Abstract
Brown adipose tissue (BAT) is specialized to burn lipids for heat generation as a natural defense against cold and obesity. Previous studies established microRNAs (miRNAs) as essential regulators of brown adipocyte differentiation, but whether miRNAs are required for the feature maintenance of mature brown adipocytes remains unknown. To address this question, we ablated Dgcr8, a key regulator of the miRNA biogenesis pathway, in mature brown as well as in white adipocytes. Adipose tissue–specific Dgcr8 knockout mice displayed enlarged but pale interscapular brown fat with decreased expression of genes characteristic of brown fat and were intolerant to cold exposure. Primary brown adipocyte cultures in vitro confirmed that miRNAs are required for marker gene expression in mature brown adipocytes. We also demonstrated that miRNAs are essential for the browning of subcutaneous white adipocytes in vitro and in vivo. Using this animal model, we performed miRNA expression profiling analysis and identified a set of BAT-specific miRNAs that are upregulated during brown adipocyte differentiation and enriched in brown fat compared with other organs. We identified miR-182 and miR-203 as new regulators of brown adipocyte development. Taken together, our study demonstrates an essential role of miRNAs in the maintenance as well as in the differentiation of brown adipocytes.
Introduction
There are two principal types of adipose tissues in mammals, white adipose tissue (WAT) and brown adipose tissue (BAT) (1). The main function of WAT is to store energy as triacylglycerol during periods of food surplus and to mobilize these stores when food is scarce. BAT is specialized to dissipate energy as heat through uncoupling of oxidative phosphorylation from ATP synthesis mediated by uncoupling protein 1 (Ucp1). Although WAT is found widespread throughout the mammalian body, BAT is localized in the interscapular region in small rodents (1) and in human infants (2). This type of BAT, known as classical BAT, has a different developmental origin from WAT and is derived from a Myf5+ lineage precursor (3). A subpopulation of subcutaneous (Sub) WAT cells has recently been identified as inducible BAT cells, also referred to as beige or brite adipocytes. Before activation, these inducible brown adipocytes contain a small number of mitochondria and have low thermogenic activity. Once activated, these cells display abundant mitochondria, express high Ucp1 levels, and exhibit thermogenesis and energy expenditure features of brown fat (4,5). Induction of classical or inducible BAT can improve the metabolic phenotype of rodent models (5,6); this is the basis for the development of BAT-based therapies for obesity.
The prevalence of BAT in human adults was not appreciated until recently. Using positron emission tomography-computed tomography, researchers detected Ucp1-positive metabolically active adipose cells in the cervical, supraclavicular, axillary, and paravertebral regions of adult human subjects (7–11). Gene expression signature comparisons indicated that BAT in adults expresses beige fat–enriched markers such as Tbx1 and Tmem26, suggesting that the BAT in these organs may be beige fat, whereas the interscapular BAT in infants resembles the classical BAT found in rodents (2,4,12). However, other reports showed that beige fat markers and classical brown-selective markers, such as Zic1 and Lhx8, can both be detected in adult BAT samples (11,13), arguing that human adult BAT may consist of a mixture of classical BAT and beige fat.
Brown adipocyte development is regulated by a cascade of protein factors, including transcriptional factors (e.g., Pparγ, Foxc2), cofactors (e.g., Prdm16, Pgc1α), and hormones (e.g., Bmp7, Fgf21) (5,14,15), but the role of microRNAs (miRNAs) in this process is not well understood. Recently, we and other groups have demonstrated that miRNAs constitute a critical regulatory layer governing brown adipocyte development and beige fat activation (16). The miR-193b-365 cluster was the first reported miRNAs that sustain brown adipocyte differentiation by repressing the myogenic potential of preadipocytes (17). On one hand, ectopic expression of miR-196a can induce a browning effect in WAT in vitro and in vivo but does not have appreciable effects on classical BAT (18). On the other hand, several miRNAs negatively regulate brown fat development. Upon cold exposure, expression of miR-133 is downregulated by Mef2 and de-represses PRDM16, thereby promoting a browning phenotype in Sub WAT (19,20) and skeletal muscle (21). miR-155 was originally identified as a key regulator of the mammalian immune system (22), but a recent study showed that miR-155–null mice exhibit enhanced BAT function and “browning” of WAT; in contrast, transgenic expression of miRNA 155 impairs BAT functions (23). Our understanding of the miRNA regulatory network in brown fat differentiation and function is incomplete, however. Although previous studies have demonstrated that miRNAs are essential for brown fat cell formation from precursors in vitro and in vivo, whether miRNAs are required to maintain brown fat features in mature brown adipocytes remains unknown.
To address this question, we crossed an adiponectin-Cre transgenic mouse strain with a Dgcr8flox/flox conditional strain to block miRNA biogenesis in mature adipocytes. The knockout (KO) mice developed BAT dysfunction and exhibited impaired thermogenesis upon cold exposure. Genome-wide miRNA profiling revealed a set of BAT-enriched miRNAs that we showed are important for brown adipocyte development or function.
Research Design and Methods
Animals, Glucose Tolerance Test, and Insulin Tolerance Test
The adiponectin-Cre transgenic mouse strain and Dgcr8flox/flox strain were gifts from Dr. Evan Rosen at Harvard University and from Dr. Robert Blelloch at the University of California, San Francisco, respectively. Adipose tissue–specific Dgcr8 KO mice were generated by crossing adiponectin-Cre with Dgcr8flox/flox mice. The sequences of genotyping primers are forward: CAGATGATCAAATGCCATCAG and reverse: CATCTCCACCTTCTCAAACCC. The size of the amplicon is 627 bp for the KO allele and 1,101 bp for wild-type allele (24).
Mice were housed in a temperature-controlled facility (21°C) with a 12-h light/12-h dark cycle. All animal experimental protocols were approved by the SingHealth Research Facilities Institutional Animal Care and Use Committee, Singapore. Body weight and food intake were measured every week. For the glucose tolerance test (GTT), mice were fasted overnight, followed by intraperitoneal glucose injection (2 g/kg). For the insulin-tolerance test (ITT), human insulin (Sigma-Aldrich) was injected (1 unit/kg) to randomly fed mice. Plasma insulin levels were measured by ELISA (Millipore).
For cold exposure, 6-week-old mice were housed at 8°C for 24 h. The rectal body temperature was recorded every hour with a probe thermometer (Advance Technology) at a constant depth.
RNA Extraction and Quantitative Real-Time PCR
Total RNA from cultured cells or tissues was isolated using the Qiagen miRNeasy kit. RNA was reverse transcribed with Moloney-murine leukemia virus (Promega). Real-time PCR for miRNA and mRNA were performed as described before (17). Briefly, mRNA SYBR green quantitative real-time PCR (Applied Biosystems) was performed to detect expression of mRNA levels in a HT7900 Fast Real-Time PCR System. 18S was used as the internal control. miRNA quantitative real-time PCR was performed according to the instruction of miRNA assay kit (Applied Biosystems) with snoRNA202 as the internal control.
miRNA Microarray
Total RNAs were extracted from brown fat tissue and sent to Exiqon (Vedbaek, Denmark). The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. Total RNA (500 ng) from both sample and reference was labeled with Hy3 and Hy5 fluorescent label, respectively, using the miRCURY LNA microRNA Hi-Power Labeling Kit Hy3/Hy5 (Exiqon), using the procedure described by the manufacturer. The Hy3-labeled samples and a Hy5-labeled reference RNA sample were mixed pairwise and hybridized to the miRCURY LNA microRNA Array, 7th Gen (Exiqon), which contains capture probes targeting all miRNAs for human, mouse, or rat registered in the miRBASE 18.0. The hybridization was performed according to the miRCURY LNA microRNA Array Instruction Manual using a Tecan HS 4800 hybridization station (Tecan, Grödig, Austria). After hybridization, the microarray slides were scanned and stored in an ozone-free environment (ozone level below 2.0 ppb) to prevent potential bleaching of the fluorescent dyes. The miRCURY LNA microRNA Array slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., Santa Clara, CA), and the image analysis was done using ImaGene 9 (miRCURY LNA microRNA Array Analysis Software; Exiqon). The quantified signals were background corrected and normalized using the global locally weighted scatterplot smoothing regression algorithm.
Western Blotting
Tissues were homogenized in radioimmunoprecipitation assay buffer (Roche Applied Science), and protein concentration was determined by bicinchoninic acid protein assay (Pierce). An even concentration of protein lysates was separated in SDS-PAGE gels, transferred to nitrocellulose membranes (Millipore), and incubated with Ucp-1, Cytochrome C, NADH dehydrogenase Fe-S protein 3 (NDUFS3), ATP5a and GAPDH antibodies (Abcam). Quantification of signals was performed using a Gel Doc XR system (Bio-Rad).
Histological Analysis and Cell Number Calculation
For histological analysis, tissues were fixed and embedded in paraffin. Hematoxylin and eosin staining was performed on 5 μm paraffin-embedded sections. Immunofluorescence (IF) staining was performed on paraffin sections, with a UCP1 antibody (Abcam), anti-mouse IgG Alexa 633 (Invitrogen), and DAPI (Invitrogen). Images were acquired with a Leica DMI 3000B microscope system (Leica Microsystems, Wetzlar, Germany) and analyzed with Leica Application Suite V4.0 and ImageJ software.
The diameter of more than 100 cells in each tissue sample was measured with ImageJ software, and the cell diameter was used to calculate cell volume. The relative cell number was estimated by the ratio between the tissue weight and average cell volume and then normalized to the cell number in control mice for presentation.
Adipose Stromal Vascular Fraction Cell Isolation and Culture
The stromal vascular fraction (SVF) cells from white fat tissue and brown fat tissue were isolated as described before (17). Briefly, tissue depots were digested in 0.2% collagenase at 37°C for 25–30 min. Digested tissues were filtered through a 100-μm membrane, and SVF cells were collected by centrifugation at 1,500 rpm for 5 min. After centrifugation, the mature adipocytes floating on top were collected for genotyping analysis. The freshly isolated SVF cells were seeded and cultured in DMEM containing 10% newborn calf serum and 0.5% penicillin/streptomycin at 37°C with 5% CO2. On confluence, the cells were induced to differentiate for 2 days with DMEM containing 10% FBS, 850 nmol/L insulin, 0.5 μmol/L dexamethasone (Sigma-Aldrich), 250 μmol/L of 3-isobutyl-methylxanthine (Sigma-Aldrich) and 1 µmol/L rosiglitazone (Sigma-Aldrich) in DMEM. The induction medium was replaced with DMEM containing 10% FBS and 160 nmol/L insulin for 2 days. Then cells were incubated in DMEM with 10% FBS.
For transfection, primary preadiocytes were kept in culture medium until 80–90% confluence. Then, locked nucleic acid (LNAs) miRNA inhibitors (100 nmol/L) were transfected by Lipofectamine RNAi Max (Invitrogen), according to the manufacturer’s instructions. At 8 h after transfection, cells were recovered in full culture media and induced to differentiation. RNAs were harvested for analysis 4 days after differentiation.
Oil Red O staining
Differentiated cells were washed with PBS twice and fixed with 2% paraformaldehyde for 15 min at room temperature. After fixation, cells were washed with PBS and stained with freshly prepared Oil Red O (ORO) working solution for 1 h at room temperature. The ORO stock solution was prepared by dissolving 0.5 g ORO in 100 mL isopropanol, and 6 mL stock solution was mixed with 4 mL H2O to prepare the working solution.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was performed as described (25). For each miRNA inhibition experiment, a rank-ordered gene list was generated according to gene expression fold changes with respect to the control experiment. The ranked lists were used as input to GSEA, which mapped genes from the C2 curated gene sets from the Molecular Signatures Database to the ranked-ordered lists. A normalized enrichment score and a P value is generated for each gene set. A negative normalized GSEA score implies that the gene set is negatively correlated to the ranked gene list.
Results
Dgcr8 Adipose KO Mice Exhibit Enlarged and Pale BAT
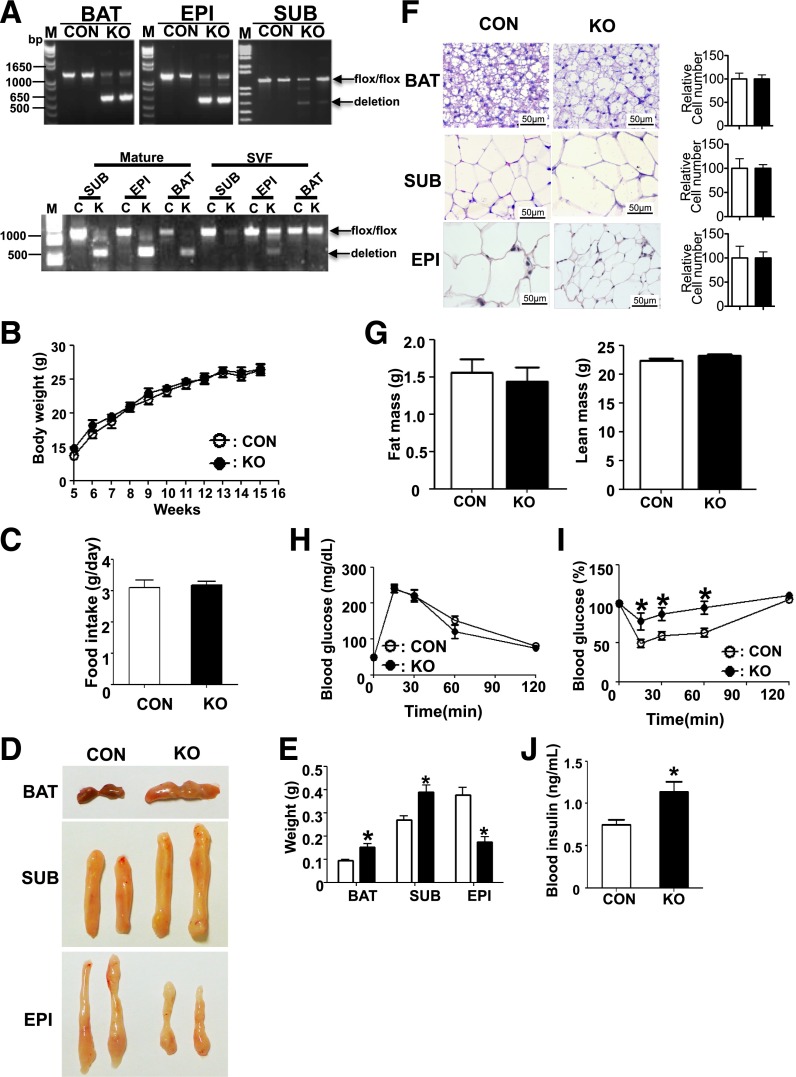
To determine the role of miRNAs in mature adipocytes in vivo, we generated adipose tissue-specific Dgcr8 KO mice by crossing Dgcr8flox/flox mice (24,26) with adiponectin-Cre transgenic mice (27). The control and KO mice were both viable and born in the expected Mendelian ratios. We observed 80–90% Dgcr8 deletion in interscapular BAT and epididymal (Epi) WAT in KO mice, but only 20–40% deletion in Sub WAT (Fig. 1A, upper panel). To determine whether adiponectin-Cre specifically deletes Dgcr8 in mature adipocytes but not in preadipocytes, we examined Dgcr8 deletion in isolated mature adipocytes as well as the SVF that is enriched for preadipocytes. We observed more than 80% deletion of Dgcr8 in mature adipocytes in all three fat tissues but no deletion in brown and Sub SVF cells, indicating a specific deletion of Dgcr8 in mature adipocytes (Fig. 1A, lower panel). A slight deletion was detected in the SVF from Epi WAT, suggesting that some cell types in the Epi SVF can express the marker AdipoQ.
Figure 1.
Dgcr8 KO mice showed increased but pale BAT. A: PCR genotyping results in interscapular BAT (BAT), Sub WAT (SUB), and Epi WAT (EPI) from 12-week-old control (CON) and KO mice (upper panel) or in mature adipocytes and SVF from each fat tissue (lower panel). B: Body weights curves of CON and KO mice at indicated times under normal chow diet (n = 8 each group). C: Food intake of CON and KO mice at 16 weeks old. D: Photographs of BAT, SUB, and EPI from 16-week-old CON and KO mice. E: Weight of each fat tissue in CON and KO mice (n = 8 each group). F: Representative data of hematoxylin and eosin staining of fat tissues from 16-week-old CON and KO mice. Scale bar: 50 µm. The relative cell number in each tissue was estimated by the ratio between tissue weight and cell volume and then normalized to the cell number in CON tissue for presentation on the right. G: Fat mass (left panel) and lean mass (right panel) from 14-week-old CON and KO mice. Blood glucose levels during GTT (H) and ITT (I) of 14-week-old mice (n = 8 each group). J: Plasma insulin levels of 16-week-old mice (n = 8 each group). *P < 0.05, Student t test. Data are shown as means ± SEM.
Control and KO mice did not exhibit significant differences in body weight or food intake (Fig. 1B and C and Supplementary Fig. 1A). The KO mice have smaller Epi WAT but enlarged BAT and Sub WAT (Fig. 1D and E and Supplementary Fig. 1B). BAT is larger in the KO mice but has a pale appearance, suggesting that the function of brown fat is impaired. Hematoxylin and eosin staining of biopsy specimens revealed that Epi white adipocytes in KO mice were smaller than normal, whereas brown adipocytes and Sub white adipocytes were larger (Fig. 1F). However, the cell numbers, estimated by the ratio of tissue weight and cell size, did not show significant difference (Fig. 1F), indicating that adipocyte recruitment was not affected. The total fat mass, as detected by magnetic resonance imaging, did not show a significant difference (Fig. 1G).
Adipose-Specific Dgcr8 KO Results in Impaired Insulin Sensitivity
Because changes in the size of adipose tissue often result in a systemic metabolic phenotype, we performed GTT and ITT. Blood glucose levels were similar between control and KO mice during the GTT (Fig. 1H and Supplementary Fig. 1C). However, the KO mice exhibited higher blood glucose levels than the control group during the ITT, indicating impaired insulin sensitivity (Fig. 1I and Supplementary Fig. 1D). Because the KO mice showed a normal GTT response but had an impaired ITT response, we suspected that the KO mice might develop hyperinsulinemia as a compensatory mechanism for insulin resistance to maintain glucose homeostasis. Indeed, ELISA showed that the insulin level was significantly higher in the KO mice (Fig. 1J). Because an enlarged Sub WAT and a smaller Epi WAT are usually associated with beneficial metabolic effects (28,29), the insulin resistance in KO mice might be due to brown fat dysfunction (30). However, the relative contribution of each adipose to the insulin resistance remains to be further investigated.
Deletion of Dgcr8 Impairs the Function of BAT In Vivo
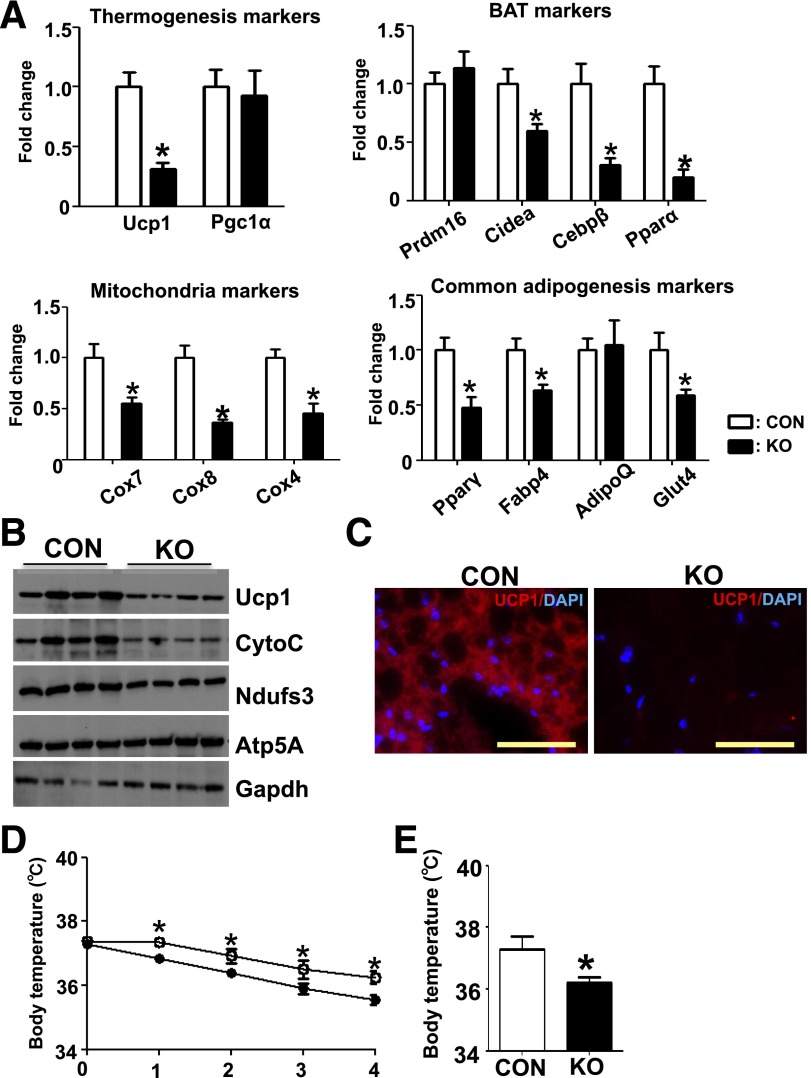
To further examine the effect of Dgcr8 deletion in BAT, we performed real-time PCR to compare the expression of selected marker genes. Deletion of Dgcr8 in BAT resulted in a significant decrease in brown fat and mitochondrial marker genes such as Ucp1, Cebpβ, Cidea, Pparα, Cox4, Cox7, and Cox8, and, to a lesser extent, common adipocyte marker genes such as Pparγ, Fabp4, and Glut4 (Fig. 2A). Western blots showed lower protein levels of Ucp1 and of the mitochondrial proteins Cytochrome C and Ndufs3 (Fig. 2B). IF staining also showed a decrease of Ucp1 expression in the BAT from KO mice (Fig. 2C). Taken together, these data indicate that miRNAs in mature brown adipocytes are essential for maintaining the expression of brown fat–important genes.
Figure 2.
Deletion of Dgcr8 in BAT results in impaired marker gene expression and thermogenic response. A: Real-time PCR to examine the expression of thermogenesis-related markers (Ucp1 and Pgc1α), brown fat markers (Prdm16, Cidea, Cebpβ, and Pparα), common adipocyte differentiation markers (Pparγ, Fabp4, AdipoQ, and Glut4), and mitochondria markers (Cox7, Cox8, and Cox4) of BAT in control (CON) and KO 12-week-old mice (n = 8 each group). B: Western blot for brown fat marker UCP1, and mitochondrial marker cytochrome C (CytoC) and Ndusf3 of BAT in CON and KO mice. C: IF of Ucp1 (red) and DAPI (blue) in the brown fat of CON and KO mice. Scale bar: 50 μm. Five-week-old CON and KO mice were exposed to 8°C (D) and body temperature was measured by rectal probe at the indicated times and at 24 h after exposure (E). *P < 0.05, Student t test. Data are shown as means ± SEM.
Because Dgcr8 deletion caused a dramatic reduction of Ucp1 at mRNA and protein levels, we suspected that the thermogenic response of these KO mice would be impaired upon cold exposure. At an ambient temperature of 21°C, control and KO mice had similar body temperatures; at 8°C, KO mice experienced a more rapid decline in body temperature than controls (Fig. 2D), and the cold intolerance phenotype in KO mice remained after 24 h cold exposure (Fig. 2E). This phenotype is likely to be attributed to the defect in BAT, but we cannot exclude the possible contribution of an indirect effect on shivering (detailed in discussion).
Deletion of Dgcr8 in Mature Brown Adipocytes Results in Impaired Gene Expression In Vitro
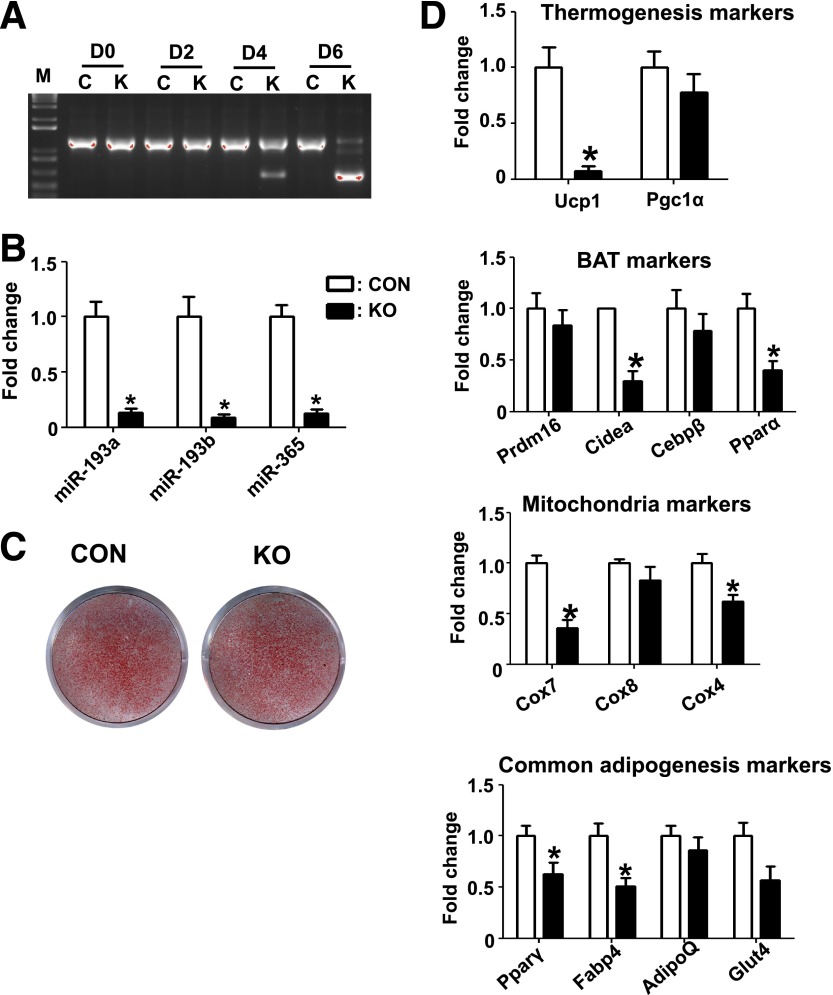
To test whether the phenotype observed in BAT is cell autonomous, we isolated the brown fat SVF cells from KO and control mice and differentiated them to brown adipocytes. To avoid activation of AdipoQ-Cre expression at early stage, we reduced the concentration of rosiglitazone from 1 µmol/L to 0.2 µmol/L. Genotyping at different time points during differentiation showed no deletion of Dgcr8 before the induction of differentiation, less than 20% deletion at day 4 when mature adipocytes had formed, but a complete deletion at day 6, indicating that the adiponectin-Cre–driven target deletion occurred only at the mature adipocyte stage (Fig. 3A).
Figure 3.
Deletion of Dgcr8 results in reduced expression of brown fat markers in primary brown adipocytes. A: Genotyping results during adipogenesis of primary brown adipocyte cultures. Genomic DNA was extracted at the indicated day and deletion efficiency of Dgcr8 was examined by PCR in KO (K) and control (C) mice, as described in research design and methods. B: Real-time PCR result of miRNA expression 8 days after differentiation (CON, control). C: ORO staining of primary brown adipocyte cultures 8 days after differentiation. D: Real-time PCR results of marker gene expression 8 days after differentiation (n = 3). *P < 0.05, Student t test. Data are shown as means ± SEM.
Eight days after differentiation, we isolated RNAs and used miRNA real-time PCR to confirm whether miRNA biogenesis was blocked. We examined the expression of miR-193a, miR-193b, and miR-365, which were previously reported as essential regulators of brown fat development (17), and found that these miRNAs were all downregulated in BAT cells from the KO mice (Fig. 3B), indicating a loss of function of Dgcr8 in our cell culture system.
No significant difference was observed in ORO staining (Fig. 3C), indicating that loss of miRNAs in mature brown adipocytes did not cause a defect in lipid accumulation. However, real-time PCR revealed a significant decrease in expression of key brown fat and mitochondria mRNAs, including Ucp1, Cidea, Pparα, Cox4, and Cox7, in KO brown fat adipocytes. KO also resulted in a slight decrease in two common adipogenic markers, Pparγ and Fabp4. Consistent with the phenotype shown in vivo in adipose tissue–specific Dgcr8 mice, these data demonstrated that miRNAs are required to maintain the expression of brown fat genes required for thermogenesis and mitochondrial biogenesis in a cell-autonomous manner.
Deletion of Dgcr8 in Brown Adipocyte Precursors Impairs the Formation of Brown Adipocytes
Because adiponectin-Cre will only delete the target gene after mature adipocytes are formed, we cannot use this system to address whether miRNAs, as a group, are required for brown adipocyte formation. To answer this question, we isolated brown fat SVF cells from Dgcr8flox/flox mice and infected these cells with adenovirus expressing Cre recombinase (Ad-Cre) or green fluorescent protein 2 days before differentiation. Four days after differentiation, Ad-Cre–infected cells showed a marked decrease of lipid accumulation (Supplementary Fig. 2A), accompanied with decreased marker gene expression, including brown fat markers and common adipogenesis markers (Supplementary Fig. 2C). Thus, as expected, miRNAs are required for lipid droplet accumulation and gene expression in the process of brown adipocyte differentiation from precursors.
Dgcr8 Deficiency Causes Attenuation of Browning of Sub WAT
In contrast to the strong phenotype observed in KO brown fat, the phenotype in WAT was mild. In Epi WAT, Dgcr8 KO resulted in a decrease in the expression levels of Pparγ and AdipoQ but not of Fabp4, Glut4, or Cebpα (Supplementary Fig. 3A). Lipolysis assay using explants from Epi WAT did not reveal a significant difference between control and KO tissues (Supplementary Fig. 3B). However, because the KO Epi WAT was much smaller, its overall capacity of lipolysis and triacylglycerol storage should be reduced, even if the remaining pads seem to be normal.
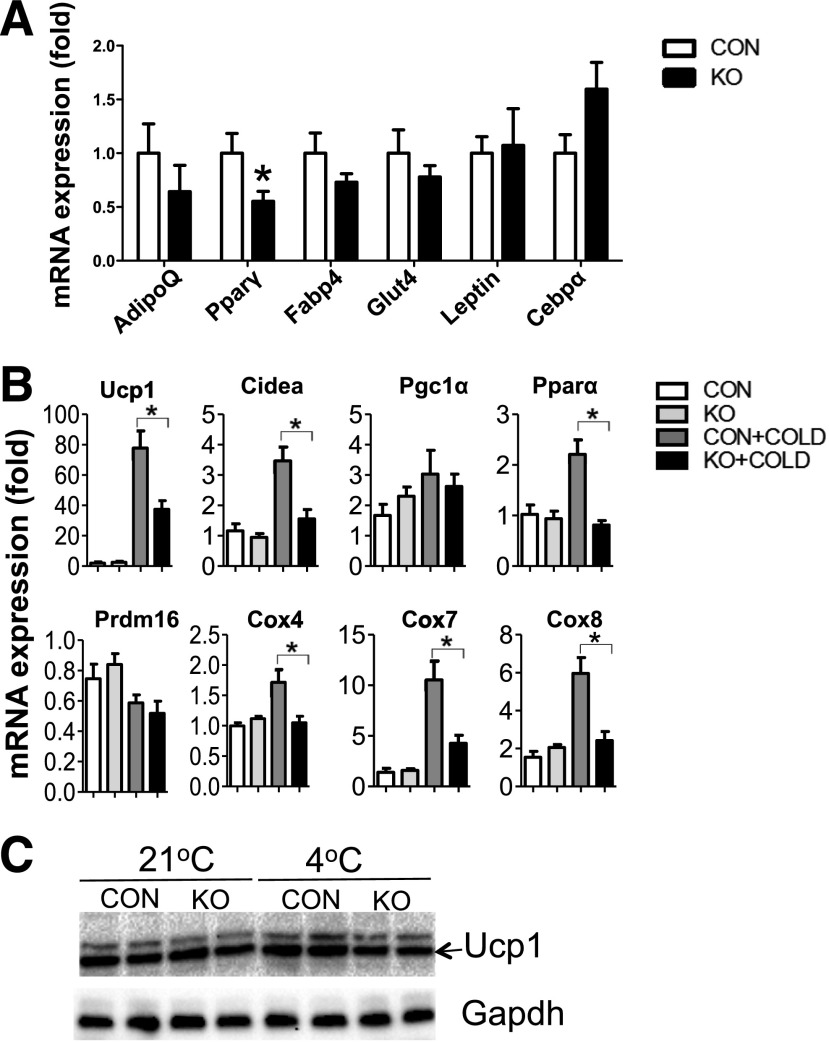
In inguinal WAT, Dgcr8 deletion did not cause significant changes in all of the examined marker mRNAs except for Pparγ (Fig. 4A). These data suggest that miRNAs have limited effects on gene expression in mature white adipocytes. Because Dgcr8 KO had a preferential influence on brown fat markers but not on the common adipogenesis marker in brown fat (Fig. 3D), we examined whether Dgcr8 deletion affected “browning” of inguinal WAT. We housed KO and control animals at 4°C for 48 h and isolated total RNAs for real-time PCR analysis. Before cold stimulation, all of the “browning” markers examined, such as Ucp1, Ppara, Cidea, and Cox7, showed little difference; after stimulation, these markers were markedly increased in control mice, but the extent of induction was significantly lower in KO mice (Fig. 4B). Western blot confirmed the blunted induction of Ucp1 protein in KO inguinal WAT upon cold exposure (Fig. 4C). These data demonstrate that miRNAs are important regulators for browning of Sub WAT.
Figure 4.
Deletion of Dgcr8 causes reduced browning of inguinal WAT. A: Real-time PCR examined the expression of common adipocyte differentiation markers in inguinal WAT from control (CON) and KO mice (n = 6 each group). B: CON and KO mice (6 weeks old) were exposed to 4°C for 48 h, and inguinal WAT total RNAs were isolated to examine BAT-selective marker expression by real-time PCR (n = 6). C: Six-week-old mice were exposed to 4°C for 48 h. Inguinal WAT was isolated for Western blot analysis. Each lane represents a pooled tissue lysate from three individual mice. *P < 0.05, Student t test. Data are shown as means ± SEM.
To test whether miRNAs are required for the browning of Sub adipocytes in vitro, we differentiated SVF cells isolated from inguinal WAT in the presence or the absence of the “browning” stimulators rosiglitazone and norepinephrine. We did not observe significant differences in lipid accumulation or common adipogenic marker gene expression (Supplementary Fig. 4A–C). However, we found that the “browning” markers induced by rosiglitazone and norepinephrine were significantly lower in KO cells (Supplementary Fig. 4D), indicating that miRNAs are essential for the browning of white adipocytes in vitro.
Microarray Study Reveals a Set of BAT-Enriched miRNAs
Because Dgcr8 is a key regulator of miRNA biogenesis, the impaired BAT function observed in vivo and in vitro should be due to the loss of some key miRNAs. Key regulators are often upregulated during adipogenesis and enriched in brown adipocytes relative to other cell types. To identify miRNAs that meet these criteria, we used miRNA microarrays to examine genome-wide miRNA expression in BAT in control and KO mice. Because BAT consists of a variety of cell types, including mature brown adipocytes, brown adipocyte precursors, endothelial cells, and some immune cells, and because adiponectin-Cre only deletes floxed genes in mature brown adipocytes (Fig. 1A), this miRNA gene expression analysis should reveal miRNAs that are enriched in mature brown adipocytes and whose formation depends on Dgcr8.
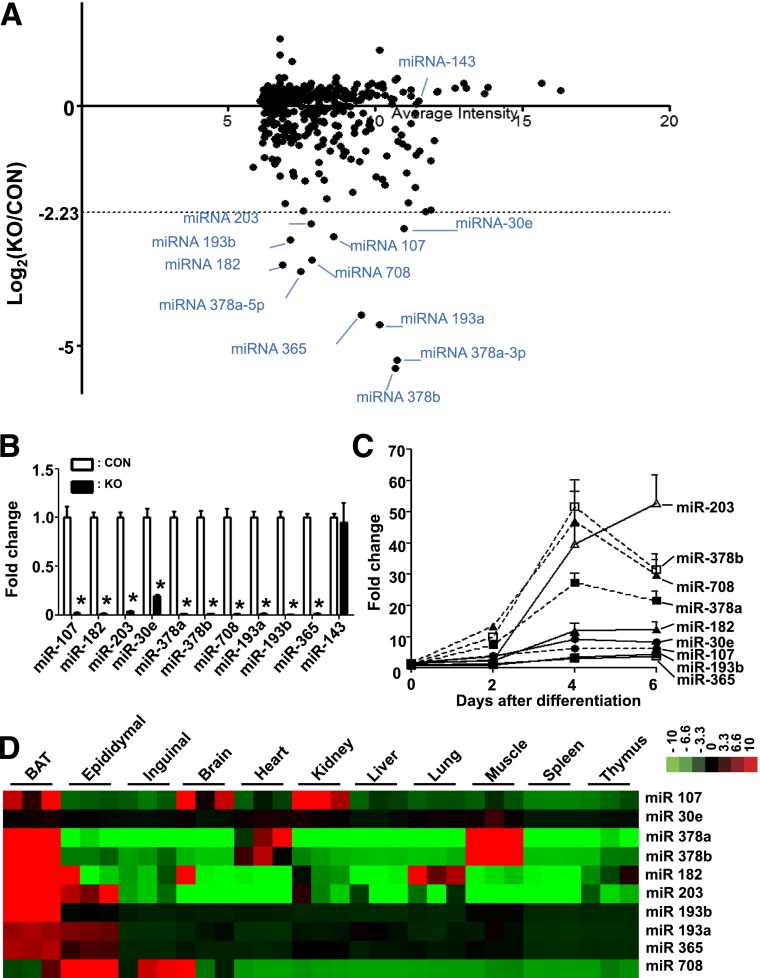
We selected the 10 most downregulated miRNAs in the Dgcr8 KO brown adipocytes for further investigation: miR-107, miR-182, miR-203, miR-378a, miR-378b, miR 708, miR-193a, miR-193b, miR-365, and miR-30e (Fig. 5A). Using quantitative PCR, we confirmed the downregulation of these 10 miRNAs in the Dgcr8 KO brown adipocytes (Fig. 5B). We also found that these miRNAs were upregulated during primary BAT adipogenesis (Fig. 5C) and that most of them, except miR-30e and miR-708, were enriched in BAT compared with other tissues (Fig. 5D). These results imply that these identified miRNAs play an important role in BAT development or function.
Figure 5.
Microarray study reveals a set of brown fat-enriched miRNAs in KO and control (CON) mice. A: Scatter plot of the logarithmic maximum intensity (x axis) vs. the fold change of the miRNAs expression (y axis). B: Real-time PCR results of selected miRNAs expression in the BAT of CON vs. KO mice (n = 4 each group). C: Expression of miRNAs during adipogenesis of primary brown adipocyte cultures. D: Heat map of real-time PCR result shows the expression of selected miRNAs in BAT and other organs. Red denotes higher and green denotes lower relative to the mean of the samples for each miRNA. *P < 0.05, Student t test. Data are shown as means ± SEM.
To test whether these BAT-enriched miRNAs are sufficient to maintain brown fat marker gene expression in the absence of other miRNAs, we introduced RNA mimics of these miRNAs into mature brown adipocytes from KO mice maintained in cell culture. The overexpression of these miRNAs could not rescue the molecular phenotype in Dgcr8 KO brown adipocyte cells (Supplementary Fig. 5). This result suggests that other miRNAs are necessary to constitute the miRNA regulatory core for marker gene expression.
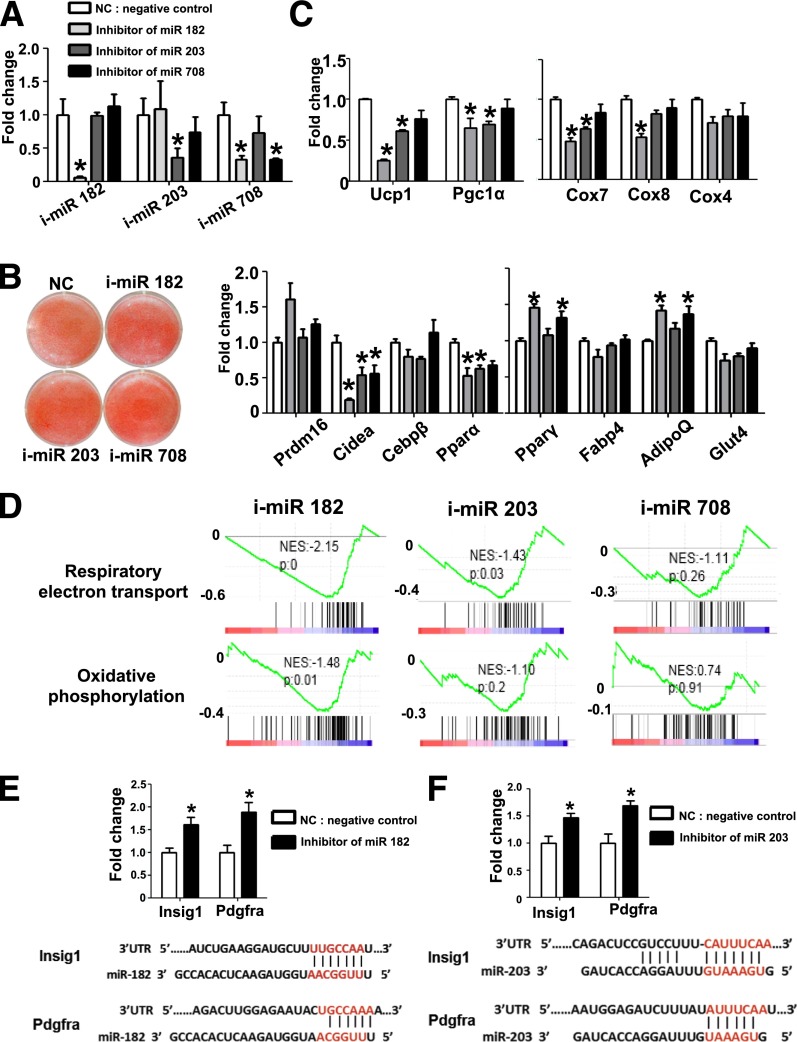
miR-203 and miR-182 Are Required for Brown Adipocyte Development
Among the BAT-specific miRNA that were identified from the microarray analysis, miR-107 (31) and the miR-193a/b-365 cluster (17) have been reported as regulators of adipogenesis; miR-378a/b are involved in mitochondrial fatty acid metabolism and oxygen consumption (32,33), but the role of miR-182, miR-203, and miR-708 in BAT remains unknown. To test their functions, we transfected LNA-miRNA inhibitors into brown adipocyte precursors and then induced the transfected cells to differentiate. Four days after differentiation, we performed real-time PCR to confirm their inhibition of target miRNA expression (Fig. 6A). We did not observe a significant difference in lipid accumulation detected by ORO staining, indicating that these miRNAs were not essential for lipid accumulation per se (Fig. 6B). Interestingly, we found that blocking miR-182 or miR-203 caused a reduction of brown fat marker mRNAs, such as Ucp1, Pgc1α, Cidea, and Pparα, and mitochondrial markers, such as Cox 7 and Cox8, but not the common adipogenic markers, including Pparγ, Fabp4, and AdipoQ (Fig. 6C). To determine the effects of miRNA knockdown on global gene expression, we performed RNA sequencing, followed by GSEA (Supplementary Data; accession number, GSE60165). The expression of gene sets controlling respiratory electron transport and oxidative phosphorylation was significantly downregulated upon blocking miR-182 and miR-203 but not miR-708 (Fig. 6D). Taken together, these data indicate that miR-182 and miR-203 are required for the full differentiation of brown adipocytes.
Figure 6.
Knockdown of miR-182 or miR-203 causes reduction of BAT markers in primary brown adipocytes. A: LNA miRNA inhibitors for miR-182 (i-miR 182), miR-203 (i-miR 203), and miR-708 (i-miR 708) were transfected into brown preadipocytes before differentiation. Real-time PCR was performed 4 days after differentiation to examine the expression of the indicated markers. B: Brown preadipocytes were transfected with miRNA inhibitors before differentiation, and ORO staining was performed 4 days after differentiation. C: Real-time PCR was performed to examine indicated marker gene expression at 4 days after differentiation (n = 3). D: Profiles from GSEA. Respiratory electron transport (upper panel) and oxidative phosphorylation pathway (lower panel). Genes were ranked based on the expression fold change of miRNA-inhibited cells vs. control. The black lines represent genes “hits” with the specified annotation. Shown on each is the normalized enrichment score (NES) and nominal P value. Real-time PCR of miR-182 targets Insig-1 and Pdgfra (E) and miR-203 target Pdgfra (F); UTR, untranslated region. *P < 0.05, Student t test. Data are shown as means ± SEM.
To identify mRNA targets of miR-182 and miR-203, we examined the expression levels of TargetScan-predicted candidates. Among the conserved miRNA targets are insulin induced gene 1 (Insig1) and platelet-derived growth factor receptor α (Pdgfrα), targeted by miR-182 and miR-203. Insig1 and Pdgfrα were both reported as inhibitors of adipocyte differentiation (34,35). Real-time PCR analysis showed that the expression of Insig1 and Pdgfrα was upregulated upon miR-182 and miR-203 knockdown (Fig. 6E and F), suggesting that miR-182 and miR-203 function partially by targeting two common targets, Insig1 and Pdgfrα.
To test whether miR-182 and miR-203 are sufficient to promote brown adipocyte differentiation, we transfected miR-182 and miR-203 mimics into brown adipocyte culture but did not observe any alteration in lipid accumulation and BAT marker expression (data not shown). Thus, miR-182 and miR-203 are not sufficient for a full brown adipocyte differentiation but are required.
Discussion
Because of the energy expenditure feature of brown fat, researchers are interested in understanding the detailed regulation of brown fat development and function. Several recent studies have demonstrated that miRNAs comprise an important regulatory network for brown fat differentiation (16). However, whether miRNAs are essential for the maintenance of brown fat function is still unclear. To address this question, we generated an adipose tissue–specific Dgcr8 KO mouse model using adiponectin-Cre mice in which miRNA biogenesis was ablated specifically in mature adipocytes but not preadipocytes. The KO mice developed brown fat dysfunction and cold intolerance, with reduced expression of brown fat markers. In vivo and in vitro data both support a key role of miRNAs in the maintenance of brown fat marker expression. In addition, deletion of Dgcr8 by Adv-Cre infection during brown adipocyte differentiation clearly impaired lipid accumulation and marker expression (Supplementary Fig. 2), indicating that miRNAs are also key regulators during brown adipocyte differentiation. To identify miRNAs that are important for brown fat, we performed miRNA arrays, followed by LNA inhibitor–mediated functional analysis. We identified miR-182 and miR-203 as two novel miRNAs regulators for brown fat development.
We noticed that although Dgcr8 was well deleted in mature adipocytes from different adipose depots, these adipocytes displayed distinct phenotypes. In Epi WAT adipocytes, Dgcr8 deletion had a limited effect on marker gene expression but caused a significant reduction in cell size. Because the deletion of Dgcr8 mainly occurred in mature adipocytes (Fig. 1A), the reduced size of Epi WAT is more likely due to the impaired lipid accumulation but not adipocyte differentiation. The exact mechanism of how miRNAs regulate lipid accumulation in Epi WAT remains to be further investigated. In Sub WAT adipocytes, Dgcr8 KO had a mild effect on gene expression but resulted in an increase of cell size and impaired browning; in BAT adipocytes, Dgcr8 KO led to much stronger molecular and cellular phenotypes. These results indicate that miRNAs play different roles in different types of adipocytes.
Although Dgcr8 deletion resulted in little effect on common adipogenic marker expression in inguinal WAT, it blunted the full induction of browning (Fig. 4). Browning could be due to de novo beige adipocyte adipogenesis or activation of resident beige adipocytes in Sub WAT. Despite many studies on the subject of browning resources, a definitive and consensus view has yet to emerge. These two points of view are not mutually exclusive; that both of them are contributing to the overall “browning” phenotype is likely (36). Because the time of cold exposure (48 h) does not allow much de novo adipogenesis to occur (37) in our mouse model, impaired activation of resident beige adipocytes likely accounts for the blunted “browning” in Sub WAT.
The defect in brown fat is an obvious cause of the cold tolerance phenotype, but we cannot exclude the possible contribution from other resources. Recent studies demonstrated significant similarities between classical brown adipocytes and activated beige adipocytes (38), and the Ucp1 protein in both cell types is functionally thermogenic (39). Theoretically, the reduction in browning of Sub WAT (Fig. 4B) could be a contributing factor to the cold intolerance. However, the acute cold exposure (4 h) in our experiments is too short for activation of beige adipocytes, so the defect in Sub WAT should make, if any, little contribution to the cold intolerance in our mouse model. In addition, the cold tolerance phenotype might result from decreased capacity of lipolysis in Epi WAT, because the fatty acid released from WAT is a major resource for thermogenic muscle shivering in response to acute cold exposure. Whether muscle shivering is impaired due to lack of fatty acid supply and, if yes, what is the relative contribution to the overall phenotype remains to be further investigated.
To determine the role of miRNAs in mature brown or white adipocytes, it is necessary to use a transgenic Cre mouse strain that can specifically delete the target genes in mature adipocytes. In a previous attempt, we bred Dgcr8flox/flox mice with aP2-Cre and found that all the aP2-Cre–driven KO mice died 2 weeks after birth (data not shown), which limited a detailed characterization of these animals. Consistently, in another independent study, Mudhasani et al. (40) deleted Dicer in adipose tissue by breeding Dicerflox/flox mice with an aP2-Cre mouse strain, and the KO animals also died postnatally. This is likely due to nonspecific deletion of target genes in other organs by aP2-Cre (41,42). Here, we solved this issue by breeding Dgcr8flox/flox mice with adiponectin-Cre mice, because all animals were born at an expected ratio and the KO animals grew normally but with a defect in brown adipocytes. To the best of our knowledge, our study is the first to clearly illustrate the role of miRNAs in mature brown adipocytes.
Although our studies and those of others have demonstrated that miRNAs are necessary for BAT development and function, an outstanding question is whether there exist a set of miRNAs sufficient to maintain brown fat features. In this study, we identified a set of BAT-enriched miRNAs that are enriched in mature brown adipocytes. Interestingly, most of these miRNAs are functionally important based on previous work and on this study (17,19,32,33). We tested whether the top brown fat enriched miRNAs were sufficient to support brown fat development by introducing miRNA mimics of these genes into Dgcr8 KO brown adipocytes in primary cell culture, but the results were negative (Supplementary Fig. 5). Thus, more efforts are required to identify the full core set of miRNAs that sustain key BAT functions. Nonetheless, our Dgcr8 KO primary cell culture has provided a unique platform to address this question. Our identification of these miRNAs may lead to new methods to activate BAT and develop new therapies for obesity.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Robert Blelloch, of the University of California, San Francisco, for the Dgcr8flox/flox. The adiponectin-Cre mouse strain was a generous gift from Dr. Evan Rosen of Beth Israel Deaconess Medical Center, Harvard University.
Funding. This work was supported by National Institutes of Health (NIH) grant K08-GM-102718 and an NIH Institutional Training Grant (T32-HL-007627) to H.C.P., and by NIH grants DK-047618-25 and DK-068348-07 to H.L. This work was also supported in part by grants from the Singapore Ministry of Health's National Medical Research Council CBRG/0012/2012 to D.L.S., and by Singapore National Research Foundation (NRF) Fellowship (NRF-2011NRF-NRFF 001-025) to L.S.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. H.-J.K. and L.S. researched data and wrote the manuscript. H.C., R.A., H.C.P., M.G., K.A.L., D.X., V.J.G., L.N.N., X.C., C.X.H., and S.G. researched data. J.-P.K., M.T., D.L.S., and H.L. contributed to discussion and edited the manuscript. L.S. is the guarantor of this work, and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-0466/-/DC1 and GSE60165.
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277–359 [DOI] [PubMed] [Google Scholar]
- 2.Lidell ME, Betz MJ, Dahlqvist Leinhard O, et al. Evidence for two types of brown adipose tissue in humans. Nat Med 2013;19:631–634 [DOI] [PubMed] [Google Scholar]
- 3.Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008;454:961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Boström P, Sparks LM, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012;150:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev 2013;27:234–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng Y-H, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov 2010;9:465–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med 2009;360:1518–1525 [DOI] [PubMed] [Google Scholar]
- 8.Cypess AM, Lehman S, Williams G, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009;360:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009;360:1500–1508 [DOI] [PubMed] [Google Scholar]
- 10.Zingaretti MC, Crosta F, Vitali A, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J 2009;23:3113–3120 [DOI] [PubMed] [Google Scholar]
- 11.Cypess AM, White AP, Vernochet C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med 2013;19:635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharp LZ, Shinoda K, Ohno H, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE 2012;7:e49452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jespersen NZ, Larsen TJ, Peijs L, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab 2013;17:798–805 [DOI] [PubMed] [Google Scholar]
- 14.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function—of mice and men. Genes Dev 2009;23:788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo KA, Sun L. Turning WAT into BAT: a review on regulators controlling the browning of white adipocytes. Biosci Rep 2013;33:art:e00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab 2013;24:442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Xie H, Mori MA, et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 2011;13:958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol 2012;10:e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol 2012;14:1330–1335 [DOI] [PubMed] [Google Scholar]
- 20.Liu W, Bi P, Shan T, et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet 2013;9:e1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin H, Pasut A, Soleimani VD, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 2013;17:210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science 2007;316:604–608 [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Siegel F, Kipschull S, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun 2013;4:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao PK, Toyama Y, Chiang HR, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 2009;105:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007;39:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eguchi J, Wang X, Yu S, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 2011;13:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra A, Garg A, Abate N, Peshock RM, Stray-Gundersen J, Grundy SM. Relationship of anterior and posterior subcutaneous abdominal fat to insulin sensitivity in nondiabetic men. Obes Res 1997;5:93–99 [DOI] [PubMed] [Google Scholar]
- 29.Snijder MB, Dekker JM, Visser M, et al. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am J Clin Nutr 2003;77:1192–1197 [DOI] [PubMed] [Google Scholar]
- 30.Lowell BB, S-Susulic V, Hamann A, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 1993;366:740–742 [DOI] [PubMed] [Google Scholar]
- 31.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011;474:649–653 [DOI] [PubMed] [Google Scholar]
- 32.Eichner LJ, Perry MC, Dufour CR, et al. miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab 2010;12:352–361 [DOI] [PubMed] [Google Scholar]
- 33.Carrer M, Liu N, Grueter CE, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A 2012;109:15330–15335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitter S, Vandyke K, Gronthos S, Zannettino AC. Suppression of PDGF-induced PI3 kinase activity by imatinib promotes adipogenesis and adiponectin secretion. J Mol Endocrinol 2012;48:229–240 [DOI] [PubMed] [Google Scholar]
- 35.Li J, Takaishi K, Cook W, McCorkle SK, Unger RH. Insig-1 “brakes” lipogenesis in adipocytes and inhibits differentiation of preadipocytes. Proc Natl Acad Sci U S A 2003;100:9476–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013;19:1252–1263 [DOI] [PubMed] [Google Scholar]
- 37.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 2013;19:1338–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long JZ, Svensson KJ, Tsai L, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab 2014;19:810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shabalina IG, Petrovic N, de Jong JM, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Reports 2013;5:1196–1203 [DOI] [PubMed] [Google Scholar]
- 40.Mudhasani R, Puri V, Hoover K, Czech MP, Imbalzano AN, Jones SN. Dicer is required for the formation of white but not brown adipose tissue. J Cell Physiol 2011;226:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urs S, Harrington A, Liaw L, Small D. Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res 2006;15:647–653 [DOI] [PubMed] [Google Scholar]
- 42.Lee KY, Russell SJ, Ussar S, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 2013;62:864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.