Abstract
Insulin resistance is central to diabetes and metabolic syndrome. To define the consequences of genetic insulin resistance distinct from those secondary to cellular differentiation or in vivo regulation, we generated induced pluripotent stem cells (iPSCs) from individuals with insulin receptor mutations and age-appropriate control subjects and studied insulin signaling and gene expression compared with the fibroblasts from which they were derived. iPSCs from patients with genetic insulin resistance exhibited altered insulin signaling, paralleling that seen in the original fibroblasts. Insulin-stimulated expression of immediate early genes and proliferation were also potently reduced in insulin resistant iPSCs. Global gene expression analysis revealed marked differences in both insulin-resistant iPSCs and corresponding fibroblasts compared with control iPSCs and fibroblasts. Patterns of gene expression in patients with genetic insulin resistance were particularly distinct in the two cell types, indicating dependence on not only receptor activity but also the cellular context of the mutant insulin receptor. Thus, iPSCs provide a novel approach to define effects of genetically determined insulin resistance. This study demonstrates that effects of insulin resistance on gene expression are modified by cellular context and differentiation state. Moreover, altered insulin receptor signaling and insulin resistance can modify proliferation and function of pluripotent stem cell populations.
Introduction
Induced pluripotent stem cells (iPSCs) are a unique tool for studying human disease (1–3). iPSCs can be derived from multiple cell types and differentiated into all three germ layer–derived tissues, thus providing an opportunity to develop patient- and tissue-specific models for molecular analysis (4,5). iPSCs and their differentiated derivatives also provide a means to dissect gene–environment interactions central to complex human diseases, such as type 2 diabetes (T2D).
Insulin resistance is a key feature of T2D, obesity, and metabolic syndrome. Longitudinal studies indicate that insulin resistance is heritable and occurs in individuals at risk for T2D many years before glucose intolerance (6,7). Both genetic and environmental factors, including overnutrition and inactivity, can contribute to insulin resistance in individuals at risk for T2D. However, the precise molecular mechanisms underlying insulin resistance and the extent to which genes versus environment determines risk remain unknown.
Rare inherited syndromes of severe insulin resistance due to mutations in the insulin receptor (INSR), such as Donohue syndrome and type A insulin resistance (8–10), have provided important insights into insulin action and insulin resistance. Studies in fibroblasts and transformed lymphocytes from individuals with these conditions have demonstrated altered INSR signaling and provided key information about receptor structure and function (11–15). In general, clinical manifestations and signaling defects are more severe in Donohue syndrome as a result of the homozygous or compound heterozygous mutations compared with the heterozygous mutations in type A insulin resistance (16). However, because patients with these mutations are seriously ill, studying insulin action in classical target tissues, such as muscle, fat, or liver, is difficult. Thus far, in vitro studies have largely used skin fibroblasts or lymphocytes, limiting the generalizability of these findings to more-relevant metabolic tissues.
To define the impact of genetically determined insulin resistance in pluripotent cells, we generated iPSCs from fibroblasts of three patients with INSR mutations and three healthy controls of similar age. Both receptor and postreceptor signaling are similarly disrupted in both iPSCs and fibroblasts. Insulin receptor mutations also alter gene expression, but the nature of these changes depends on the cellular context. Thus, iPSCs are a powerful new tool in the study of insulin resistance that uncovers interactions between genetics and cellular environment in the pathogenesis of T2D.
Research Design and Methods
Animal protocols were approved by the Joslin Diabetes Center institutional animal use and care committee. Generation and usage of iPSCs was approved by the Joslin Committee on Human Studies.
Reagents
Primary antibodies included anti-phospho-IGF1R-β (Tyr1135/1136)/INSR-β (Tyr1150/1151), phospho-AKT (Ser473), AKT, phospho-ERK1/2 (Thr202/Tyr204), ERK1/2, and phospho-GSK3α/β (Ser21/9) (Cell Signaling); INSR-β and IGF1R-β (Santa Cruz); GSK3α/β (Millipore); phospho-insulin receptor substrate 1 (IRS1) (Y612) (Life Technologies); IRS1 (BD Biosciences); NANOG and OCT4 (Abcam); and SSEA4 and TRA-1-60 (Millipore).
Fibroblasts and iPSC Derivation
C1 fibroblasts (BJ) were from ATCC (Manassas, VA); C2 (GM05400), C3 (GM00409), IR-M2 (GM10277), and IR-M3 (GM20034) were from Coriell (Camden, NJ). IR-M1 (Minn1) and IR-M3 were previously characterized (9,17). Fibroblasts were reprogrammed with individual retroviral constructs (OCT4, KLF4, c-MYC, SOX2) and plated on irradiated mouse embryonic fibroblasts (1,2,5). Individual clones were analyzed for pluripotency and teratoma formation using standard techniques (1,2,5). Chromosomal analysis was performed on colcemid-arrested iPSCs at the Dana-Farber/Harvard Cancer Center Cytogenetics Core (Boston, MA) and Cell Line Genetics (Madison, WI).
Western Blot Analysis
Fibroblasts were serum starved overnight in DMEM (25 mmol/L glucose) and 0.1% BSA and then treated with 0 or 100 nmol/L insulin for 10 min and lysed in radioimmunoprecipitation assay buffer. iPSCs were starved in DMEM (25 mmol/L glucose) and 0.5% BSA for 3 h before stimulation with 0 or 100 nmol/L insulin. Protein concentration was determined by BCA Protein Assay (Thermo Scientific). Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween, incubated with primary antibodies overnight, and washed before incubation with horseradish peroxidase–conjugated secondary antibodies (18). Proteins were detected by enhanced chemiluminescence (PerkinElmer Life Sciences). Band intensities were quantified on scanned images using Adobe Photoshop on inverted images; bands were selected using the rectangular marquee tool, and histogram function was used to determine the average pixel intensity.
DNA Sequencing
Genomic DNA was isolated (DNeasy; Qiagen), and INSR exons were amplified by PCR (19) (GoTaq PCR; Promega) and sequenced (Molecular Biology Core, Dana-Farber Cancer).
Proliferation and EdU Incorporation
Ten thousand iPSCs were grown on 12-well plates in mTesR1 media. Cells were counted at 2-day intervals (Nexcelom). 5-Ethynil-2′-deoxyuridine (EdU) incorporation into DNA was measured using Alexa 488 Click-It HCS assay (Life Technologies) after incubation for 12–14 h with 100 nmol/L insulin or 100 nmol/L IGF-I.
Expression Analysis
RNA was extracted using RiboZol (AMRESCO). Complementary RNA was prepared and hybridized to Affymetrix PrimeView microarrays. Data were normalized using Robust Multichip Average (20). Heat maps were created using GENE-E (GenePattern; Broad Institute). Ontology analysis was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) (21). Putative transcription factor binding sites were identified using hypergeometric enrichment (Molecular Signatures Database) (22). For PCR, cDNA was synthesized (High Capacity cDNA Reverse Transcription Kit; Life Technologies) and amplified (iTaq Universal SYBR Green Supermix [Bio-Rad], ABI 7900HT [Life Technologies]). GAPDH and 36B4 were used for normalization, using the 2-ΔΔCT method (5).
Statistical Analysis
Data are presented as mean ± SEM. Between-group differences were examined by Student t test (P values) and false discovery rate (q values). P < 0.05 and q < 0.1 were considered significant.
Results
Insulin-Resistant Human Fibroblasts
Skin fibroblasts were obtained from three healthy control subjects (BJ, GM05400, and GM00409 hereafter designated C1, C2, and C3) and three individuals with severe insulin resistance (IR-Mut) due to mutations in the insulin receptor gene (INSR) (Minn1, GM10277, and GM20034 hereafter designated IR-M1, IR-M2, and IR-M3). DNA sequencing confirmed INSR mutations in all three patients (Table 1). IR-M1 was a compound heterozygote, with a mutation in one allele generating a stop codon at amino acid 897 in the extracellular domain of the β-subunit, yielding a nonfunctional INSR. Previous studies indicated the presence of a second mutation in IR-M1 that results in a 95% reduction in INSR expression (23). DNA sequencing of IR-M2 identified a missense mutation in exon 1 (5C→G), leading to an alanine-to-glycine substitution at the second amino acid in the signal peptide, which would be expected to alter membrane trafficking. In IR-M3, sequencing revealed a homozygous mutation in exon 3 (T→C, codon 233), resulting in a leucine-to-proline mutation and impaired INSR trafficking and autophosphorylation (17).
Table 1.
Summary of fibroblasts, including donor age and sex, mutation analysis, and functional domain of INSR mutations
| Cell line | Age | Sex | Mutation | Domain |
|---|---|---|---|---|
| C1 (BJ) | Neonatal | M | No mutation | — |
| C2 (GM05400) | 6 years | M | No mutation | — |
| C3 (GM00409) | 7 years | M | No mutation | — |
| IR-M1 (Minn1) | 1 month | F | Arg897→stop (one allele) Unknown mutation (second allele) Exon 14 | β-Subunit (one allele) Decreased expression (second allele) |
| IR-M2 (GM10277) | 15 years | F | Ala2→Gly (both alleles) Exon 1 | α-Subunit Signal sequence L1 domain |
| IR-M3 (GM20034) | 3 months | M | Leu233→Pro (both alleles) Exon 3 | α-Subunit L2 domain |
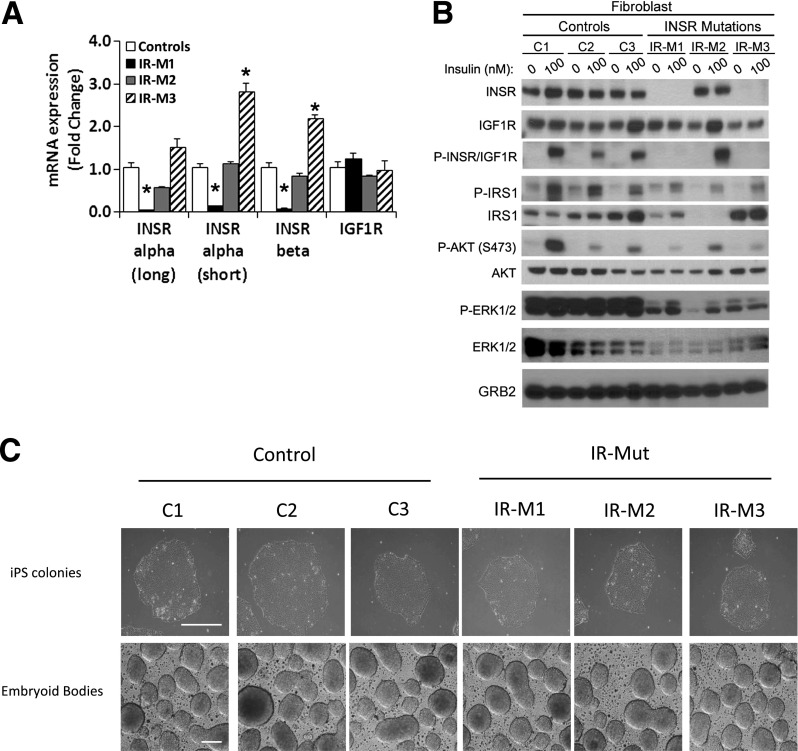
INSR expression and downstream signaling were reduced in fibroblasts from IR-Mut patients. Quantitative PCR (qPCR) with primers specific for the A (−exon 11, short) and B (+exon 11, long) isoforms of the α- and β-subunits of INSR revealed a >85% decrease in expression in IR-M1 cells (P < 0.05) (Fig. 1A), consistent with prior studies (23,24). INSR expression in IR-M2 fibroblasts did not differ from controls, whereas IR-M3 fibroblasts had a twofold increase in INSR mRNA (Fig. 1A). IGF1R mRNA did not differ between control and any IR-Mut fibroblasts (Fig. 1A, right).
Figure 1.
Insulin signaling in fibroblast cell lines and generation of iPSCs. A: Gene expression analysis of INSR α- and β-subunits and IGF1R expressed relative to the average of controls (n = 3). B: Fibroblasts were serum starved overnight before 10-min stimulation with 100 nmol/L insulin. Western blot analysis of insulin signaling in control and IR-Mut fibroblasts are shown. Specific antibodies are indicated adjacent to the respective image. Images are representative of two independent experiments. C: Bright-field images of control and patient iPSC colonies and embryoid bodies (scale bar = 100 μm). Data are mean ± SEM. *P < 0.05 vs. controls.
Consistent with previous studies (23,24) and mRNA expression, INSR protein was decreased by 95% in IR-M1 fibroblasts. In IR-M2, expression did not differ from controls (Fig. 1B, Supplementary Fig. 1A). Mature INSR protein was also decreased by 88% in IR-M3, despite increased INSR mRNA, consistent with impaired proreceptor processing (17). By contrast, IGF1R protein was unchanged in IR-Mut cells (Fig. 1B, Supplementary Fig. 1B).
Western blotting with an antibody that recognizes both the phosphorylated INSR and IGF1R (P-INSR/IGF1R) revealed a robust eightfold increase in insulin-stimulated phosphorylation in control fibroblasts (Fig. 1B, left, third row). In contrast, P-INSR/IGF1R was undetectable in IR-M1 and IR-M3 fibroblasts, but similar to control in IR-M2 (Fig. 1B, right; Supplementary Fig. 1C). Nonetheless, insulin-stimulated phosphorylation of IRS1 was reduced by 30–40% in all three IR-Mut fibroblasts. In IR-M2 cells, this was at least partly due to a marked decrease in IRS1 protein. Insulin-stimulated phosphorylation of AKT was also decreased in all IR-Mut fibroblasts by ∼70%, with no change in AKT protein or the loading control GRB2. Basal and insulin-stimulated phosphorylation of ERK1/2 was decreased by >50% in all IR-Mut.
Creation of iPSC Lines and Analysis of Insulin Signaling
To isolate the impact of genetic insulin resistance from the effects of cellular differentiation, we derived iPSCs from control and IR-Mut fibroblasts by introduction of OCT4, KLF4, SOX2, and c-MYC (1) (Fig. 1C). When cultured on low-adherence plates, these cells formed embryoid bodies (Fig. 1C, bottom). For each iPSC line, we confirmed pluripotency using three independent criteria: 1) expression of pluripotency factors OCT4, TRA1-60, NANOG, and SSEA4 (Supplementary Fig. 2A); 2) alkaline phosphatase staining (Supplementary Fig. 2B); and 3) in vivo teratoma formation with histological identification of ectodermal, mesodermal, and endodermal layers (Supplementary Fig. 3A). Additionally, all iPSCs had normal karyotype (Supplementary Fig. 3B) and no difference in reprogramming efficiency.
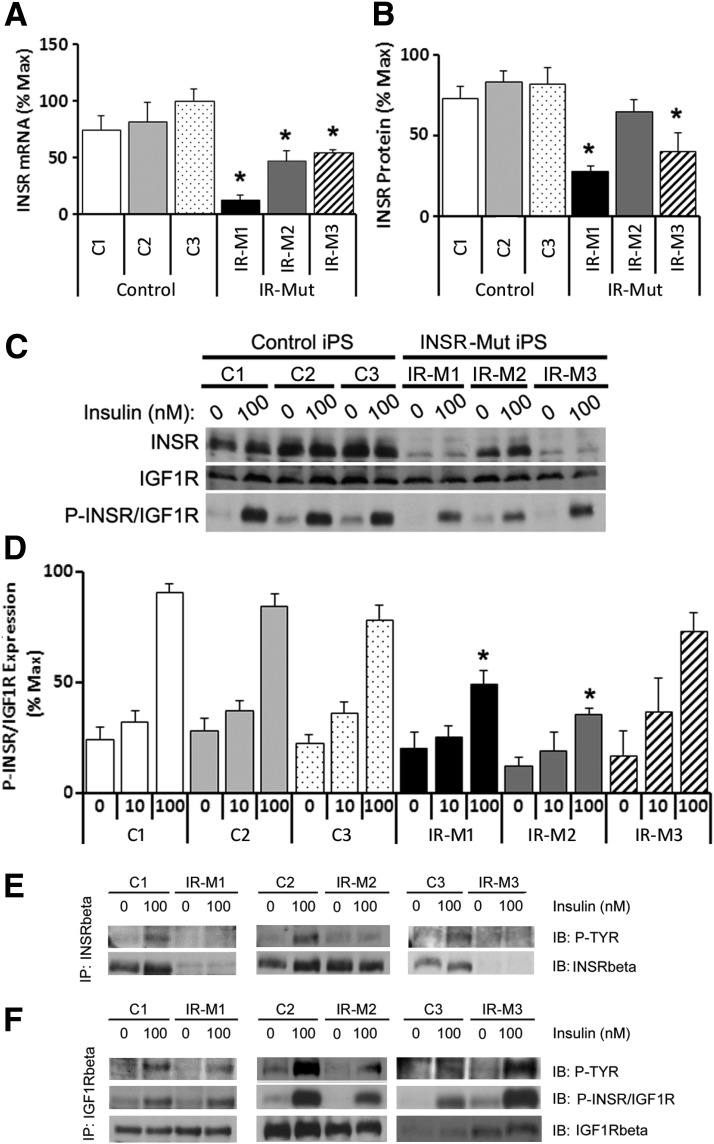
INSR mRNA was decreased by 95% in IR-M1 and by 60% in IR-M2 and IR-M3 iPSCs (Fig. 2A). Mature INSR protein was also decreased in IR-M1 and IR-M3 iPSCs by 73% and 59%, respectively, but unchanged in IR-M2 (Fig. 2B and C), similar to the pattern in fibroblasts. In IR-M3 cells, there was also a sixfold increase in proreceptor expression (Supplementary Fig. 3A and B), consistent with the known impairment in proreceptor processing (17).
Figure 2.
Expression and phosphorylation of INSR in iPSCs. A: mRNA expression of INSR by qRT-PCR, expressed as percent of maximum value (n = 3 experiments). B: Quantification of INSR protein level from Western blot analysis, expressed as percentage of maximum value (n = 3). C: iPSCs were serum starved for 3 h before 10-min stimulation with 0, 10, or 100 nmol/L insulin. Representative Western blot analysis of INSR and IGF1R expression and phosphorylation are shown (n = 3). D: Quantification of Western blot analysis of P-INSR/IGF1R, represented as percent of maximum value (n = 3). E: iPSCs were serum starved for 3 h before 5-min stimulation with 0 or 100 nmol/L insulin. Immunoprecipitation (IP) of INSR using a β-subunit–specific anti-INSR antibody followed by immunoblotting (IB) with an antiphosphotyrosine antibody (top row) and anti-INSR antibody (bottom row) was performed. Representative blots are shown (n = 2). F: iPSCs were serum starved for 3 h before 5-min stimulation with 0 or 100 nmol/L insulin. IP of IGF1R using a β-subunit–specific anti-IGF1R antibody and IB with an antiphosphotyrosine antibody (top row), anti-P-INSR/IGF1R (middle row), and anti-IGF1R antibody (bottom row) was performed. Representative blots are shown (n = 2). For panels A, B, and D, data are mean ± SEM. *P < 0.05 vs. controls.
There were no differences in IGF1R protein (Fig. 2C, Supplementary Fig. 3C) or mRNA (Supplementary Fig. 3D). Western blotting using P-INSR/IGF1R antibody demonstrated robust insulin stimulation in controls (Fig. 2C and D, right). By contrast, insulin-stimulated P-INSR/IGF1R was decreased by 50% in IR-M1 and IR-M2 but unchanged in IR-M3 iPSCs (Fig. 2C, right, and D).
Given that the anti-P-INSR/IGF1R antibody represents a composite of phosphorylation and abundance of both INSR and IGF1R, these receptors were individually immunoprecipitated and then blotted with anti-phosphotyrosine antibody. All three control iPSCs showed increased tyrosine phosphorylation of INSR upon insulin stimulation (Fig. 2E), whereas negligible phosphorylation was observed in the three IR-Mut iPSC lines. Anti-INSR Western blots of these anti-INSR immunoprecipitates revealed reduced mature INSR expression in IR-M1 and IR-M3, with normal INSR protein in IR-M2 (Fig. 2E, bottom row). Furthermore, anti-INSR immunoprecipitation showed negligible insulin-stimulated phosphorylation of the proreceptor (Supplementary Fig. 4E). Immunoprecipitation of IGF1R revealed similar levels of IGF1R protein in all lines (Fig. 2F, bottom row) and robust insulin-stimulated phosphorylation of IGF1R in both control and IR-M1 and IR-M3 iPSCs (Fig. 2F). Of note, basal and insulin-stimulated phosphorylation of IGF1R was reduced ∼20% in IR-M2 (Fig. 2F), suggesting an effect of the mutant INSR to inhibit IGF1R.
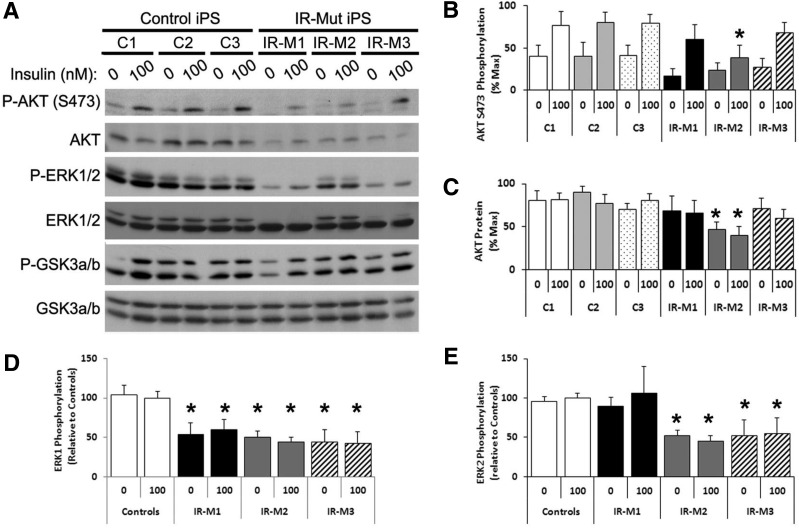
Western blotting demonstrated that insulin-stimulated phosphorylation of AKT (Ser473) was reduced in all IR-Mut iPSCs, reaching significance in IR-M2 (Fig. 3A and B) in parallel with an ∼60% reduction in AKT protein (P < 0.05) Fig. 3A and C). Likewise, both basal and insulin-stimulated phosphorylation of ERK1/2 was decreased in insulin-resistant cells by 40–60% (Fig. 3A, D, and E). Protein expression of ERK1 (but not ERK2) was also significantly decreased in IR-M1 and IR-M3, with a similar trend in IR-M2 (Supplementary Fig. 5A and B). These changes in protein expression were present despite unaltered mRNA expression of these molecules (Supplementary Fig. 4D).
Figure 3.
Regulation of downstream insulin signaling in iPSCs. A: iPSCs were serum starved for 3 h before 10-min stimulation with 0 or 100 nmol/L insulin. Western blot analysis was performed for key insulin signaling pathway proteins, as indicated. Representative blots are shown (n = 3). Quantitation of Western blot analysis for AKT Ser473 phosphorylation (B) and expression (C) shown as percent of maximum value (n = 3). D and E: Quantification of ERK1 and ERK2 phosphorylation expressed relative to the average of controls (n = 3). Data are mean ± SEM. *P < 0.05 vs. controls.
Proliferation and Expression of Early Response Genes Are Impaired in iPSCs From Individuals With Severe Insulin Resistance
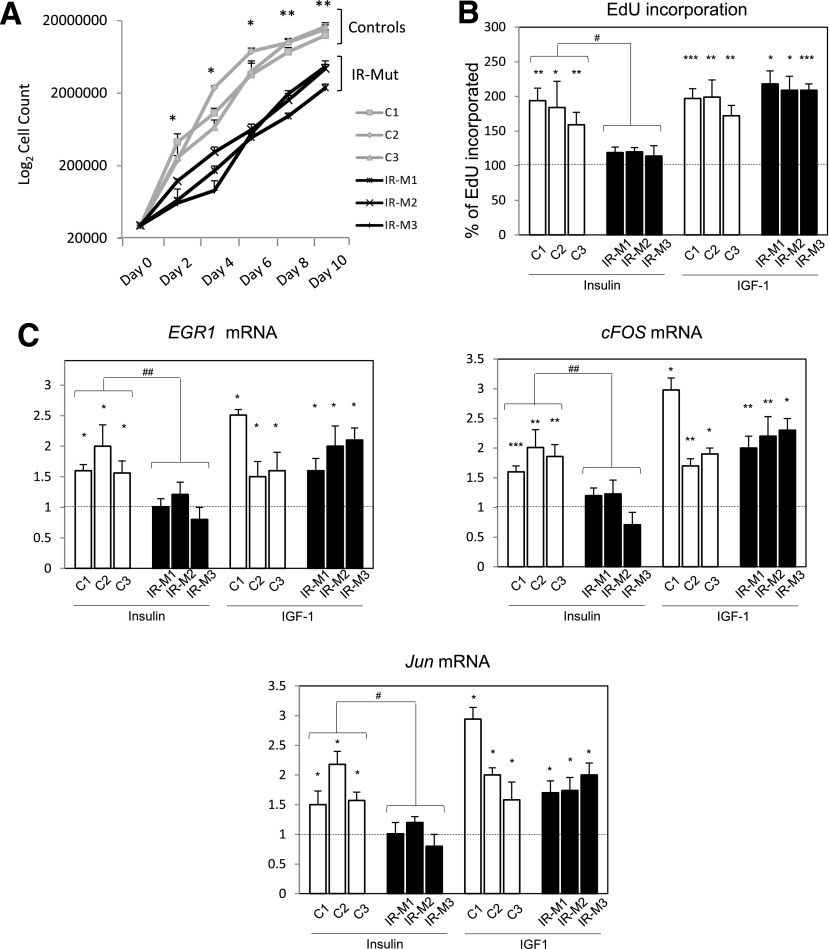
Insulin exerts potent effects on cell proliferation and transcriptional regulation (25–27). All IR-Mut iPSCs had significant reductions in growth (Fig. 4A) and reduced insulin-stimulated incorporation of EdU into DNA compared with controls (Fig. 4B, left). These differences were not due to reduced cell attachment (Supplementary Fig. 6) but were associated with dysregulation of mitogenic gene expression. Indeed, control iPSCs responded to insulin with 1.7–1.8-fold increases in expression of EGR1, cFOS, and JUN (P < 0.05) (Fig. 4C, left). By contrast, these insulin-stimulated transcriptional responses were nearly absent in IR-Mut (Fig. 4C). IGF-I–stimulated EdU incorporation and expression of mitogenic genes were robust and similar in both control and IR-Mut (Fig. 4B and C, right).
Figure 4.
Mitogenesis of iPSCs. A: Proliferation in iPSC lines was assessed every 2 days over 10 days by cell counting. Data are the mean of triplicates for each of the three control and three IR-Mut lines; all data are mean ± SEM. *P < 0.05, **P < 0.01 vs. controls (n = 2). B: EdU incorporation in iPSC lines after insulin and IGF-1 stimulation (100 nmol/L). Bars represent percentage of incorporated EdU compared with unstimulated iPSC (dash line). Data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. unstimulated; #P < 0.05 vs. controls. C: qRT-PCR analysis for insulin-stimulated EGR1, cFOS, and JUN mRNA expression relative to unstimulated cells (dash line). Data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. unstimulated; #P < 0.05; ##P < 0.01 vs. controls.
Global Gene Expression Is Altered in Insulin-Resistant iPSCs
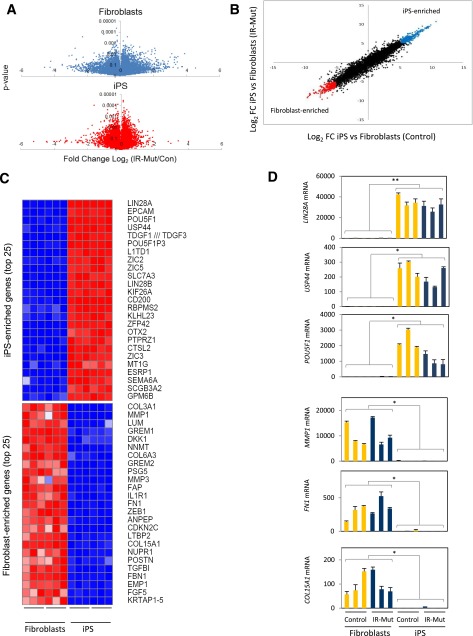
To investigate the impact of genetic insulin resistance on transcriptional regulation in iPSCs, we analyzed global gene expression using microarrays in both fibroblasts and iPSCs. All cells were studied at confluence in serum-free medium without added insulin or growth factors. Volcano plots (Fig. 5A) demonstrate the distribution of gene expression differences between IR-Mut and control (x-axis) for fibroblasts and iPSCs plotted against corresponding P values (y-axis). In general, gene expression differences were less variable in iPSCs, as demonstrated by 290 probes with P < 0.01 vs. 87 in fibroblasts.
Figure 5.
Microarray analysis of fibroblast and iPSC lines. A: Volcano plots showing distribution of differential expression in fibroblasts (top) and iPSCs (bottom), with ratio of IR-Mut/control on x-axis and P values on y-axis. B: Comparison of gene expression (iPSCs/fibroblasts ratio) for the IR-Mut (y-axis) and control (x-axis) lines. Colored dots at each end of the distribution mark genes with an expression ratio greater than or less than fivefold (log2 scale). C: Heat maps of expression differences for the top 25 genes in either iPSC- or fibroblast-enriched groups (log2 fold change [FC] >5 or <5), normalized by row. D: Expression of LIN28A, USP44, POU5F1, MMP1, FN1, and COL15A1 was analyzed by qRT-PCR, using GAPDH as the housekeeping gene. *P < 0.05, **P < 0.01 average of iPSCs vs. fibroblasts.
We next compared gene expression in iPSCs and their paired fibroblasts for both control (x-axis) and IR-Mut cells (y-axis) (Fig. 5B). Probes for which expression is increased in iPSCs by >32-fold (5-fold on log2 scale) are shown in blue, whereas downregulated genes are shown in red. The top 25 genes within these two groups are shown in Fig. 5C (iPS enriched, top; fibroblast enriched, bottom). Differential expression of representative genes was confirmed by qPCR (Fig. 5D).
Analysis of transcripts overexpressed in both control and IR-Mut iPSCs versus paired fibroblasts (Fig. 5B, top right) revealed mRNAs that encode proteins involved in pluripotency, self-renewal, and development, including LIN28A, OCT4, and NANOG (Fig. 5C and D). Conversely, transcripts enriched in both control and IR-Mut fibroblasts (Fig. 5B, bottom left) represent genes typically highly expressed in fibroblasts (e.g., related to extracellular matrix, fibroblast growth factors, and metalloproteases) (Fig. 5C and D). These patterns were confirmed using DAVID ontology analysis (21) (Supplementary Table 1).
Transcription factor binding motif analysis (using 2-kb upstream sequences) of genes within both fibroblast- and iPSC-enriched groups (22) demonstrated enrichment of binding site motifs for several transcription factors related to insulin action and iPSC physiology (Supplementary Table 2). Of note, forkhead box protein O (FOXO4), lymphoid enhancer-binding factor-1 (LEF1), and NFATc were common to the two gene groups, consistent with their important role in both somatic and iPSC specification.
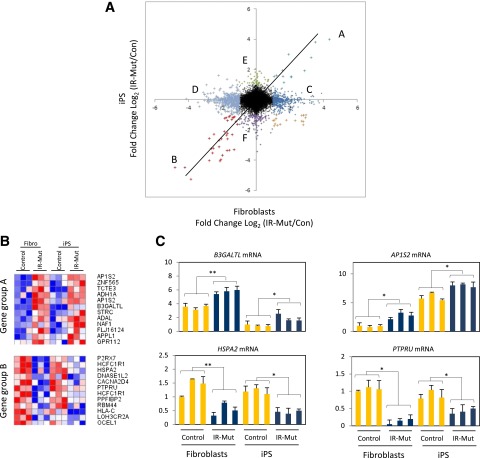
To assess the impact of INSR mutations on expression patterns in both cell types, we plotted the ratio of gene expression in IR-Mut to control in fibroblasts (x-axis) versus the corresponding ratio in iPSCs (y-axis) (Fig. 6A), with colored points indicating expression differences greater than or equal to twofold. mRNAs along the diagonal represent genes regulated by insulin resistance similarly in fibroblasts and iPSCs (i.e., independent of cellular context). Of note, only 46 genes met these criteria, with 12 upregulated in both IR-Mut fibroblasts and iPSCs (group A) and 34 downregulated in both IR-Mut cell types (group B). Thus, groups A and B represent genes regulated by insulin resistance in fibroblasts and iPSCs independent of differentiation state (heat maps [Fig. 6B]). We validated selected genes using quantitative RT-PCR (qRT-PCR) (Fig. 6C). For example, insulin signaling–related gene adapter protein 1 sigma 2 (AP1S2) is upregulated in IR-Mut fibroblasts and iPSCs, whereas receptor-type tyrosine-protein phosphatase PCP-2 (PTPRU) is downregulated. Transcription factor binding motif analysis for the top 100 genes (P < 0.05) with similar expression patterns to those in groups A and B revealed an enrichment of binding sites for transcription factors involved in glucose homeostasis and/or insulin action, such as E2F1, FOXF2, SP1, JUN, and CREB1 (Supplementary Table 3).
Figure 6.
Gene expression analysis of gene groups A and B. A: Dot plot shows distribution of the ratio of gene expression (IR-Mut/control) in iPSCs (y-axis) vs. that in fibroblasts (x-axis). Letters indicate gene groups with expression ratio greater than twofold (log2 fold change [FC] >1 or <1). B: Heat maps of top 12 significant genes within gene groups A and B normalized by row (P < 0.05). C: Expression of B3GALTL, AP1S2, HSPA2, and PTPRU was measured by qRT-PCR, with GAPDH as the housekeeping gene. *P < 0.05, **P < 0.01 average of controls vs. mutants.
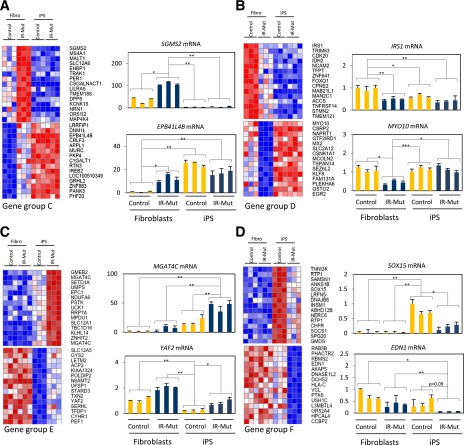
In contrast to the modest number of genes coordinately regulated by insulin resistance in both fibroblasts and iPSCs, the majority of insulin resistance–regulated genes were differentially expressed in only fibroblasts (Fig. 6A, groups C and D) or iPSCs (groups E and F). Indeed, many mRNAs showed robust differences in expression as a function of insulin resistance in fibroblasts (groups C and D) but were unaltered in iPSCs or vice versa (groups E and F).
To clarify potential regulatory and functional differences between subgroups of genes altered in response to insulin resistance in a cell context–dependent manner, we compared those genes altered in fibroblasts but unchanged in iPSCs (groups C and D, 294 and 262 mRNAs, respectively) with genes altered in iPSCs but unchanged in fibroblasts (groups E and F, 44 and 85 mRNAs, respectively). For each group, the top 30 differentially expressed mRNAs (all P < 0.05, false discovery rate <0.1) are presented as heat maps in Fig. 7, with qPCR validation of representative genes in adjacent graphs. Group C was upregulated by insulin resistance in fibroblasts but unaltered in iPSCs. This pattern was often linked to either constitutively low expression (e.g., SGMS2) or high expression (e.g., EPB41L4B) in iPSCs. Likewise, group D was downregulated by insulin resistance in fibroblasts but unregulated in iPSCs (e.g., IRS1, MYO10). Ontology analysis of genes regulated by insulin resistance specifically in fibroblasts (groups C and D) revealed genes involved in development (homeobox, cell proliferation, development), signaling (IGF, FGF2R, WNT), and fibroblast function (extracellular matrix) (Supplementary Table 4). Ontology analysis of genes regulated by insulin resistance in iPSCs (groups E and F) also revealed enrichment of genes in IGF, FGF2R, and WNT signaling pathways.
Figure 7.
Gene expression analysis of gene groups C–F. A–D: Heat maps and mRNA expression of gene groups C–F. Heat maps represent the top 30 significant genes of gene groups C–F normalized by row (P < 0.05, q < 0.1), with qRT-PCR analysis in adjacent graphs. *P < 0.05, **P < 0.01, ***P < 0.001 average of controls vs. mutants.
Transcription factor binding motifs enriched in promoters of insulin resistance–regulated genes revealed several with regulatory roles in development (e.g., MYC-associated zinc finger protein [MAZ], paired box gene 4 [PAX4], paired-like homeodomain transcription factor 2 [PITX2]) or insulin action (e.g., FOXO4, LEF1, myocyte enhancer factor 2A [MEF2A]) (Supplementary Table 5). Of note, several motifs were identified in both fibroblast and iPSC-specific groups (FOXO4, NFAT/NFATc, TAF, REPIN1, MYOD1, and MEF2A), suggesting the potential for insulin resistance to confer unique transcriptional regulation (either suppression or activation), depending on the cell context or differentiation state.
Discussion
Insulin resistance is central to the pathogenesis of T2D, obesity, and metabolic syndrome. Insulin resistance precedes and predicts the onset of T2D and is even present in offspring of individuals with T2D, indicating a hereditary component (6,7). Determining the contributions of genetic factors to insulin resistance in humans has been challenging given the limited access to relevant tissues, especially during preclinical stages of disease. Although there has been some success in studying human insulin resistance using cultured myoblasts (28–30), the majority of cellular studies have used fibroblasts (24,31) or circulating blood cells (32,33), which show little or no metabolic response to insulin. iPSCs are a new tool for studying human disease in that they can be derived from a variety of cells, passaged indefinitely in culture, and studied either in the pluripotent state or after differentiation into muscle, fat, or other tissues (1–5,34).
To explore the utility of iPSCs in the study of insulin resistance, we have created and characterized iPSCs from patients with severe insulin resistance due to mutations in the INSR and compared these with the fibroblasts from which they were derived. These IR-Mut iPSCs exhibit major defects in signaling, proliferation, and gene expression but exhibit important differences from the fibroblasts of the same individuals, indicating that cellular context is a potent modifier, even in genetically determined insulin resistance.
Each patient had a distinct INSR mutation, resulting in different defects in iPSCs. IR-M1 is a compound heterozygote in which a nonsense mutation in one allele produces a truncated receptor lacking the tyrosine kinase, whereas the other allele reduces INSR transcription. This receptor cannot signal but can bind insulin and form hybrids with normal insulin/IGF-I receptors (24). IR-M2 has a mutation in the INSR signal sequence, which impairs membrane trafficking. Of note, we observed more severe reductions in receptor expression and phosphorylation in IR-M2 iPSCs than in fibroblasts. This is similar to the previously reported Asp15Lys mutation in this region in which defects in insulin binding differed between fibroblasts and lymphoblasts (35). IR-M3 has a homozygous missense mutation in the α-subunit that results in impaired proreceptor processing and membrane transport (17). Consistent with this, we observed accumulation of the proreceptor in IR-M3 iPSCs, although it was not phosphorylated upon insulin stimulation, suggesting that it may not have been normally transported to the plasma membrane. Of note for this patient, levels of INSR mRNA were reduced in iPSCs but increased in fibroblasts, consistent with cell-specific INSR expression patterns previously observed (35) and indicating differentiation dependence.
Despite differing mutations, insulin-stimulated phosphorylation of INSR was significantly reduced in all IR-Mut iPSCs. However, insulin was able to mediate some downstream signaling, in part through IGF1R phosphorylation, which was similar in IR-M1 cells and increased in IR-M3 cells. In IR-M2 cells, IGF1R phosphorylation was reduced, suggesting that the mutated INSR functions in a dominant-negative manner to modulate IGF1R signaling (36), potentially contributing to more severe downstream signaling defects. The importance of IGF1R signaling to compensate for reduced INSR activity in IR-Mut iPSCs also supports a greater reliance of iPSCs on IGF1R-mediated signaling.
Different INSR mutations also cause both cell type– and pathway-specific impairments in downstream signaling. Phosphorylation of AKT, the kinase linked to many metabolic actions of insulin, was reduced in all three IR-Mut fibroblast lines. In iPSCs, AKT phosphorylation was significantly impaired only in IR-M2, with similar trends in the other two lines. Phosphorylation of GSK3 was unaffected by INSR mutations, suggesting compensation from other signaling pathways. By contrast, ERK expression and basal phosphorylation were altered in both IR-Mut iPSCs and fibroblasts, with more dramatic decreases in ERK1 than in ERK2. Although ERK1 and ERK2 are often considered functionally redundant, isoform-specific and differentiation-dependent differences have been observed, including preferential roles of ERK2 in myogenic differentiation (37) and regulation of epithelial-to-mesenchymal transitions (38). Differential regulation of ERK signaling in iPSCs is interesting because ERK regulates transcription, stem cell proliferation, and differentiation (39,40). Together these data indicate that INSR mutations can selectively perturb specific downstream components of the insulin signaling pathway and that the cellular context adds a further layer of regulation by modulating expression and function of insulin signaling molecules.
Given that insulin is a potent regulator of transcription (27), we assessed the impact of this genetic insulin resistance by analyzing gene expression in control and IR-Mut fibroblasts and iPSCs. Not surprisingly, expression patterns differ markedly in fibroblasts versus iPSCs. Fibroblasts strongly express genes related to extracellular matrix synthesis and wound healing, whereas iPSCs express pluripotency-related genes, such as OCT4, NANOG, ZIC, and the micro-RNA–binding protein LIN28 (41). LIN28 itself modulates insulin action by inhibiting microRNA let-7, which represses multiple components of the insulin/PI 3-kinase/mTOR pathway (42).
Insulin resistance imparted robust effects on gene expression. A small subset of genes was concordantly highly regulated by insulin resistance in both fibroblasts and iPSCs (groups A and B). These genes were predominantly related to cell growth and signaling, including IGF1 and 2, IGFBPs, EGFR, bFGFR, BMP2, and SERPINE1. However, most effects on gene expression were cell-type specific, with major dysregulation of expression present either only in IR-Mut fibroblasts (groups C and D) or only in iPSCs (groups E and F). In many cases, loss of regulation in either fibroblasts or iPSCs appears to be linked to constitutively high or low expression of the regulated gene, which in turn may be linked to cellular differentiation state or epigenetic regulation of transcription. Analysis suggested that insulin resistance may affect regulation of developmental pathways (HOX, MEIS), potentially affecting the capacity of iPSCs to differentiate along specific developmental lineages (43–45).
At a functional level, insulin resistance resulted in a significant reduction in iPSC proliferation and insulin-stimulated DNA synthesis. Reduced proliferation is particularly important in iPSCs because self-renewal is a critical and defining characteristic of stem cells, and altered growth rates could have a major impact on differentiation (46). Impairment in insulin signaling through the ERK pathway and reductions in insulin-stimulated expression of the early growth response genes are likely contributors to reduced proliferation. Importantly, these defects were specifically due to defective insulin signaling because mutant lines remained responsive to IGF-1 stimulation. Thus, these data underscore the importance of insulin signaling for proliferation of stem cells. Although PI 3-kinase has been shown to be important for pluripotency in iPSCs (46), we did not observe significant differences in markers of pluripotency, despite reduced insulin action within this pathway.
Collectively, the current data support the hypothesis that insulin resistance in stem cells may be a novel mechanism contributing to diabetes pathogenesis. Reductions in insulin signaling could decrease the size of stem cell populations, disrupt developmental trajectories, alter differentiation, and potentially reduce tissue-resident stem cell numbers, thus reducing regenerative responses to injury during adult life (47). Consistent with this hypothesis, we have previously demonstrated that nutritional signals can reduce the size and functional capacity of stem cell pools in mice at risk for diabetes (48). Going forward, iPSCs will be an important tool in dissecting pathways contributing to diabetes by leveraging their potential capacity to differentiate into tissue-specific insulin resistant cell lines.
In summary, iPSCs generated from fibroblasts from patients with severe insulin resistance have defects in signaling, gene expression, and proliferation. Thus, genetically determined insulin resistance is an important modifier of stem cell function and could contribute to disease pathogenesis. iPSCs are also a potent tool for studying the impact of cellular context on insulin resistance and dissecting the contributions of genetics and differentiation-dependent effects of insulin resistance to disease risk.
Supplementary Material
Article Information
Acknowledgments. The authors thank C. Cowan and L. Daheron of the Harvard Stem Cell Institute iPS Core Facility for valuable support and assistance with generation of human iPSCs.
Funding. A.M.B. was supported by grant T32-DK-007260 and the American Diabetes Association mentor-based fellowship (to M.-E.P.). This work was supported by a Joslin Diabetes Center Diabetes and Research Center grant (P30-DK-036836), funding from the Novo Nordisk Foundation (to M.-E.P. and C.R.K.), National Institutes of Health grant R01-DK-31036 (to C.R.K.), and pilot funding from the Harvard Stem Cell Institute.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. S.I. and A.M.B. designed, researched, and analyzed data and wrote the manuscript. K.K., L.W., K.J.H., and M.M. researched data. Y.-K.L. provided materials. M.-E.P. and C.R.K. oversaw the project and contributed to the discussion and writing of the manuscript. C.R.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-0109/-/DC1.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872 [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 3.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature 2012;481:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahfeldt T, Schinzel RT, Lee YK, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol 2012;14:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu C, Tabebordbar M, Iovino S, et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 2013;155:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin BC, Warram JH, Rosner B, Rich SS, Soeldner JS, Krolewski AS. Familial clustering of insulin sensitivity. Diabetes 1992;41:850–854 [DOI] [PubMed] [Google Scholar]
- 7.Shulman RG. Nuclear magnetic resonance studies of glucose metabolism in non-insulin-dependent diabetes mellitus subjects. Mol Med 1996;2:533–540 [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn CR, Flier JS, Bar RS, et al. The syndromes of insulin resistance and acanthosis nigricans. Insulin-receptor disorders in man. N Engl J Med 1976;294:739–745 [DOI] [PubMed] [Google Scholar]
- 9.Taylor SI, Hedo JA, Underhill LH, Kasuga M, Elders MJ, Roth J. Extreme insulin resistance in association with abnormally high binding affinity of insulin receptors from a patient with leprechaunism: evidence for a defect intrinsic to the receptor. J Clin Endocrinol Metab 1982;55:1108–1113 [DOI] [PubMed] [Google Scholar]
- 10.Savage DB, Semple RK, Chatterjee VK, Wales JK, Ross RJ, O’Rahilly S. A clinical approach to severe insulin resistance. Endocr Dev 2007;11:122–132 [DOI] [PubMed] [Google Scholar]
- 11.Taylor SI, Cama A, Accili D, et al. Mutations in the insulin receptor gene. Endocr Rev 1992;13:566–595 [DOI] [PubMed] [Google Scholar]
- 12.Longo N, Wang Y, Smith SA, Langley SD, DiMeglio LA, Giannella-Neto D. Genotype-phenotype correlation in inherited severe insulin resistance. Hum Mol Genet 2002;11:1465–1475 [DOI] [PubMed] [Google Scholar]
- 13.Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocr Rev 2011;32:498–514 [DOI] [PubMed] [Google Scholar]
- 14.Kadowaki H, Kadowaki T, Cama A, et al. Mutagenesis of lysine 460 in the human insulin receptor. Effects upon receptor recycling and cooperative interactions among binding sites. J Biol Chem 1990;265:21285–21296 [PubMed] [Google Scholar]
- 15.Nobile S, Semple RK, Carnielli VP. A novel mutation of the insulin receptor gene in a preterm infant with Donohue syndrome and heart failure. J Pediatr Endocrinol Metab 2012;25:363–366 [DOI] [PubMed] [Google Scholar]
- 16.Elsas LJ, 2nd, Longo N, Langley S, Griffin LD, Shuster RC. Molecular genetics of severe insulin resistance. Yale J Biol Med 1989;62:533–547 [PMC free article] [PubMed] [Google Scholar]
- 17.Maassen JA, Van der Vorm ER, Van der Zon GC, Klinkhamer MP, Krans HM, Möller W. A leucine to proline mutation at position 233 in the insulin receptor inhibits cleavage of the proreceptor and transport to the cell surface. Biochemistry 1991;30:10778–10783 [DOI] [PubMed] [Google Scholar]
- 18.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 1979;76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple RK, Soos MA, Luan J, et al. Elevated plasma adiponectin in humans with genetically defective insulin receptors. J Clin Endocrinol Metab 2006;91:3219–3223 [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 2005;434:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadowaki T, Kadowaki H, Taylor SI. A nonsense mutation causing decreased levels of insulin receptor mRNA: detection by a simplified technique for direct sequencing of genomic DNA amplified by the polymerase chain reaction. Proc Natl Acad Sci U S A 1990;87:658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller-Wieland D, Taub R, Tewari DS, et al. Insulin-receptor gene and its expression in patients with insulin resistance. Diabetes 1989;38:31–38 [DOI] [PubMed] [Google Scholar]
- 25.Boulton TG, Nye SH, Robbins DJ, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 1991;65:663–675 [DOI] [PubMed] [Google Scholar]
- 26.Ueki K, Yamamoto-Honda R, Kaburagi Y, et al. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem 1998;273:5315–5322 [DOI] [PubMed] [Google Scholar]
- 27.O’Brien RM, Streeper RS, Ayala JE, Stadelmaier BT, Hornbuckle LA. Insulin-regulated gene expression. Biochem Soc Trans 2001;29:552–558 [DOI] [PubMed] [Google Scholar]
- 28.Bouzakri K, Koistinen HA, Zierath JR. Molecular mechanisms of skeletal muscle insulin resistance in type 2 diabetes. Curr Diabetes Rev 2005;1:167–174 [DOI] [PubMed] [Google Scholar]
- 29.Ciaraldi TP, Mudaliar S, Barzin A, et al. Skeletal muscle GLUT1 transporter protein expression and basal leg glucose uptake are reduced in type 2 diabetes. J Clin Endocrinol Metab 2005;90:352–358 [DOI] [PubMed] [Google Scholar]
- 30.Jiang LQ, Duque-Guimaraes DE, Machado UF, Zierath JR, Krook A. Altered response of skeletal muscle to IL-6 in type 2 diabetic patients. Diabetes 2013;62:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longo N, Langley SD, Griffin LD, Elsas LJ, 2nd. Reduced mRNA and a nonsense mutation in the insulin-receptor gene produce heritable severe insulin resistance. Am J Hum Genet 1992;50:998–1007 [PMC free article] [PubMed] [Google Scholar]
- 32.Kadowaki T, Kadowaki H, Accili D, Taylor SI. Substitution of lysine for asparagine at position 15 in the α-subunit of the human insulin receptor. A mutation that impairs transport of receptors to the cell surface and decreases the affinity of insulin binding. J Biol Chem 1990;265:19143–19150 [PubMed] [Google Scholar]
- 33.Twig G, Afek A, Shamiss A, et al. White blood cells count and incidence of type 2 diabetes in young men. Diabetes Care 2013;36:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua H, Shang L, Martinez H, et al. iPSC-derived β cells model diabetes due to glucokinase deficiency. J Clin Invest 2013;123:3146–3153 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Taylor SI, Underhill LH, Hedo JA, Roth J, Rios MS, Blizzard RM. Decreased insulin binding to cultured cells from a patient with the Rabson-Mendenhall syndrome: dichotomy between studies with cultured lymphocytes and cultured fibroblasts. J Clin Endocrinol Metab 1983;56:856–861 [DOI] [PubMed] [Google Scholar]
- 36.Hovnik T, Bratanič N, Podkrajšek KT, et al. Severe progressive obstructive cardiomyopathy and renal tubular dysfunction in Donohue syndrome with decreased insulin receptor autophosphorylation due to a novel INSR mutation. Eur J Pediatr 2013;172:1125–1129 [DOI] [PubMed] [Google Scholar]
- 37.Sarbassov DD, Jones LG, Peterson CA. Extracellular signal-regulated kinase-1 and -2 respond differently to mitogenic and differentiative signaling pathways in myoblasts. Mol Endocrinol 1997;11:2038–2047 [DOI] [PubMed] [Google Scholar]
- 38.Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell 2010;38:114–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol 1999;210:30–43 [DOI] [PubMed] [Google Scholar]
- 40.Göke J, Chan YS, Yan J, Vingron M, Ng HH. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol Cell 2013;50:844–855 [DOI] [PubMed] [Google Scholar]
- 41.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013;12:395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Shyh-Chang N, Segrè AV, et al. DIAGRAM Consortium. MAGIC Investigators . The Lin28/let-7 axis regulates glucose metabolism. Cell 2011;147:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bami M, Episkopou V, Gavalas A, Gouti M. Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors. PLoS One 2011;6:e20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oriente F, Iovino S, Cabaro S, et al. Prep1 controls insulin glucoregulatory function in liver by transcriptional targeting of SHP1 tyrosine phosphatase. Diabetes 2011;60:138–147 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Orvis GD, Hartzell AL, Smith JB, et al. The engrailed homeobox genes are required in multiple cell lineages to coordinate sequential formation of fissures and growth of the cerebellum. Dev Biol 2012;367:25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLean AB, D’Amour KA, Jones KL, et al. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 2007;25:29–38 [DOI] [PubMed] [Google Scholar]
- 47.Woo M, Patti ME. Diabetes risk begins in utero. Cell Metab 2008;8:5–7 [DOI] [PubMed] [Google Scholar]
- 48.Woo M, Isganaitis E, Cerletti M, et al. Early life nutrition modulates muscle stem cell number: implications for muscle mass and repair. Stem Cells Dev 2011;20:1763–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.