Abstract
Disulfide-bond A oxidoreductase-like protein (DsbA-L) possesses beneficial effects such as promoting adiponectin multimerization and stability, increasing insulin sensitivity, and enhancing energy metabolism. The expression level of DsbA-L is negatively correlated with obesity in mice and humans, but the underlying mechanisms remain unknown. To address this question, we generated reporter gene constructs containing the promoter sequence of the mouse DsbA-L gene. Deletion analysis showed that the proximal promoter of mouse DsbA-L is located between −186 and −34 bp relative to the transcription start site. In silico analysis identified a putative Sp1 transcription factor binding site in the first intron of the DsbA-L gene. Electrophoretic mobility shift assay and chromatin immunoprecipitation analysis indicated that Sp1 bound to this intron region in vitro and in intact cells. Overexpression of Sp1 or suppressing Sp1 expression by siRNA reduced or increased DsbA-L promoter activity, respectively. The binding activity of Sp1 was gradually decreased during 3T3-L1 cell differentiation and was significantly increased in adipose tissues of obese mice. Our results identify Sp1 as an inhibitor of DsbA-L gene transcription, and the Sp1-mediated inhibition of DsbA-L gene expression may provide a mechanism underlying obesity-induced adiponectin downregulation and insulin resistance.
Introduction
Disulfide-bond A oxidoreductase-like protein (DsbA-L) has been demonstrated to play a key role in the biosynthesis and multimerization of adiponectin (1), an adipocyte-derived hormone with multiple salutary effects including insulin sensitizing, anti-inflammatory, antiatherogenic, and cardioprotective (2). DsbA-L promotes adiponectin multimerization and stability in 3T3-L1 cells and in mice (1,3) and provides a protective effect against endoplasmic reticulum stress–mediated adiponectin downregulation in obesity (4). The level of DsbA-L is negatively correlated with obesity in mice and humans (1). Recently, it was demonstrated that the fat-specific overexpression of DsbA-L protected mice against diet-induced obesity, insulin resistance, and hepatic steatosis (3). These findings suggest that DsbA-L has beneficial effects on metabolism and the regulation of DsbA-L expression could be an effective therapeutic regiment for the treatment of obesity and its associated metabolic disorders.
DsbA-L is expressed ubiquitously but at variable levels (5,6). However, little is known on the mechanisms regulating DsbA-L gene expression. Here, we have characterized the promoter activity of mouse DsbA-L and identified Sp1 as a negative regulator of DsbA-L gene transcription.
Research Design and Methods
Plasmids
A 2593 bp fragment containing the 5′-flanking region and DNA encoding exon 1 and the first intron region of the mouse DsbA-L gene (−2004 to +588 relative to the transcription start site [TSS]) was inserted into the luciferase reporter vector pGL3-basic (Promega) to create construct p(−2004 to +588)Luc. Then, a series of 5′ or 3′ deletion reporter constructs and mutant construct was generated. The primers used for various constructs are listed in Table 1.
Table 1.
Sequences of primers and oligonucleotides used for generation of constructs, mutagenesis, EMSA probes, ChIP, and quantitative real-time PCR
| Primer/oligonucleotide | Sequence |
|---|---|
| Primers for constructs | |
| p(−2004 to +588)Luc | F1: 5′-CTAGCTAGCAACTGCCTCAGCTGCTTCAC-3′; R1: 5′-CCGCTCGAGAAGAAGGGGAGCTTGTGTGG-3′ |
| p(−2004 to +41)Luc | F1; R2: 5′-CCGCTCGAGGGGCTGCCGAAGAGCTTTAG-3′ |
| p(−1591 to +41)Luc | F2: 5′-CTAGCTAGCCAGCCAGTGACTCCAGTAAC-3′; R2 |
| p(−894 to +41)Luc | F3: 5′-CTAGCTAGCCACCATTGACCCACTAAACC-3′; R2 |
| p(−186 to +41)Luc | F4: 5′-CTAGCTAGCGCTGGCTGCACACACTTGAG-3′; R2 |
| p(−34 to +41)Luc | F5: 5′-CTAGCTAGCTTAGCAGACCCGCCTGCAAT-3′; R2 |
| p(−186 to +588)Luc | F4; R1 |
| p(−186 to +432)Luc | F4; R3: 5′-CCGCTCGAGTCCCGTAGAGCATCCTAGCG-3′ |
| p(−186 to +391)Luc | F4; R4: 5′-CCGCTCGAGCTTGACCTAGCTTCATTGGAG-3′ |
| p(−186 to +371)Luc | F4; R5: 5′-CCGCTCGAGGTTGCCTCTGTTCTTTCTAG-3′ |
| p(−186 to +227)Luc | F4; R6: 5′-CCGCTCGAG AATTTCTGCAGCCCGGTGAT-3′ |
| siRNA | F: 5′- GGATGGTTCTGGTCAAATACA-3′ |
| Scrambled | F: 5′- GGGTGCATATACAGATTGACT-3′ |
| Probes for site-directed mutagenesis and EMSA | |
| p(−186 to 432)Luc mut | 5′-CGAGTCCCGTAGAGCATCCTAGCGCCCCTGATGAACTTGACCTAGCTTCATTGGAGTTGCCTCTG-3′ |
| DsbA-L | 5′-GTCAAGTTCGGGGAGGGGGATCAG-3′ |
| Sp1C | 5′-GGGTCTGGGCGGGGGGGAGGGGACC-3′ |
| Primers for ChIP | |
| DsbA-L | F: 5′-CAGAGGCAACTCCAATGAAG-3′; R: 5′-CTAAGTGCTGGACAGAGATG-3′ |
| Primers for quantitative real-time PCR | |
| DsbA-L | F: 5′-GAATGTCCACAGCGCAAGCC-3′; R: 5′-AGTGGTGGGTAGCCCAAAGG-3′ |
| GAPDH | F: 5′-GGATTTGGCCGTATTGGG-3′; R: 5′-GTTGAGGTCAATGAAGGGG-3′ |
NheI and XhoI restriction sites are underlined. mut, mutant; siRNA, small interfering RNA.
Luciferase Reporter Assays
The various firefly luciferase reporter plasmids and the Renilla luciferase–expressing plasmid (pRL-TK; Promega) were transiently transfected into human embryonic kidney (HEK)293 cells and 3T3-L1 preadipocytes cells. Forty-eight hours after transfection, cells were harvested and luciferases were assessed using the Dual Luciferase Assay System (Promega) according to the manufacturer’s protocols. For overexpression or knockdown experiments, Sp1 expression plasmid or Sp1 shRNA plasmid was cotransfected, respectively.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was carried out using the LightShift EMSA Optimization and Control kit (Pierce). Probes and competitors are listed in Table 1.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed by using the Agarose ChIP kit (Pierce) according to the manufacturer’s instructions. Primers are listed in Table 1.
Quantitative Real-Time PCR
Experiments were performed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR Green, and primers are listed in Table 1.
Statistical Analysis
All data were analyzed by Student t tests or ANOVA, followed by Dunnett post-hoc test using GraphPad Prism (GraphPad Software). P values <0.05 were considered statistically significant.
Results
Identification of DsbA-L Proximal Promoter Region and Potential Transcription Factor Binding Sites
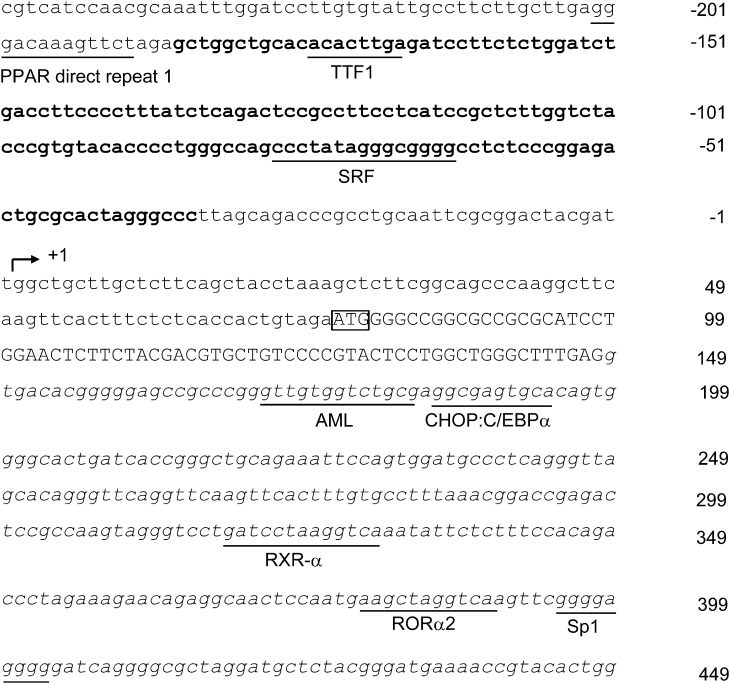
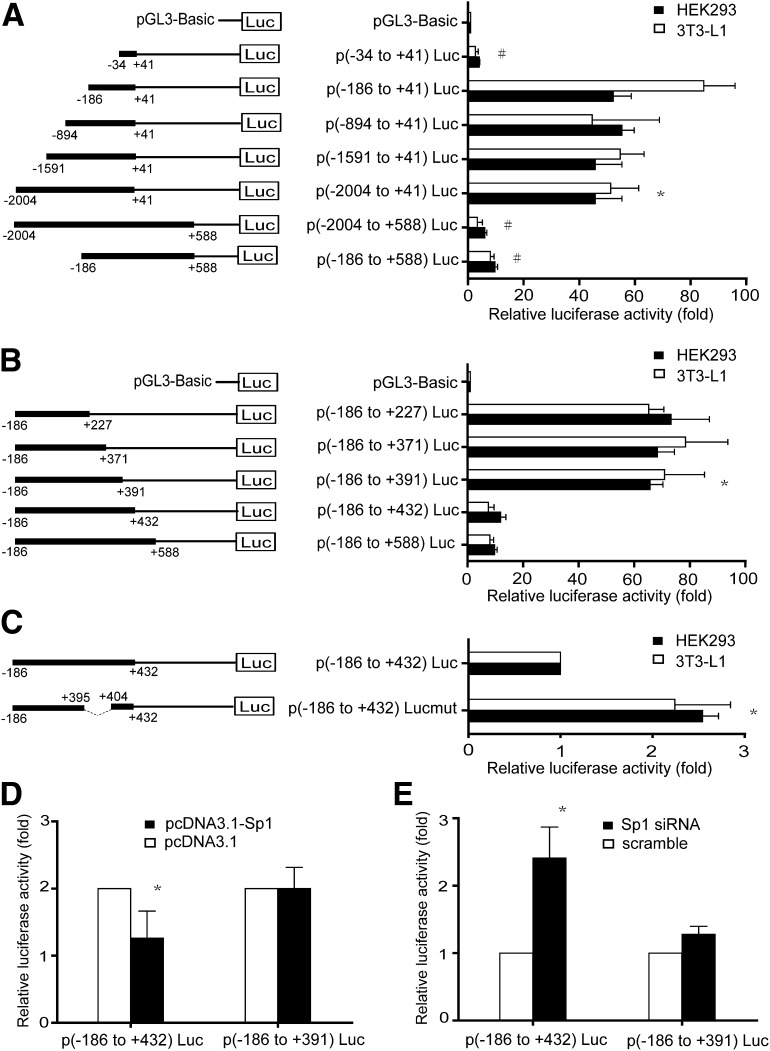
Nucleotide sequence of mouse DsbA-L promoter was obtained from the National Center for Biotechnology Information (GenBank accession no. NC_000072.5). The TSS for the DsbA-L gene was determined from a search of the transcriptional regulatory element database (7) and was defined as +1 (Fig. 1). For identification of the proximal promoter region, a series of 5′ deletion promoter reporter constructs with a common 3′-terminus at nucleotide +41 was generated and transient expressed into HEK293 or 3T3-L1 cells. In HEK293 cells, the p(−2004 to +41)Luc showed a 50-fold increase in promoter activity compared with the luciferase reporter vector pGL3-basic (Fig. 2A). Deletion from −2004 to −186 (p[−186 to +41]Luc) had no significant effect on reporter gene activities, but further deletion to −34 (p[−34 to +41]Luc) dramatically reduced the luciferase levels to ∼5% of that of the p(−2004 to +41)Luc, implying that the proximal promoter is located between −186 and −34 relative to TSS of DsbA-L. Similar results were observed in 3T3-L1 cells (Fig. 2A).
Figure 1.
Nucleotide sequence of the 5′-flanking region and DNA encoding exon 1 and intron 1 region of the mouse DsbA-L gene. The numbering of the sequence is relative to the TSS (+1) indicated by the arrow. The translation start site (ATG) is boxed. Sequence of the proximal promoter appears in boldface type. The potential binding sites predicted by MAPPER software are underlined. Coding nucleotides are in capital letters, and intronic sequences are in italics.
Figure 2.
Analysis of the mouse DsbA-L promoter activities in HEK293 cells and 3T3-L1 cells. Luciferase reporter constructs containing the indicated promoter fragments of the mouse DsbA-L gene were transiently transfected into HEK293 cells or 3T3-L1 cells. Forty-eight hours after transfection, cells were harvested and luciferase activity was determined. Relative luciferase activities were normalized to pRL-TK luciferase activity and expressed as means ± SD in fold of activity obtained with the luciferase reporter vector pGL3-basic. All data are presented as three individual transfection experiments. A: 5′ deletion analysis of the mouse DsbA-L promoter reveals the proximal DsbA-L promoter located between nt −186 and −34 relative to the TSS. Schematic structures of the reporter constructs are shown on the left. *P < 0.05 vs. pGL3-basic, #P < 0.05 vs. p(−2004 to +41)Luc. B: Mapping of the cis-acting elements in the first intron responsible for the mouse DsbA-L promoter activity. Schematic representation of the reporter construct including the −186/+588 region of the mouse DsbA-L gene. *P < 0.05 vs. p(−186 to +588)Luc. C: Sp1-binding motif is important for mouse DsbA-L promoter activity. Schematic representation of the reporter constructs with internal deletion mutation of Sp1. *P < 0.05 vs. wild-type construct. D and E: Analysis of the influence of Sp1 on DsbA-L promoter activity. HEK293 cells were cotransfected with the reporter constructs p(−186 to +432)Luc and p(−186 to +391)Luc and Sp1 expression construct or Sp1 shRNA plasmid. *P < 0.05 vs. p(−186 to +391)Luc.
Since the first intron often contains cis-acting regulatory elements, we tested whether the DNA sequence encoding the first intron of mouse DsbA-L gene confers DsbA-L promoter activity by two report constructs, p(−2004 to +588)Luc and p(−186 to +588)Luc, both containing the first intron region in addition to the promoter region. As shown in Fig. 2A, compared with p(−2004 to +41)Luc, the presence of the first intron resulted in a dramatic reduction in DsbA-L promoter activity. For mapping of the essential regulatory region within the first intron, a series of truncated report constructs, which contain a fixed 5′ end at the −186 position but different 3′ ends, was generated. In both cell lines, deletion of the sequence from +588 to +432 did not alter the luciferase activity (Fig. 2B). However, deletion from +432 to +391 led to recovery of luciferase reporter activity (Fig. 2B). Further deletion to +371 or to +227 did not further increase the promoter activity (Fig. 2B), suggesting that the region between +391 and +432 contains important cis-acting repressor elements that suppress DsbA-L transcription.
Putative Sp1-Binding Site Within the First Intron Region of the Mouse DsbA-L Gene Is Responsible for Regulating the Promoter Activity
To elucidate the mechanisms underlying the regulation of the transcriptional activity of the DsbA-L promoter, we searched for transcription factor binding sites using the MAPPER software. Several putative transcription factor binding sites were identified (Fig. 1). Notably, a single Sp1-binding site was found in between +391 to +432, the region involved in regulating the promoter activity (Fig. 2B). To estimate the contribution of this binding site to the negative regulation of the DsbA-L promoter activity, we generated a mutant reporter construct by removing the sequence between +395 and +404 that harbors the putative Sp1-binding site. Deletion of the putative Sp1-binding site increased DsbA-L promoter activities by ∼2.6- and ∼2.0-fold compared with the control construct (p[−186 to +432]Luc) in HEK293 cells and 3T3-L1 cells, respectively (Fig. 2C). Overexpression of Sp1 reduced the luciferase activities of the reporter gene containing the putative Sp1 site (p[−186 to +432]Luc) by ∼40% compared with the control construct that does not contain the putative Sp1 site (p[−186 to +391]Luc) (Fig. 2D). In contrast, suppressing Sp1 expression by siRNA increased luciferase activity of p(−186 to +432)Luc by approximately twofold (Fig. 2E). Taken together, these results implied that DsbA-L promoter transcriptional activity might be negatively regulated by Sp1 through the putative Sp1-binding sites.
Sp1 Binds to the Sequences Encoding the First Intron of the DsbA-L Gene
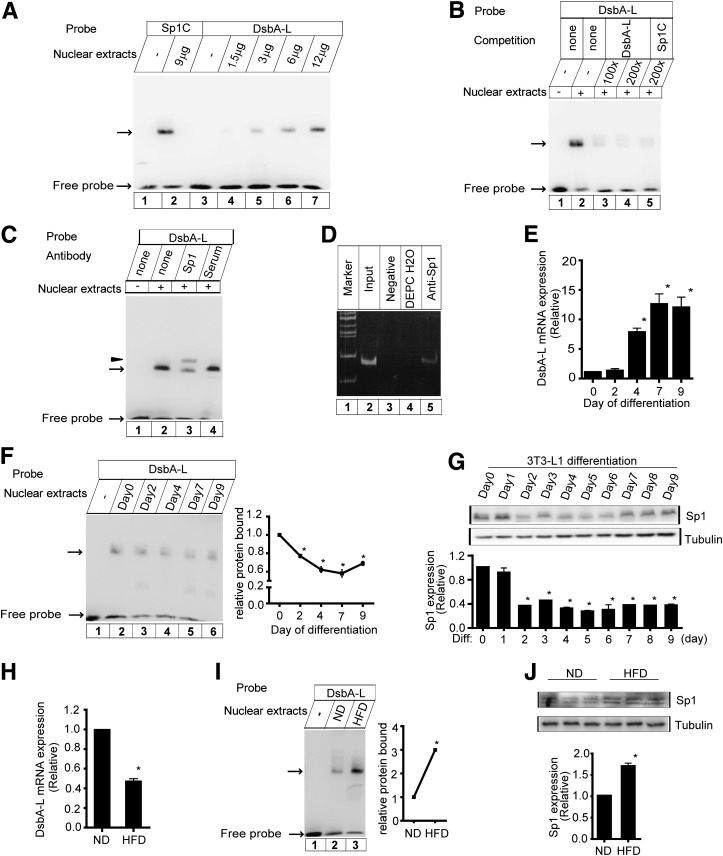
To determine the interaction of Sp1 with the putative binding sites, we performed EMSA using the DNA probe derived from the DsbA-L intron (+386 to +409) that encompasses the putative Sp1-binding site. The oligonucleotide formed a prominent complex with the nuclear extracts from HEK293 cells, and the intensity of the complex increased with the increasing amounts of the nuclear extracts (Fig. 3A, lanes 4–7). The mobility of DNA-protein complex is similar to that of the complex with the Sp1C probe (Fig. 3A, lane 2), which contains the consensus Sp1-binding site and a similar core sequence to that of the potential Sp1-binding site in the intron of DsbA-L. Similar results were observed using the nuclear extracts from 3T3-L1 cells (data not shown). The formation of the complex of DsbA-L probe was competed using excess of unlabeled DsbA-L probe (Fig. 3B, lanes 3–4) or unlabeled Sp1C probe (Fig. 3B, lane 5). Furthermore, the formation of the DNA-protein complex was prevented by an anti-Sp1 antibody, which led to the production of a slow migrating band in supershift assays (Fig. 3C, lane 3). Besides, ChIP assays were performed in 3T3-L1 cells. The protein-DNA complex coimmunoprecipitated with Sp1 was recovered, and the purified DNA was used as a template for PCR using primers corresponding to the potential Sp1-binding site. A 184-bp PCR product was amplified from the DNA fragment immunoprecipitated by Sp1 antibody (Fig. 3D, lane 5). These results indicated that Sp1 bound to the potential Sp1 site within the intron region of the DsbA-L in vitro and in intact cells.
Figure 3.
Specific binding of Sp1 to the first intron of the DsbA-L gene. A: EMSA. The DsbA-L probe encompassed the putative Sp1-binding site, and Sp1C probe contained the consensus site for Sp1. In binding reactions, the labeled probes were incubated in the absence or presence of increasing amounts of nuclear extracts from HEK293 cells. B: For the competition, the unlabeled competitors in 100- and 200-fold molar excesses of the labeled probe were included in the reaction prior to the addition of the labeled probes. C: For supershift analysis, nuclear extracts were incubated with the antibody against Sp1 or serum before the addition of the labeled DsbA-L probe. The arrow and an arrowhead indicate the specific DNA-protein complexes and supershift band, respectively. D: ChIP analysis. ChIP analysis was performed as described in research design and methods. As a negative control, the chromatin was incubated with nonspecific IgG. DNA immunoprecipitated through the antibody against Sp1 was analyzed by PCR with primers specific for the DsbA-L. Input DNA and diethylpyrocarbonate-treated water (DEPC H2O) were used as a positive or negative control for the PCR reaction, respectively. E: DsbA-L transcription is increased during 3T3-L1 differentiation to adipocytes. 3T3-L1 fibroblasts were differentiated into adipocytes, and cells were collected on the indicated days. Total RNA was isolated and subjected to quantitative real-time PCR analysis for DsbA-L mRNA. Data are means ± SEM, n = 4. *P < 0.05 vs. day 0 of differentiation. F: The binding activity of Sp1 to the DsbA-L intronic sequence was reduced during 3T3-L1 adipocyte differentiation. Nuclear extracts from 3T3-L1 cells during different stages of differentiation were incubated with labeled DsbA-L oligonucleotide. The specific DNA-protein complexes are indicated by the arrow. Quantification of the relative change in protein bound (expressed as percentage of day 0 protein bound, arbitrarily set as 1.0) was performed by analyzing EMSA data using the Scion Image program (Scion Corp.). Data are presented as means ± SEM from three independent experiments with similar results. *P < 0.05 vs. day 0. G: Sp1 expression during 3T3-L1 cell differentiation (Diff). Sp1 gives rise to two bands of 95 and 106 kDa on SDS gels. Quantification of the relative protein levels (expressed as percentage of day 0 protein levels, arbitrarily set as 1.0) was performed by analyzing Western blot data from three independent experiments using the Scion Image program. Tubulin was used as a loading control. Data are means ± SEM. *P < 0.05 vs. day 0. H: DsbA-L mRNA is decreased in HFD-fed mice. Data are means ± SEM, n = 5. *P < 0.05 vs. ND-fed mice. I: The binding activity of Sp1 is increased in adipose tissues of diet-induced obese mice. Nuclear proteins were prepared from the adipose tissues of ND-fed or HFD-fed C57BL/6 mice. The specific DNA-protein complexes are indicated by the arrow. Quantification of the relative change in protein bound (expressed as percentage of ND-fed mice protein bound, arbitrarily set as 1.0) was performed as described for F. *P < 0.05 vs. ND-fed mice. J: Sp1 expression in diet-induced obese mice. Quantification of the relative protein levels (expressed as percentage of ND-fed mice protein level, arbitrarily set as 1.0) was performed as described in G. *P < 0.05 vs. ND-fed mice.
Expression of DsbA-L mRNA in 3T3-L1 Cells During Differentiation and in Diet-Induced Obese Mice
Since DsbA-L is reported to be highly expressed in adipose tissue and the protein expression is activated during 3T3-L1 cells differentiation (1), we examined how DsbA-L transcription is regulated during 3T3-L1 differentiation. As shown in Fig. 3E, DsbA-L mRNA was barely detectable in preadipocytes, remained low at day 2 of differentiation and increased markedly from day 4 to day 9 during the late stage of differentiation. Given that Sp1 acted repressively on the DsbA-L promoter activity, we investigated whether the transcriptional role of Sp1 might be influenced during differentiation by EMSA using nuclear extracts from adipocytes during different differentiation stages. EMSA detected a band in 3T3-L1 preadipocytes (Fig. 3F, lane 2). The intensity of this band was decreased during differentiation compared with preadipocytes (Fig. 3F), indicating a gradual reduction in the binding activity of Sp1 to the DsbA-L intronic sequence during adipocyte differentiation, which coincided with the increases in DsbA-L mRNA levels in the course of 3T3-L1 adipogenesis. Consistent with this result, Sp1 expression was reduced during 3T3-L1 differentiation (Fig. 3G), which was reflected in the decrease in Sp1 binding activity. Moreover, the expression and the binding activity of Sp1 were markedly increased in adipose tissues of high-fat diet (HFD)-induced obese mice compared with normal diet (ND)-fed mice (Fig. 3I and J) concurrently with a significant decrease in DsbA-L mRNA levels in adipose tissues of the HFD-fed mice (Fig. 3H).
Discussion
In this study, we characterized the DsbA-L promoter region and analyzed potential transcription factors involved in the regulation of DsbA-L promoter activity. Our results demonstrate the presence of a Sp1-binding site in the first intron of the DsbA-L gene. We also show that Sp1 functions as a transcriptional repressor to regulate DsbA-L gene expression.
Sp1, a ubiquitously expressed mammalian transcription protein, is known to interact directly or indirectly with several nuclear proteins such as transcription-associated proteins, sequence-specific DNA-binding proteins, transcriptional regulators, and chromatin remodeling factors in a dynamic complex to regulate transcription (8–10). Depending upon the proteins with which it interacts, Sp1 activates or represses transcription of many genes in response to physiological and pathological stimuli (11–14). Sp1 has been shown to repress gene expression through the recruitment of histone deacetylase or DNA methyltransferase (11,12,15). However, Sp1 has also been shown to regulate gene expression by binding to the same sites where some other transcription factors such as MAZ, YY1, and c/EBP bind, leading to overlapping specificities and affinities (16–19). Our results show that the Sp1-binding site within the intron 1 is important for the negative regulation of DsbA-L. However, removing 9 bp within the Sp1-binding site did not completely recover luciferase reporter activity, suggesting that Sp1 may only be part of the transcriptional machinery that cooperatively inhibited DsbA-L gene expression. Other transcriptional factors or transcription components may also be involved in the regulation of DsbA-L. Further studies will be needed to address these questions.
DsbA-L is an evolutionarily conserved protein, and its transcript is widely expressed among various kinds of tissues (5). A recent study showed that DsbA-L is highly expressed in adipose tissues and the protein level increased dramatically during 3T3L-1 differentiation (1). Coincidentally, we found that DsbA-L transcript was greatly induced in the course of 3T3-L1 adipogenesis. Interestingly, the binding activity of Sp1 to the DsbA-L intronic sequence was reduced during 3T3-L1 adipocyte differentiation, which is in line with the increase in the mRNA level of DsbA-L during the differentiation process. Taken together, our results demonstrate that Sp1 functions as a transcription repressor of the DsbA-L gene in preadipocytes and reduced Sp1 expression and its binding capacity to the intron region of DsbA-L gene may account for the increased expression of DsbA-L in adipocytes. These results are in agreement with the findings that the cellular levels and/or the binding activity of Sp1 decreased after exposure of preadipocytes to the differentiation inducers (19,20). In accordance with the previously observation that HFD feeding significantly reduced the protein level of DsbA-L (1), Sp1 expression and its binding activity are increased in adipose tissues of diet-induced obese mice, suggesting that increased Sp1 levels and its action may provide a mechanism underlying obesity-induced downregulation of DsbA-L and thus adiponectin multimerization and stability.
In summary, our study has demonstrated that the proximal promoter of DsbA-L is located from −186 to −34 relative to TSS. The +391 to +432 region encompassing the Sp1-binding site is essential for the negative regulation of DsbA-L gene expression. Overexpression of DsbA-L has been shown to protect mice against diet-induced obesity and metabolic dysfunction (3). Identification of Sp1 as a negative regulator of DsbA-L gene expression should provide useful information on the development of therapeutic treatment for obesity and its associated metabolic disorders.
Article Information
Acknowledgments. The authors thank Dr. Jianping Ye (Louisiana State University) for providing the Sp1 expression construct. The authors thank Wei Ren (Shanghai Jiao Tong University Affiliated Sixth People’s Hospital) for discussion.
Funding. This work was supported by the National 973 project of China (2011CB504001) to W.J., Major Program of Shanghai Municipality for Basic Research (10JC1412400) to Q.F., National Institutes of Health RO1 grant DK76902 and National Nature Science Foundation of China (81130015) to F.L., and Young Scientists Fund of National Natural Science Foundation of China (81200292) to H.L.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Q.F. designed the research, analyzed data, and wrote the manuscript. W.Y., H.L., W.H., L.C., S.J., K.D., and Q.S. performed research. C.W. was involved in experiment design in the study of HFD-induced obese mice. S.C. contributed to discussion and reviewed the manuscript. F.L. was involved in study design and revised the manuscript. W.J. designed the study and interpreted data. W.J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Liu M, Zhou L, Xu A, et al. A disulfide-bond A oxidoreductase-like protein (DsbA-L) regulates adiponectin multimerization. Proc Natl Acad Sci U S A 2008;105:18302–18307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest 2006;116:1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M, Xiang R, Wilk SA, et al. Fat-specific DsbA-L overexpression promotes adiponectin multimerization and protects mice from diet-induced obesity and insulin resistance. Diabetes 2012;61:2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L, Liu M, Zhang J, Chen H, Dong LQ, Liu F. DsbA-L alleviates endoplasmic reticulum stress-induced adiponectin downregulation. Diabetes 2010;59:2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morel F, Rauch C, Petit E, et al. Gene and protein characterization of the human glutathione S-transferase kappa and evidence for a peroxisomal localization. J Biol Chem 2004;279:16246–16253 [DOI] [PubMed] [Google Scholar]
- 6.Jowsey IR, Thomson RE, Orton TC, Elcombe CR, Hayes JD. Biochemical and genetic characterization of a murine class Kappa glutathione S-transferase. Biochem J 2003;373:559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao F, Xuan Z, Liu L, Zhang MQ. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res 2005;33:D103–D107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YC, Illenye S, Heintz NH. Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol Cell Biol 2001;21:1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu S. Transcriptional regulation by post-transcriptional modification—role of phosphorylation in Sp1 transcriptional activity. Gene 2012;508:1–8 [DOI] [PubMed] [Google Scholar]
- 10.Wierstra I. Sp1: emerging roles—beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun 2008;372:1–13 [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Kang JK, Kim YK, et al. Histone deacetylase inhibitor apicidin induces cyclin E expression through Sp1 sites. Biochem Biophys Res Commun 2006;342:1168–1173 [DOI] [PubMed] [Google Scholar]
- 12.Doetzlhofer A, Rotheneder H, Lagger G, et al. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 1999;19:5504–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banchio C, Schang LM, Vance DE. Activation of CTP:phosphocholine cytidylyltransferase alpha expression during the S phase of the cell cycle is mediated by the transcription factor Sp1. J Biol Chem 2003;278:32457–32464 [DOI] [PubMed] [Google Scholar]
- 14.Roder K, Kim KH, Sul HS. Induction of murine H-rev107 gene expression by growth arrest and histone acetylation: involvement of an Sp1/Sp3-binding GC-box. Biochem Biophys Res Commun 2002;294:63–70 [DOI] [PubMed] [Google Scholar]
- 15.Song J, Ugai H, Kanazawa I, Sun K, Yokoyama KK. Independent repression of a GC-rich housekeeping gene by Sp1 and MAZ involves the same cis-elements. J Biol Chem 2001;276:19897–19904 [DOI] [PubMed] [Google Scholar]
- 16.Ebert SN, Wong DL. Differential activation of the rat phenylethanolamine N-methyltransferase gene by Sp1 and Egr-1. J Biol Chem 1995;270:17299–17305 [DOI] [PubMed] [Google Scholar]
- 17.Her S, Claycomb R, Tai TC, Wong DL. Regulation of the rat phenylethanolamine N-methyltransferase gene by transcription factors Sp1 and MAZ. Mol Pharmacol 2003;64:1180–1188 [DOI] [PubMed] [Google Scholar]
- 18.Dong XP, Pfister H. Overlapping YY1- and aberrant SP1-binding sites proximal to the early promoter of human papillomavirus type 16. J Gen Virol 1999;80:2097–2101 [DOI] [PubMed] [Google Scholar]
- 19.Tang QQ, Jiang MS, Lane MD. Repressive effect of Sp1 on the C/EBPalpha gene promoter: role in adipocyte differentiation. Mol Cell Biol 1999;19:4855–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole LK, Vance DE. A role for Sp1 in transcriptional regulation of phosphatidylethanolamine N-methyltransferase in liver and 3T3-L1 adipocytes. J Biol Chem 2010;285:11880–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]