence of acrylamide and thus the ratio Fo/F increases with increased quenching. For reference, acrylamide induced fluorescence quenching of equal molar amounts of N-acetyl tryptophanamide (NATA) was included. Quenching increased with higher temperatures (Figure S2), indicating a dynamic quenching mechanism (i.e., collisional frequency increases with added heat). If fluorescence quenching was caused by a static (i.e., binding) mechanism, then temperature increases would reduce the formation of bound complexes, thus reducing quenching, which was not observed for any of the p53(1–93) variants.
Figure 6. p53(1–93) fluorescence spectrum and acrylamide induced quenching of tryptophan fluorescence.
Panel A shows the fluorescence spectrum of p53(1–93) measured at 20°C using excitation wavelengths of 280 nm (filled markers) and 295 nm (open markers). Panel B provides Stern-Volmer plots of acrylamide induced quenching of tryptophan at 20°C. Both panels used circles for wild type, squares for ALA−, triangles for PRO−, and diamonds for ALA−PRO−. NATA is shown in panel B using (X). In both panels, fluorescence was measured using 0.5 µM p53(1–93) or 1.5 µM NATA buffered at pH 7 with 10 mM sodium phosphate, 100 mM sodium chloride.
The extent of acrylamide induced fluorescence quenching, and thus tryptophan solvent accessibility, was assessed using the Stern-Volmer equation and the resulting Stern-Volmer constant, KS-V (61). Briefly, fluorescence intensity can be described as,
| (6) |
where [Q] is the quencher concentration. KS-V was 18.9 M−1 for wild type, 17.1 M−1 for ALA−, 19.0 M−1 for PRO−, 23.3 M−1 for ALA−PRO−, and 28.2 M−1 for the NATA reference. Disordered proteins (56) and proteins denatured by guanidine hydrochloride with reduced disulfides (62) characteristically have KS-V values ≥ 12 M−1, while values of 1–2 M−1 are typical for folded proteins with tryptophan groups that are protected from solvent (55,56,62). KS-V measured for each p53(1–93) variant at 20°C thus was consistent with exposed tryptophan groups. Also, the substitution effects on KS-V values indicated mostly minor changes in tryptophan solvent accessibility. In general, the trends from PRO-to-GLY and ALA-to-GLY substitutions on KS-V were slight increases in tryptophan solvent accessibility, possibly owing to increased chain disorder from decreased PPII content. The fluorescence quenching results did not seem to indicate significant burial of hydrophobic surface associated with Rh compaction owing to the PRO-to-GLY and ALA-to-GLY substitutions.
Rh trends among intrinsically disordered proteins
Rh that was measured for wild type p53(1–93) is similar in character to Rh measured for other IDPs, which is demonstrated in Figure 7 panel A. Here, Rh values for IDPs (26,63–80) and folded proteins (32,54,81) were plotted according to chain length. This dataset excluded IDPs with histidine tags, since histidine tags are known to compact Rh in IDPs (52). For reference, Rh for statistical coils were shown in this figure and provide a defining separation between folded and intrinsically disordered proteins. The folded proteins were all below the coil line and IDPs were all at or, more commonly, above this line.
Figure 7. Rhcomparisons to number of residuesNfor folded and intrinsically disordered proteins.
This set was limited to N > 50 and used Rh from published reports (26,32,54,63–81). In panel A, the line labeled “coil” shows Rh calculated using equations 1 and 2 with SPPII = 0. Filled circles are IDPs and open circles are folded proteins. In panel B Rh values for IDPs were converted to RSCPPII RSCPPII was determined for each IDP by first calculating the statistical coil ∂Rh/∂fPPII (which varies with N) and IDP ∂Rh/∂fPPII using the method given in Figure 1 for wild type p53(1–93). These two ∂Rh/∂fPPII values were then divided (IDP/coil) to give RSCPPII. For p53(1–93), the averaged Rh (32.4 Å) was used to determine RSCPPII rather than the DLS-measured Rh (32.8 Å). The dashed line shows the averaged RSCPPII (2.3 ± 0.8) from the set of IDPs in panel A.
To test if ALA content contributes to Rh for IDPs in general, as it does for p53(1–93), linear regression analysis was used to correlate fractional alanine content (fALA) with increases in Rh relative to the coil value (Rh,rel = Rh/Rh,coil). No correlation was observed using the set of IDPs shown in Figure 7 (coefficient of determination, R2 = 0.0001, Table S1 in Supporting Information). Fractional proline content (fPRO) and net charge (Qnet) each had positive correlations with Rh,rel in this set (R2 = 0.242 and R2 = 0.194, respectively), in agreement with the report by Marsh and Forman-Kay (52). Since ALA effects on Rh for p53(1–93) were detected only in the variant that had the largest Rh value (i.e., wild type with high fPRO and Qnet), multiple linear regression was used to determine if fALA in combination with fPRO and Qnet could increase sequence-based correlations to Rh,rel. Regression analysis showed that Rh,rel correlated with fPRO and Qnet with R2 = 0.539. Adding fALA as a fitting parameter increased R2 to 0.542. The increase in R2 owing to fALA was not meaningful, however, since adding random numbers from 0 to 1 as the additional fitting parameter rather than fALA increased R2 on average to 0.546 ± 0.008. Of note, if the set of IDPs used in regression analysis was limited to those with fPRO above 10%, then fALA correlated strongly with Rh,rel (R2 = 0.472). Similarly, if regression analysis was limited to IDPs with Qnet > 10, fALA again correlated with Rh,rel with high confidence (R2 = 0.200). These data (tabulated in Table S1) suggest that ALA contributions to Rh may be minor for IDPs overall, but significant in combination with high fPRO and/or high Qnet.
Figure 7A implies that, on average, IDPs are structurally expanded above coil estimates. Panel B in Figure 7 provides a possible selective advantage for disordered structures with expanded Rh – to regulate the coupling of local and global structure changes in IDPs. For example, the subtle effects of ALA-to-GLY substitutions on Rh were observed in wild type p53(1–93) and not in a structurally compacted variant. In Figure 7B, the reported Rh values were used to estimate ∂Rh/∂fPPII (using equations 1–3) for each IDP and then divided by ∂Rh/∂fPPII for a statistical coil to give a relative metric of how changes in fPPII affect Rh (referred to as Relative Chain Sensitivity, RCSPPII). An RCSPPII of 1 represents the baseline effect of changes in fPPII on Rh for a statistical coil. The IDP averaged RCSPPII (dashed line in Figure 7B) was more than double the baseline coil value but the standard deviation was large (averaged RCSPPII = 2.3 ± 0.8). Wide variations in RCSPPII among IDPs could be related to protein-specific requirements for coordinating global structure changes to discrete local structure perturbations, such as phosphorylation, though experiments are needed to quantify the relationships between Qnet, fPRO, fALA, and Rh. Rh,rel correlated with Qnet (R2 = 0.194), as noted above, suggesting that charge effects on IDP structure may be intertwined with fPRO and fALA.
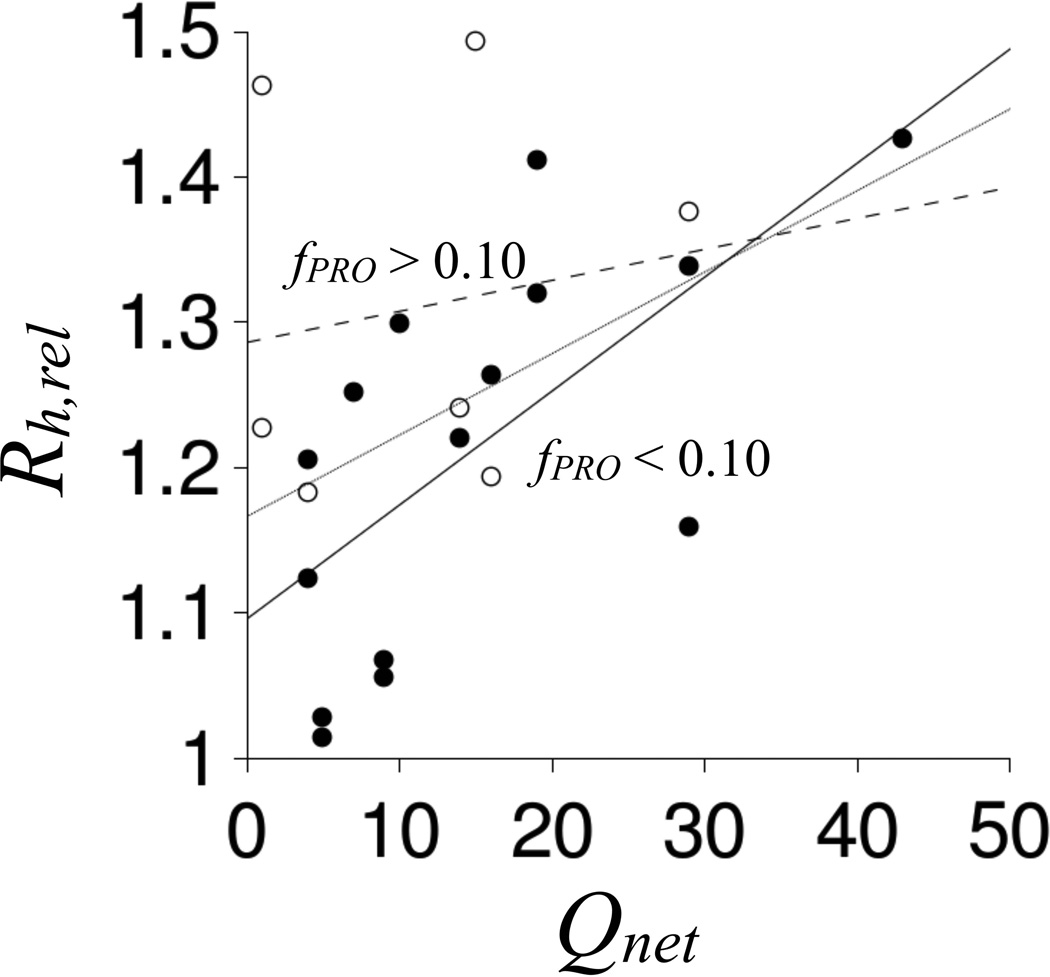
To test if charge effects on Rh depend on proline content, the set of IDPs in Figure 7 was split according to fPRO and the relationships between Rh,rel and Qnet were re-assessed. The data in Figure 8 show that the correlation between Rh,rel and Qnet decreased to negligible levels (R2 = 0.028 with ∂Rh,rel/∂Qnet = 0.002) in IDPs with fPRO > 10%. In contrast, Rh,rel correlated strongly with Qnet in IDPs with fPRO < 10% (R2 = 0.439 with ∂Rh,rel/∂Qnet = 0.008). These results predict variable charge effects, and presumably phosphorylation effects, in IDP structures dependent on proline content. A simple mechanism could account for this observation – proline promotes structural expansion in IDPs to weaken electrostatic-based interactions between charged groups. Accordingly, IDPs with high proline content, in general, should exhibit reduced coupling of changes in charge state with changes in Rh. IDPs with low proline content should demonstrate the opposite – increased coupling of changes in charge state with changes in Rh. Each IDP in Figure 7 was predicted to contain at least one phosphorylation site using the DISPHOS algorithm (Table S2 in Supporting Information), an algorithm that shows 80% accuracy in predicting protein phosphorylation sites from sequence (10). Owing to variable proline and alanine content among these IDPs, variable phosphorylation effects on Rh should be expected.
Figure 8. Comparison ofRh,relandQnetfor intrinsically disordered proteins.
IDPs with fPRO > 0.10 are shown using open circles. IDPs with fPRO < 0.10 are shown with filled circles. Lines represent linear fits of Rh,rel to Qnet for IDPs with fPRO > 0.10 (dashed), IDPs with fPRO < 0.10 (solid), and all IDPs (stippled).
Conclusions
Proteins that are structurally dynamic often exhibit random coil characteristics (82–84), but IDPs under normal conditions seem to show non-random structural organization (23,26,28) that is sensitive to sequence (52), charge (27,85,86), and temperature (32,87). To investigate the structural relationships between the global parameter Rh and intrinsic backbone conformational propensities in a disordered polypeptide chain, PRO-to-GLY and ALA-to-GLY substitutions were applied to the intrinsically disordered N-terminal region of the p53 protein, p53(1–93). The effects of GLY substitutions on Rh in p53(1–93) were compared to the results of molecular simulations of PPII effects on disordered protein structures. The experimental results qualitatively matched the simulation predictions, but lacked quantitative agreement. Differences between computational and experimental data in our study may be from non-PPII effects on disordered structure that were not included in the simulation. For example, charge-based interactions are known to affect the structural properties of IDPs (27,85,86). An analysis of IDP Rh values presented here indicated that proline content, and thus PPII structure, and charge may act in concert to affect Rh. Specifically, Rh compaction (e.g., owing to glycine substitutions) should decrease the contributions of PPII content to Rh and increase the effects of charge. ALA effects on Rh also seemed to be enhanced by high Qnet in IDPs.
The experimental results showed clearly that ALA content contributes to IDP Rh values, promoting structures larger than statistical coil estimates. Analysis of CD spectra indicated that ALA-to-GLY substitutions decreased PPII content in p53(1–93), suggesting that ALA effects on Rh were primarily owing to changes in intrinsic PPII propensities. The magnitude of Rh compaction in p53(1–93) from the ALA-to-GLY substitutions implied per-residue PPII propensities of 48% for ALA in the wild type. Likewise, CD spectra indicated that PRO-to-GLY substitutions decreased PPII content in p53(1–93), causing Rh compaction that was consistent with per-residue PPII propensities of 76% for PRO. These results demonstrated that for IDPs, Rh compaction may trend with increased chain disorder, which, for p53(1–93), was caused by decreased PPII content. This contrasts with Rh compaction associated with folded proteins (32,54), which typically follow disorder-to-order transitions (i.e., cooperative folding).
Overall, the results seemed to show that Rh sensitivity to discrete structural perturbations could vary significantly among IDPs. For example, the PRO-to-GLY substitutions caused a 5 Å compaction in Rh when applied to the wild type, but only a 3 Å compaction in Rh when applied to ALA−. Also, the effects of ALA-to-GLY substitutions on Rh were context specific, causing a 2 Å compaction of the wild type, but no effect on Rh for PRO−. In addition, correlations of Rh,rel with Qnet in IDPs showed differences owing to proline content. These results predict that IDPs, though structurally dynamic and lacking in global conformational stability, can exhibit varied global structural responses to local structural perturbations. Accordingly, the effects of phosphorylation or cofactor interactions on disordered structure may differ substantially among IDPs, but likely in a predictable and sequence dependent manner.
Supplementary Material
Acknowledgements
This work was supported by grants from the Research Corporation for Science Advancement 20039, the Texas Higher Education Coordinating Board 003615-0003-2011, and the NSF DMR-1205670 to STW and the National Heart Lung and Blood Institute of the National Institutes of Health HL109109 to MA. RBP was supported in part by NIGMS funds from the South Texas Doctoral Bridge program R25-GM-102783.
References
- 1.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inf Ser. 2000;11:161–171. [PubMed] [Google Scholar]
- 4.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Kjaergaard M, Poulsen FM. Disordered proteins studied by chemical shifts. Prog Nucl Magn Reson Spectrosc. 2012;60:42–51. doi: 10.1016/j.pnmrs.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules. 2013;18:10802–10828. doi: 10.3390/molecules180910802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 12.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98:85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, Lee JC, Bolen DW, Thompson EB. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem. 2001;276:18146–18152. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- 15.Wu P, Bolen DW. Osmolyte-induced protein folding free energy changes. Proteins. 2006;63:290–296. doi: 10.1002/prot.20868. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Motlagh HN, Chakuroff C, Thompson EB, Hilser VJ. Thermodynamic dissection of the intrinsically disordered N-terminal domain of human glucocorticoid receptor. J Biol Chem. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motlagh HN, Hilser VJ. Agonism/antagonism switching in allosteric ensembles. Proc Natl Acad Sci USA. 2012;109:4134–4139. doi: 10.1073/pnas.1120519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Pino A, Balasubramanian S, Wyns L, Gazit E, De Greve H, Magnuson RD, Charlier D, van Nuland NA, Loris R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010;142:101–111. doi: 10.1016/j.cell.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 21.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vise PD, Baral B, Latos AJ, Daughdrill GW. NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res. 2005;33:2061–2077. doi: 10.1093/nar/gki336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence CW, Showalter SA. Carbon-Detected 15N NMR spin relaxation of an intrinsically disordered protein: FCP1 dynamics unbound and in complex with RAP74. J Phys Chem Lett. 2012;3:1409–1413. doi: 10.1021/jz300432e. [DOI] [PubMed] [Google Scholar]
- 24.Rogers JM, Steward A, Clarke J. Folding and binding of an intrinsically disordered protein: fast, but not 'diffusion-limited'. J Am Chem Soc. 2013;135:1415–1422. doi: 10.1021/ja309527h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence CW, Kumar S, Noid WG, Showalter SA. Role of ordered proteins in the folding-upon-binding of intrinsically disordered proteins. J Phys Chem Lett. 2014;5:833–838. doi: 10.1021/jz402729x. [DOI] [PubMed] [Google Scholar]
- 26.Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19:566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 29.Vise P, Baral B, Stancik A, Lowry DF, Daughdrill GW. Identifying long-range structure in the intrinsically unstructured transactivation domain of p53. Proteins. 2007;67:526–530. doi: 10.1002/prot.21364. [DOI] [PubMed] [Google Scholar]
- 30.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 31.Müller-Tiemann BF, Halazonetis TD, Elting JJ. Identification of an additional negative regulatory region for p53 sequence specific DNA binding. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langridge TD, Tarver MJ, Whitten ST. Temperature effects on the hydrodynamic radius of the intrinsically disordered N-terminal region of the p53 protein. Proteins. 2014;82:668–678. doi: 10.1002/prot.24449. [DOI] [PubMed] [Google Scholar]
- 33.Schaub LJ, Campbell JC, Whitten ST. Thermal unfolding of the N-terminal region of p53 monitored by circular dichroism spectroscopy. Protein Sci. 2012;21:1682–1688. doi: 10.1002/pro.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitten ST, García-Moreno EB. pH dependence of stability of staphylococcal nuclease: evidence of substantial electrostatic interactions in the denatured state. Biochemistry. 2000;39:14292–14302. doi: 10.1021/bi001015c. [DOI] [PubMed] [Google Scholar]
- 35.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 36.Fitzkee NC, Rose GD. Reassessing random-coil statistics in unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 38.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Z, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Polyproline II structure in a sequence of seven alanine residues. Proc Natl Acad Sci USA. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Z, Chen K, Liu Z, Ng A, Bracken WC, Kallenbach NR. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proc Natl Acad Sci USA. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woody RW. Circular dichroism and conformation of unordered polypeptides. Adv Biophys Chem. 1992;2:37–79. [Google Scholar]
- 42.Rucker AL, Creamer TP. Polyproline II helical structure in protein unfolded states: lysine peptides revisited. Protein Sci. 2002;11:980–985. doi: 10.1110/ps.4550102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittington SJ, Chellgren BW, Hermann VM, Creamer TP. Urea promotes polyproline II helix formation: implications for protein denatured states. Biochemistry. 2005;44:6269–6275. doi: 10.1021/bi050124u. [DOI] [PubMed] [Google Scholar]
- 44.Chen K, Liu Z, Kallenbach NR. The polyproline II conformation in short alanine peptides is noncooperative. Proc Natl Acad Sci USA. 2004;101:15352–15357. doi: 10.1073/pnas.0406657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreerama N, Woody RW. Estimation of protein secondary structure from CD spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 46.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 47.Rucker AL, Pager CT, Campbell MN, Qualls JE, Creamer TP. Host-guest scale of left-handed polyproline II helix formation. Proteins. 2003;53:68–75. doi: 10.1002/prot.10477. [DOI] [PubMed] [Google Scholar]
- 48.Stein PE, Leslie AG, Finch JT, Turnell WG, McLaughlin PJ, Carrell RW. Crystal structure of ovalbumin as a model for the reactive centre of serpins. Nature. 1990;347:99–102. doi: 10.1038/347099a0. [DOI] [PubMed] [Google Scholar]
- 49.Saito R, Sato T, Ikai A, Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 Å resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:792–795. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- 50.Hynes TR, Fox RO. The crystal structure of Staphylococcal nuclease refined at 1.7 Å resolution. Proteins. 1991;10:92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- 51.Zahran ZN, Chooback L, Copeland DM, West AH, Richter-Addo GB. Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations. J Inorg Biochem. 2008;102:216–233. doi: 10.1016/j.jinorgbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferreon JC, Hilser VJ. The effect of the polyproline II (PPII) conformation on the denatured state entropy. Protein Sci. 2003;12:447–457. doi: 10.1110/ps.0237803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 55.Eftink MR, Ghiron CA. Dynamics of a protein matrix revealed by fluorescence quenching. Proc Natl Acad Sci USA. 1975;72:3290–3294. doi: 10.1073/pnas.72.9.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 57.Calhoun DB, Vanderkooi JM, Holtom GR, Englander SW. Protein fluorescence quenching by small molecules: protein penetration versus solvent exposure. Proteins. 1986;1:109–115. doi: 10.1002/prot.340010202. [DOI] [PubMed] [Google Scholar]
- 58.Eftink MR, Hagaman KA. Fluorescence quenching of the buried tryptophan residue of cod parvalbumin. Biophys Chem. 1985;22:173–180. doi: 10.1016/0301-4622(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 59.Beechem JM, Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 60.Eftink MR. Fluorescence techniques for studying protein structure. Methods Biochem Anal. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- 61.Lakowicz JR. Principles of Fluorescence Spectroscopy Third Edition. New York: Springer; 2006. p. 954. [Google Scholar]
- 62.Tischer A, Madde P, Blancas-Mejia LM, Auton M. A molten globule intermediate of the von Willebrand factor A1 domain firmly tethers platelets under shear flow. Proteins. 2014;82:867–878. doi: 10.1002/prot.24464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chong PA, Ozdamar B, Wrana JL, Forman-Kay JD. Disorder in a target for the smad2 mad homology 2 domain and its implications for binding and specificity. J Biol Chem. 2004;279:40707–40714. doi: 10.1074/jbc.M404375200. [DOI] [PubMed] [Google Scholar]
- 64.Sivakolundu SG, Nourse A, Moshiach S, Bothner B, Ashley C, Satumba J, Lahti J, Kriwacki RW. Intrinsically unstructured domains of Arf and Hdm2 form bimolecular oligomeric structures in vitro and in vivo. J Mol Biol. 2008;384:240–254. doi: 10.1016/j.jmb.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danielsson J, Liljedahl L, Bárány-Wallje E, Sønderby P, Kristensen LH, Martinez-Yamout MA, Dyson HJ, Wright PE, Poulsen FM, Mäler L, Gräslund A, Kragelund BB. The intrinsically disordered RNR inhibitor Sml1 is a dynamic dimer. Biochemistry. 2008;47:13428–13437. doi: 10.1021/bi801040b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi S, Boys BL, Brickenden A, Konermann L, Choy WY. Effects of zinc binding on the structure and dynamics of the intrinsically disordered protein prothymosin alpha: evidence for metalation as an entropic switch. Biochemistry. 2007;46:13120–13130. doi: 10.1021/bi7014822. [DOI] [PubMed] [Google Scholar]
- 67.Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT, Jr., Fernandez CO, Eliezer D, Zweckstetter M, Lashuel HA. J Biol Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker JMR. Structural characterization and interactions of the CFTR regulatory region (PhD Thesis) Toronto: Department of Biochemistry, University of Toronto; 2009. [Google Scholar]
- 69.Soragni A, Zambelli B, Mukrasch MD, Biernat J, Jeganathan S, Griesinger C, Ciurli S, Mandelkow E, Zweckstetter M. Structural characterization of binding of Cu(II) to tau protein. Biochemistry. 2008;47:10841–10851. doi: 10.1021/bi8008856. [DOI] [PubMed] [Google Scholar]
- 70.Lowry DF, Stancik A, Shrestha RM, Daughdrill GW. Modeling the accessible conformations of the intrinsically unstructured transactivation domain of p53. Proteins. 2008;71:587–598. doi: 10.1002/prot.21721. [DOI] [PubMed] [Google Scholar]
- 71.Adkins JN, Lumb KJ. Intrinsic structural disorder and sequence features of the cell cycle inhibitor p57Kip2. Proteins. 2002;46:1–7. doi: 10.1002/prot.10018. [DOI] [PubMed] [Google Scholar]
- 72.Uversky VN, Permyakov SE, Zagranichny VE, Rodionov IL, Fink AL, Cherskaya AM, Wasserman LA, Permyakov EA. Effect of zinc and temperature on the conformation of the gamma subunit of retinal phosphodiesterase: a natively unfolded protein. J Proteome Res. 2002;1:149–159. doi: 10.1021/pr0155127. [DOI] [PubMed] [Google Scholar]
- 73.Donaldson L, Capone JP. Purification and characterization of the carboxyl-terminal transactivation domain of Vmw65 from herpes simplex virus type 1. J Biol Chem. 1992;267:1411–1414. [PubMed] [Google Scholar]
- 74.Haaning S, Radutoiu S, Hoffmann SV, Dittmer J, Giehm L, Otzen DE, Stougaard J. An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro. J Biol Chem. 2008;283:31142–31152. doi: 10.1074/jbc.M805024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geething NC1, Spudich JA. Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J Biol Chem. 2007;282:21518–21528. doi: 10.1074/jbc.M701932200. [DOI] [PubMed] [Google Scholar]
- 76.Permyakov SE, Millett IS, Doniach S, Permyakov EA, Uversky VN. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- 77.Magidovich E, Orr I, Fass D, Abdu U, Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K+ channel modulates its interaction with scaffold proteins. Proc Natl Acad Sci USA. 2007;104:13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell KM1, Terrell AR, Laybourn PJ, Lumb KJ. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 2000;39:2708–2713. doi: 10.1021/bi9923555. [DOI] [PubMed] [Google Scholar]
- 79.Sánchez-Puig N, Veprintsev DB, Fersht AR. Binding of natively unfolded HIF-1alpha ODD domain to p53. Mol Cell. 2005;17:11–21. doi: 10.1016/j.molcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 80.Sánchez-Puig N, Veprintsev DB, Fersht AR. Human full-length securin is a natively unfolded protein. Protein Sci. 2005;14:1410–1418. doi: 10.1110/ps.051368005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tcherkasskaya O, Davidson EA, Uversky VN. Biophysical constraints for protein structure prediction. J Proteome Res. 2003;2:37–42. doi: 10.1021/pr025552q. [DOI] [PubMed] [Google Scholar]
- 82.Chan HS, Dill KA. Polymer principles in protein structure and stability. Annu Rev Biophys Biophys Chem. 1991;20:447–490. doi: 10.1146/annurev.bb.20.060191.002311. [DOI] [PubMed] [Google Scholar]
- 83.Tanford C, Kawahara K, Lapanje S. Proteins in 6-M guanidine hydrochloride. Demonstration of random coil behavior. J Biol Chem. 1996;241:1921–1923. [PubMed] [Google Scholar]
- 84.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Wiley; 1969. p. 432. [Google Scholar]
- 85.Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wuttke R1, Hofmann H, Nettels D, Borgia MB, Mittal J, Best RB, Schuler B. Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc Natl Acad Sci USA. 2014;111:5213–5218. doi: 10.1073/pnas.1313006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.