Abstract
Antisaccade deficits reflect abnormalities in executive function linked to various disorders including schizophrenia, externalizing psychopathology, and neurological conditions. We examined the genetic bases of antisaccade error in a sample of community-based twins and parents (N = 4,469). Biometric models showed that about half of the variance in the antisaccade response was due to genetic factors and half due to nonshared environmental factors. Molecular genetic analyses supported these results, showing that the heritability accounted for by common molecular genetic variants approximated biometric estimates. Genome-wide analyses revealed several SNPs as well as two genes—B3GNT7 and NCL—on Chromosome 2 associated with antisaccade error. SNPs and genes hypothesized to be associated with antisaccade error based on prior work, although generating some suggestive findings for MIR137, GRM8, and CACNG2, could not be confirmed.
Descriptors: Endophenotypes, Antisaccade, Heritability, Genome-wide association study, Molecular genetics, Gene-based tests, GCTA
The hunt for endophenotypes that may index the genetic liability for mental disorders is currently of great interest in psychology and psychiatry. Nowhere is this search more marked than schizophrenia, and with good reason given its high heritability and the significant public health impact associated with its often-chronic course and consequent psychosocial debilitation. Indeed, countless studies have been published documenting various deleterious outcomes associated with schizophrenia such as developmental abnormalities, impaired role functioning, cognitive deficits, and deviations in brain structure and processes. More recently, stimulated by progress stemming from the human genome project, scientists have applied new, potentially powerful molecular genetic strategies to identify genetic variants underpinning schizophrenia.
While some intriguing results have been published (e.g., Greenwood et al., 2013; Smoller et al., 2013), much work is needed to flesh out the genetic influence on schizophrenia. Of considerable interest is what additional etiological insights might be gained by examining the molecular genetic basis of associated psychophysiological measures that appear to tap into the genetic liability for schizophrenia. Indeed, there have already been notable attempts along these lines (e.g., Consortium on the Genetics of Schizophrenia [COGS]; Greenwood et al., 2011) using candidate gene and whole genome linkage studies (Greenwood et al., 2011, 2013), as well as initiatives that provide the stimulus for such research (e.g., Research Domain Criteria [RDoC]; Insel et al., 2010). However, genome-wide association studies (GWAS) of schizophrenia-related endophenotypes represent an underexploited opportunity to identify genetic variants linked to the disorder. This is due in part to the difficulty in collecting laboratory data from the large sample sizes required for genetic analyses. However, it remains unclear how large such samples need to be. Because endophenotypes are linked to brain processes that are posited to be more proximal to the effects of genes influencing psychopathology, there is a priori reason to believe that the effects of genetic variants may be detectable using samples considerably smaller than those used in GWAS with clinical diagnostic phenotypes (for examples, see the concluding comments in Iacono, Malone, Vaidyanathan, & Vrieze, 2014). Among the various criteria proposed for endophenotypes, of particular importance is evidence of stability over time, heritability, cosegregation with the illness, appearance in unaffected relatives, and ability to predict the development of disorder in longitudinal research (Braff et al., 2008; Gottesman & Gould, 2003; Iacono, 1985; Iacono & Malone, 2011)—which several psychophysiological indices satisfy easily. Thus, a GWAS of endophenotypes seems an appropriate next step in attempting to understand potential causal factors implicated in mental disorders.
Here, we take this next step by testing one schizophrenia-related endophenotype—antisaccade error—using GWAS methodology in a community-based sample of 4,469 individuals. In the antisaccade task, subjects are required to suppress the natural tendency to direct their gaze in pursuit of a centered fixation stimulus that appears to move abruptly to the right or left of their visual field. Their task is to inhibit the natural tendency to generate prosaccades towards the target while directing their gaze in the opposite direction (producing an antisaccade). The subject thus has to suppress a prepotent response and make an appropriate eye movement in the opposite direction. The inability to suppress this response is posited to be an indicator of deficits in executive control (Munoz & Everling, 2004).
Antisaccade Neurobiology and Neuropsychology
Various regions of the brain have been implicated in the generation of the antisaccade response. The cerebral cortex, thalamus, basal ganglia, superior colliculus, brainstem reticular formation, and cerebellum are all involved in visual fixation and saccadic eye movements (Munoz & Everling, 2004). One region of the brain that has been theorized to be crucial for the generation of the antisaccade is the dorsolateral prefrontal cortex (DLPFC), a structure that has long been associated with executive function indexed by tasks such as those involving working memory (Hutton & Ettinger, 2006). Bolstering this hypothesis, poor inhibitory control and thus poor performance (in the form of increased antisaccade error rate) occurs when there is damage to the DLPFC (Clementz, 1998; Curtis, Calkins, Grove, Feil, & Iacono, 2001). Other indices of executive function have also been shown to be related to antisaccade performance. For example, Hellmuth et al. (2012) showed that antisaccade performance is associated with overall executive function, while others consider it an important aspect of executive function in latent variable models (Miyake & Friedman, 2012). Likewise, working memory tasks have been shown previously to correlate with antisaccade error rate in psychiatric groups (Gooding & Tallent, 2002). Thus, the antisaccade endophenotype itself is of interest, apart from its relationship with psychiatric disorders, as it sheds light on basic brain function and processes, including executive function processes that are likely to be important in schizophrenia.
Various parameters of the antisaccade response such as error rates and latency have also been linked to neurological disorders including Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, Lewy body dementia, Wilson’s disease, and progressive supranuclear palsy (Clark & Eggenberger, 2012; Hutton & Ettinger, 2006). Recent work has suggested that, at least for Parkinson’s and Alzheimer’s disease, antisaccade deficits are reflective of executive dysfunction and not simply attributable to effects on brain regions associated with oculomotor control (Cameron et al., 2012; Heuer et al., 2013). Such findings further reinforce the conclusion that impaired antisaccade performance is reflective of executive dysfunction that cuts across a wide range of psychiatric and disease states.
Antisaccade Performance as an Endophenotype
Among putative endophenotypes for schizophrenia, the antisaccade response is one of the strongest candidates (Clementz, 1998). Findings from numerous studies have shown that antisaccade dysfunction characterizes individuals with schizophrenia and their healthy relatives, is stable (intraclass correlation coefficient > .80) over a 1-year interval in schizophrenia patients and healthy individuals (Light et al., 2012), is highly heritable, and cosegregates with the disorder (Calkins, Iacono, & Curtis, 2003). Indeed, a large body of research (over 40 studies without replication failure) has shown that people with schizophrenia exhibit poor performance on the antisaccade task (Calkins, Curtis, Iacono, & Grove, 2004; Hutton & Ettinger, 2006), and with large effect sizes. Such findings put the antisaccade on a very strong footing when it comes to investigating neurobiologically informed correlates of schizophrenia.
Behavior genetic investigations have noted that anywhere from 42% to 61% of the variation in the antisaccade response is heritable (Greenwood et al., 2007; Malone & Iacono, 2002), depending on the sample used. Previous candidate gene studies have linked antisaccade error rate to a variety of genes including RGS4 (Kattoulas et al., 2012), NOS1AP (Greenwood, Light, Swerdlow, Radant, & Braff, 2012), NRG-1 (Haraldsson, Ettinger, Magnusdottir, Ingason et al., 2010), CHRNA7 (Petrovsky et al., 2009), and CACNG2 (Greenwood et al., 2012). The COMT gene, often associated with schizophrenia, has also been linked with oculomotor disturbances (Haraldsson, Ettinger, Magnusdottir, Sigmundsson et al., 2010; Rybakowski, Borkowska, Czerski, & Hauser, 2002). Of particular relevance is a recent investigation by Greenwood et al. (2013) that found a genome-wide significant linkage effect for antisaccade performance on Chromosome 3p14, a region near several neuronally expressed genes.
Given its links to executive function, it is perhaps not surprising that deficient antisaccade performance has been associated with other disorders also posited to involve problems with prefrontal inhibitory control. These include the so-called externalizing disorders, which involve substance use, aggression, and problems with impulsivity in general. For example, individuals with attention deficit hyperactivity disorder (ADHD) show deficits in voluntary eye movement control (Feifel, Farber, Clementz, Perry, & Anllo-Vento, 2004; Habeych, Folan, Luna, & Tarter, 2006; Munoz, Armstrong, Hampton, & Moore, 2003; O’Driscoll et al., 2005) as do children at risk for alcohol use disorders (Habeych et al., 2006), and those with autism (Kelly, Walker, & Norbury, 2013; Luna, Doll, Hegedus, Minshew, & Sweeney, 2007). Individuals with bipolar disorder display similar deficits as well (Gooding & Tallent, 2001), in addition to the first-degree relatives of psychotic bipolar probands (Reilly et al., 2013).
Since executive dysfunction has been implicated in these disorders, these findings are consistent with expectation. In addition, the finding that poor antisaccade performance is present in first-degree relatives of those with some of these disorders is consistent with the hypothesis that this index is an endophenotype for psychopathology associated with executive dysfunction. Such an interpretation is also directly in line with the goals motivating the development of the RDoC, which include identification of endophenotypes that tap into basic mechanisms spanning traditional diagnostic categories (Insel & Cuthbert, 2009).
Aims of the Current Study
The present study represents the first GWAS of antisaccade performance, here defined as error rate reflecting the proportion of trials where a prosaccade was generated in response to fixation target movement. Our GWAS was carried out in a general population sample comprising twins and their parents who underwent a psychophysiological assessment as participants in the Minnesota Twin Family Study (MTFS; Iacono et al., 2014; Iacono & McGue, 2002; Keyes et al., 2009). Because participants in our sample were members of twin families, our analyses began with biometric modeling designed to examine the heritability of antisaccade error in the GWAS sample. This analysis provided a benchmark against which to evaluate the amount of variance accounted for in the same sample by the molecular genetic variants.
This biometric evaluation was followed by a genome-wide complex trait analysis (GCTA; Yang, Lee, Goddard, & Visscher, 2011), a whole genome test that determined the degree to which the genetic variants used in the GWAS described below accounted for phenotypic similarity in antisaccade performance—in other words, GCTA provided a molecular genetic equivalent of an additive biometric model of heritability. GCTA was followed by a GWAS carried out on over 527,000 single nucleotide polymorphisms (SNPs), providing an indication of the degree to which each SNP was associated with antisaccade error rate. Next, we examined a set of 1,180 candidate SNPs previously identified as being of potential interest in recent meta-analyses of genetic studies of disorders such as alcohol and drug dependence, cocaine abuse, smoking and nicotine dependence, ADHD, schizophrenia, bipolar disorder, and major depression, or related phenotypes such as heavy drinking or excessive consumption, the personality characteristic of excitement seeking, and antisaccade-related SNPs that were part of those investigated by COGS (Greenwood et al., 2011) in relation to the antisaccade error rate in the current study. We also examined SNPs that had been implicated in linkage analyses in Greenwood et al. (2013). It is worth pointing out here that GWAS is predicated on the notion of an additive genetic model where thousands of common variants are thought to affect the trait in question. Thus, there is an implicit assumption that the endophenotype in question is dimensional in nature, here implying that antisaccade error rates perhaps index a dimension of psychosis.
Following these SNP-based tests, we undertook a series of gene-based tests. First, we undertook a gene-based genome-wide association study to examine the degree to which the aggregated effect of SNPs within a gene was associated with antisaccade error. This was followed by evaluation of the strength of specific candidate genes that, based on the literature, might reasonably be expected to be associated with brain neurotransmitter and neuromodulatory systems, psychiatric disorders associated with antisaccade error (in particular schizophrenia), or antisaccade error itself. Thus, our overall approach involved exploring plausibly relevant SNP and gene candidate sets, from an atheoretical GWAS perspective and motivated as well by findings from prior research.
Method
Details of the method, including the rationale that guided the development of this project and information regarding how antisaccade error was related to the other 16 endophenotypes assessed in these same participants, can be found in Iacono et al. (2014).
Participants
Data for the current study were collected as part of the MTFS, a large population-based longitudinal study of same-sex twins and their parents. Antisaccade data from twins was obtained at the age-17 assessment because all twins had in common being assessed at this age. In addition, the antisaccade performance of the parents of the twins was used for the subset of parents (all available fathers and the mothers of twins from the younger cohort) who were tested in our psychophysiology laboratory. Parents (including stepparents) ranged in age from 28–66 years old at the time of their assessment. Written informed consent or assent was obtained from all participants and guardians for both the psychophysiological testing session, and the collection and use of molecular genetic data.
Antisaccade Task
The antisaccade task was performed as part of an oculomotor battery. It was always preceded by a prosaccade task that required subjects to look in the direction of the target when it moved at unpredictable intervals from the central fixation point to either side. This ordering was carried out to strengthen the prepotent tendency to generate a prosaccade during the antisaccade task. During the antisaccade task, subjects were seated in front of a computer screen with a chin rest to support their head at a distance 82 cm away from the monitor. A circle subtending 0.4° of visual arc on the screen was used as a fixation point that lasted between 1.5–2.5 s. This fixation disappeared and a response cue, lasting 1.5 s, appeared approximately 6° to either side of the fixation point. Subjects were told to respond to the cue by moving their eyes to approximately the same location on the opposite side of the screen, following which the central fixation target would reappear on the screen, thus signaling the beginning of the next trial.
The number of trials administered for this task was increased from 8 to 20 during the course of this longitudinal study. To accommodate this change in study protocol, yet maximize sample size, we used data from both versions, treating task length as a covariate in our statistical analyses. An overwhelming majority of the subjects (78%) saw the 20-trial version of the task.
The corneal reflection technique was used to collect eye movement data. In this method, an infrared (IR) light source mounted on empty spectacle frames was reflected off the cornea and detected by a pair of sensors on either side of the IR source. An Eye Trac Model 210 (Applied Science Laboratories) monitoring system, with resolution of 0.25° of visual angle and time constant of 4 ms, was used. Eye movement was also measured using electrooculography (EOG) via two pairs of Ag/AgCl electrodes, with one pair placed above and below the eye to measure vertical eye movement, and another pair near the outer canthi of each eye for horizontal movements. An electrode on the shin served as ground. Data from these electrodes provided information on eye blinks and other artifacts that were not readily observable in the IR data. EOG signals were amplified to 5,000 times using Grass Model 12A Neurodata recording systems and filtered with a pass-band of 0.01 to 30 Hz (half-amplitude). All data were digitized at 256 Hz to 12 bits resolution.
For each trial in a given subject’s record, a trained rater determined the nature of the subject’s response from the IR and/or horizontal EOG data, categorizing each as a correct response (subject’s first response was to produce an antisaccade), incorrect response (the subject produced a prosaccade), self-corrected error (the subject’s initial eye movement was a prosaccade toward the target but then corrective antisaccade is made), no response (subject does not respond at all), or an anticipatory response (subject responds before the trial begins). Responses were scored using an in-house scoring program (Malone & Iacono, 2002). Individual trial data were not used if the direction of the response could not be ascertained by either the IR or EOG record (e.g., signals too noisy, excessive eye movements other than blinking, equipment malfunction, etc.) and was then coded as an “other” response. Incorrect responses and self-corrected errors were considered errors, while trials on which subjects did not make a response or made an anticipatory response were not counted as errors or scorable trials. The proportion of direction errors served as our measure of performance (number of errors/number of scorable trials). Operationalizing the dependent measure in this manner also helped us account for the differences in task length for the different versions of the task we used (8 or 20 trials).
We started with an initial sample of 4,595 with usable antisaccade task data and who had molecular genetic data and were Caucasian as well (Iacono et al., 2014). Participants typically spontaneously corrected themselves when making an errant response, indicating that their having first made a saccade in the direction of the cue was not due to forgetting the task instructions or to a failure to understand the task in the first place. However, 16 subjects with error rates of 100% made no self-corrective responses and were excluded from analyses. Of note, an additional 25 participants had error rates of 100% but were included because they made self-corrective adjustments. Thus, unlike the excluded 16, they clearly understood the nature of the task. Subjects with fewer than seven scorable trials were also excluded, resulting in another 21 being eliminated from these analyses. Three subjects were missing percent error information and original data, and were excluded. In addition, 86 participants were excluded for the following reasons: equipment failure; head trauma with loss of consciousness leading to hospitalization, or lasting more than a day; use of psychoactive medications, alcohol, or illicit drugs on the day of testing; lack of valid molecular genetic data (for details, see following section). In total, 126 subjects were excluded leading to a final sample of 4,469 study participants (MZ [monozygotic] twins = 1,712; DZ [dizygotic] twins = 917; mothers = 593; fathers = 1,170; stepmothers = 4; stepfathers = 73; males = 2,492; females = 1,977).
Molecular Genetic Data
We used the Illumina Human660W-Quad array (Illumina, Inc., San Diego, CA; see Miller et al., 2012, for details) for genotyping primarily of blood-based DNA; saliva samples were used in a small fraction of cases where subjects refused a blood draw. For further details on genotyping and quality control, see Miller et al. (2012) and Iacono et al. (2014). As the MTFS sample is mostly Caucasian (representing the ethnic distribution of the state of Minnesota), to preclude ethnic-related differences in allele frequencies, which may present problems for genetic analyses, we used only Caucasian subjects for this study. Further, to prevent any other differences due to ethnic variation, a principal component analysis (PCA) of genotypes was conducted using EIGENSTRAT (Price et al., 2006), and the first 10 components identified from the PCA were regressed out of all molecular genetic analyses as well—a common procedure used to eliminate minor sources of ethnic variation in genotypes. In addition to this, age, gender, generation (parent vs. child), cohort (enrichment vs. nonenrichment), and task length were used as covariates in all analyses.
Statistical Analyses
Biometric heritability
To investigate whether the proportion of errors was heritable, we used standard biometric models. These models make use of the hypothetical degree to which people in families are genetically related to estimate the phenotypic variance in a trait that is attributable to genetic and shared environmental influences (both of which act to make people in families more similar; labeled A and C effects, respectively, in such models) and nonshared environmental influences (which act to make them phenotypically dissimilar; labeled E in such models). In twin families, MZ twins share 100% of their genetic material in common compared to the 50% on average shared by DZ twins. If a trait is heritable, the MZ twin correlation should be approximately twice that of the DZ twins; in contrast, if the DZ twin correlation is greater than half the magnitude of the MZ twin correlation, this suggests that shared environmental experience is contributing to twin similarity. We fit an ADE model (D for dominance) to the twin data as well.
SNP heritability
To estimate the amount of phenotypic variance in antisaccade performance accounted for by all SNPs together simultaneously, we used GCTA (Yang et al., 2011). GCTA treats the effects of all the SNPs as random in a mixed linear model using the restricted maximum likelihood estimation method. The family structure of the sample is taken into account by incorporating a genetic relationship matrix (GRM) that indexes the degree of genetic relatedness in the sample based on shared combinations of SNPs and their frequencies in the mixed linear model. Typically GCTA is carried out on unrelated people to determine the degree to which those who are phenotypically similar share SNPs in common. There is no absolute standard as to what constitutes unrelatedness. Yang and colleagues recommend using several thresholds of relatedness and looking for stability of estimates across them (Yang, Lee, Goddard, & Visscher, 2013). We used .025, .05, and .10, which eliminates all but distant relatives. For instance, a relatedness coefficient of .025 corresponds approximately to third or fourth cousins. By convention, an arbitrary GRM cutoff of .025 is commonly used. If no cutoff is used, then the entire sample can be included, and we provide this estimate as well. We repeated the analysis with the three subsamples created by filtering the sample in this way with an alternative representation of the GRM that is based on weighting SNPs to account for linkage disequilibrium (LD) among them, which may inflate GCTA estimates (Speed, Hemani, Johnson, & Balding, 2012). To accomplish this, we used the LDAK software, which weights SNPs by local LD patterns.
We also conducted a variant of the GCTA model that incorporates the C or shared environment effect (Yang et al., 2013). This is advantageous in that it allows the use of the entire sample of related people. The amount of phenotypic variance accounted for when no GRM cutoff is specified should roughly equal the A (additive genetic) and C (shared environment) effects from the biometric model. If the estimates provided by this model match those of the raw GRM model (and the A effect from the biometric models), then this would provide indirect evidence that the shared environment (or C from biometric models) does not affect the endophenotype.
SNP effects: Genome-wide scan
To investigate whether any of the 527,829 successfully genotyped SNPs exerted a significant effect on the antisaccade error rate, we used generalized least squares regression. Analyses took into account the correlated nature of the data and family structure, using an R package, rapid feasible generalized least squares (RFGLS; Li, Basu, Miller, Iacono, & McGue, 2011). Each SNP was coded in terms of the number of minor alleles. We used a conventional GWAS statistical threshold of p < 5 × 10−8 for determining whether individual SNP effects were significant.
SNP effects: Candidate SNPs
In addition to this GWAS providing a broad atheoretical test of the effect of all 527,829 SNPs, we tested 1,180 SNPs identified from a variety of publications that examined SNPs in relation to various disorders such as schizophrenia, depression, substance use, etc. Further, we examined SNPs that in past work have been associated with antisaccade error (e.g., Greenwood et al., 2011). SNPs not on the Illumina array were imputed using minimac (Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012) with reference CEU haplotypes from the 1000 Genomes Project (2012) after having been prephased with BEAGLE (http://faculty.washington.edu/browning/beagle/beagle.html). Online supporting information Table S1 lists the candidate SNPs we tested along with the references they were drawn from. To account for multiple testing, we used a Bonferroni-corrected threshold of 4.24 × 10−5 (i.e., .05/1180). In addition to this, we examined a region on Chromosome 3 that has been linked to antisaccade error in studies of schizophrenia families (Greenwood et al., 2013).
Gene effects: Genome-wide scan
VEGAS (versatile gene-based association study) analysis software (J. Z. Liu et al., 2010) was used to assess the degree to which SNPs in 17,601 autosomal genes and their surrounding regions were associated with antisaccade error. VEGAS utilizes the p values obtained from RFGLS, adjusts for LD among SNPs related to particular genes by using simulations from the multivariate normal distribution, and yields a gene-based test statistic and a corresponding p value for each gene. It also outputs a test statistic and p value for the best SNP from each gene. These analyses used a Bonferroni-corrected threshold of p < 2.84 × 10−4.
Gene effects: Candidate genes
Next, we focused on 204 candidate genes selected from the NeuroSNP database (https://zork5.wustl.edu/nida/neurosnp.html). The selected set covered specific neurotransmitter systems including dopamine, noradrenaline, acetylcholine, GABA, glutamate, and serotonin, nicotine, drug, and alcohol-related genes, and endocannabinoid and opioid genes. For this list, the significance threshold we utilized was 2.45 × 10−4 (≈ 0.05/204). To link up with prior findings in the literature, especially given the strong association between antisaccade error and schizophrenia, we examined results for the 921\fn1\ schizophrenia-relevant candidate genes identified by the Consortium on the Genetics of Schizophrenia (see supporting information Table S3) using a p value threshold of 5.43 × 10−4 (≈ 0.05/92). These genes were chosen by COGS based on functional relevance to schizophrenia and related phenotypes, or having been implicated in published linkage, association, and model organism studies (see Greenwood et al., 2011). Finally, we examined 10 candidate genes that have been specifically associated with the antisaccade response using the Bonferroni-corrected threshold of p ≤ .005.
Results
Figure S1 in the supporting information depicts the distribution of antisaccade error rates adjusted for all covariates, which was somewhat positively skewed. The mean proportion of errors was .41 for all 4,469 participants (SD = .21).
Heritability from Biometric Models
Table 1 characterizes familial similarity for antisaccade performance. As can be seen, the MZ twin correlation for proportion of errors (after accounting for covariates) was more than twice that seen in the other pairings of first-degree relatives presented in Table 1, consistent with expectation for a genetically influenced trait. Biometric analyses yielded similar parameter estimates for both the twin-family and twin data (see Table 2\t2\) and indicated that about half of the variation in the antisaccade response was affected by additive genetic effects and the remaining half by unique environment or measurement error. Shared environment had no impact on the antisaccade error response. An ADE model was also fit using the twin data, but did not produce a significant effect for D. Confidence intervals included 0, and D could be constrained to 0 without significantly degrading model fit (all likelihood ratio test p values > .05).
Table 1.
Correlations Among Family Members for Antisaccade Proportion of Errors
| Family member | Correlation |
|---|---|
| Mother-father | .045 |
| Offspring-mother | .235 |
| Offpsring-father | .175 |
| MZ twins | .525 |
| DZ twins | .186 |
Table 2.
ACE Model Fitting Results Characterizing the Heritability of Antisaccade Error
| Data | A | C | E |
|---|---|---|---|
| Family | .489 (.432–.534) | .000 (.000–.034) | .511 (.466–.558) |
| Twins | .510 (.419–.557) | .000 (.000–.076) | .490 (.443–.541) |
Note. Point estimates of the corresponding variance components and 95% confidence intervals are given in parentheses. These are standardized and sum to 1. Data = whether the model was estimated based on the entire family or only the MZ and DZ twins; A = additive genetic influence; C = common or shared environmental influence; D = dominance influence; E = unique or unshared environmental influence.
SNP Heritability
The combined effect of all SNPs, as estimated in GCTA, accounted for almost half of the variance in the antisaccade response regardless of the genetic relatedness of those included in the sample and the GCTA method used (see Table 3\t3\). As expected, standard errors tended to be larger at the smaller cutoffs, reflecting the smaller sample size, due to restrictions on relatedness. The GCTA model with a parameter for C (shared environmental influences) yielded an estimate of .502, almost identical to that for the model without C, and, in turn, similar to the biometric model estimate of .48 for A. Thus, combining a biometric and molecular genetic perspective, antisaccade error appears to be a polygenic trait influenced by the additive effects of genes; shared environment appears to play no role.
Table 3.
SNP Heritability of Antisaccade Proportion of Errors from GCTA Analyses
| \tch\Threshold | ||||
|---|---|---|---|---|
| .025 | .050 | .100 | None | |
| Unweighted | .433 (.172) | .467 (.167) | .439 (.166) | .503 (.023) |
| Weighted, all | .540 (.224) | .499 (.216) | .468 (.215) | – |
| Family C | – | – | – | .502 (.041) |
Note. Sample sizes range from 2,045 to 2,112 for the three subsets and equaled 4,469 for the full sample. Point estimates of variance accounted for at each threshold are provided along with standard errors in parentheses. Threshold = the genetic relatedness threshold used for selecting unrelated individuals; None = no threshold was imposed and all subjects were included; Unweighted GRM = raw GRM; Weighted GRM = weights based on LD patterns to discount those SNPs in high LD (Speed, Hemani, Johnson, & Balding, 2012). This is not used in the full sample, because the method was designed for samples of unrelated individuals or samples containing a small number of large pedigrees (Doug Speed, e-mail communication, May 4, 2014). Family C = all subjects while simultaneously modeling shared environmental influences.
SNP Effects: Genome-Wide Scan
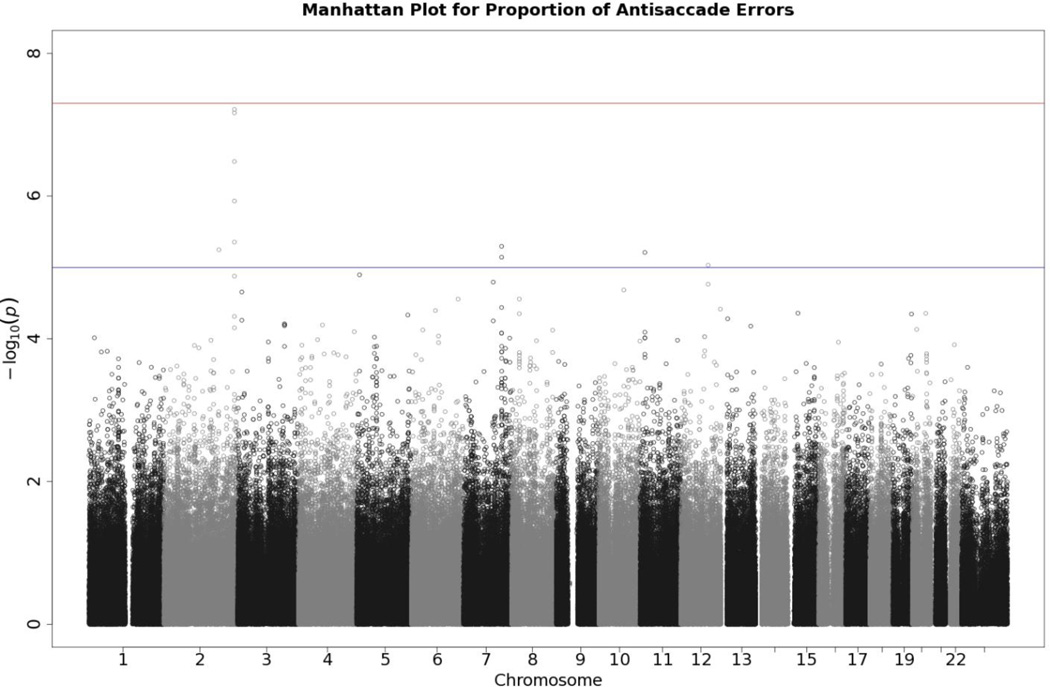
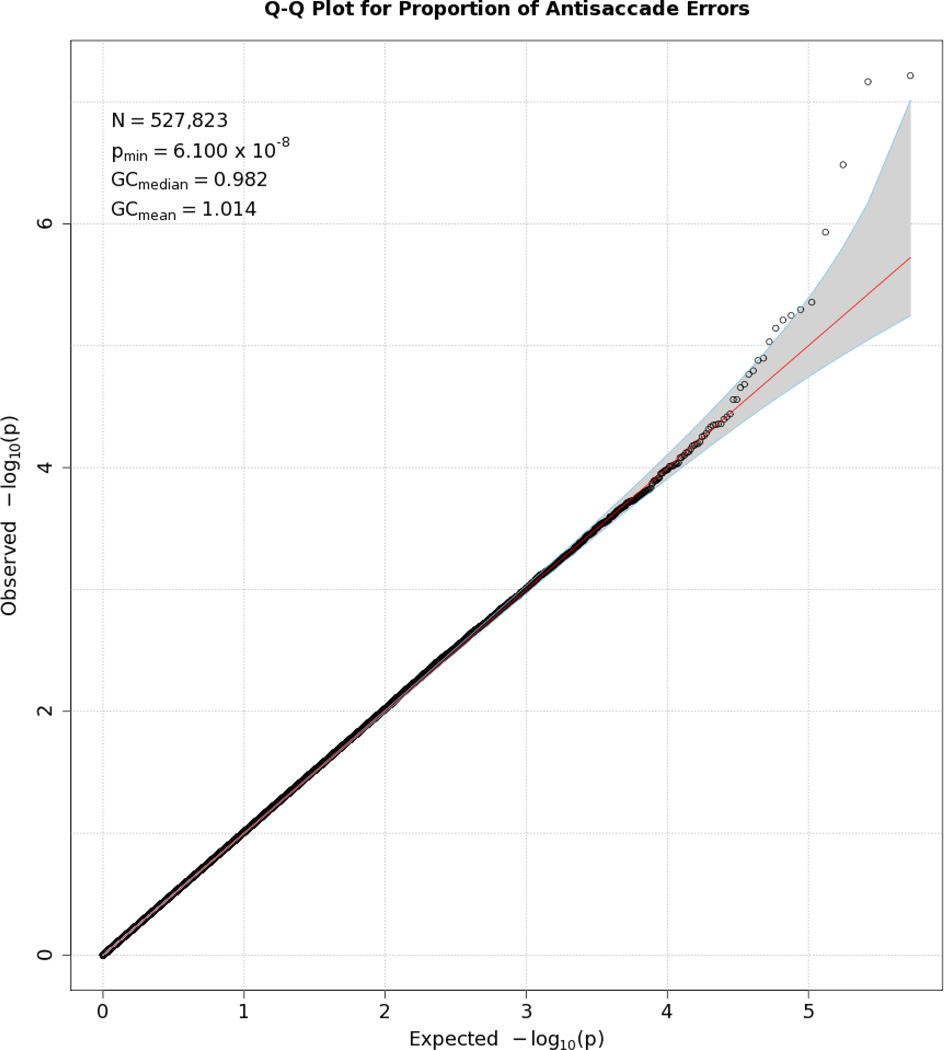
Figures 1 and 2, depicting Manhattan and Q-Q plots, respectively, summarize the results of the GWAS analysis. No single SNP crossed the Bonferroni-corrected threshold for statistical significance of 5 × 10−8 for the antisaccade response. The Manhattan plot shows several SNPs with elevated p values on Chromosome 2, albeit that they are not statistically significant. The Q-Q plot shows the expected distribution of p values for SNPs plotted against the actual observed p values. A few SNPs deviated from expectation towards the extreme end of the Q-Q plot distribution, but no anomalies were observed, suggesting that no artifacts (e.g., population stratification) affected the results. This is confirmed by the genomic control statistics appearing in the inset to Figure 3\f3\, which are close to 1.
Figure 1.
Manhattan plot of individual SNP associations with antisaccade error rates. Manhattan plots depict the distribution of −log10(p values) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p values. The horizontal upper line indicates the genome-wide significance level (5E-08). The horizontal lower line indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 2.
Q-Q plot for SNP associations with antisaccade error rates. The line bisecting the graph gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. Q-Q plots in GWAS give the observed p values against the expected p values under the null distribution of no association, although the additive inverse of the common log of p values (−log10[p value]) is used in order to emphasize small p values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p values should conform closely to their expected values, falling on or very close to a 45° line, which is plotted in the center.
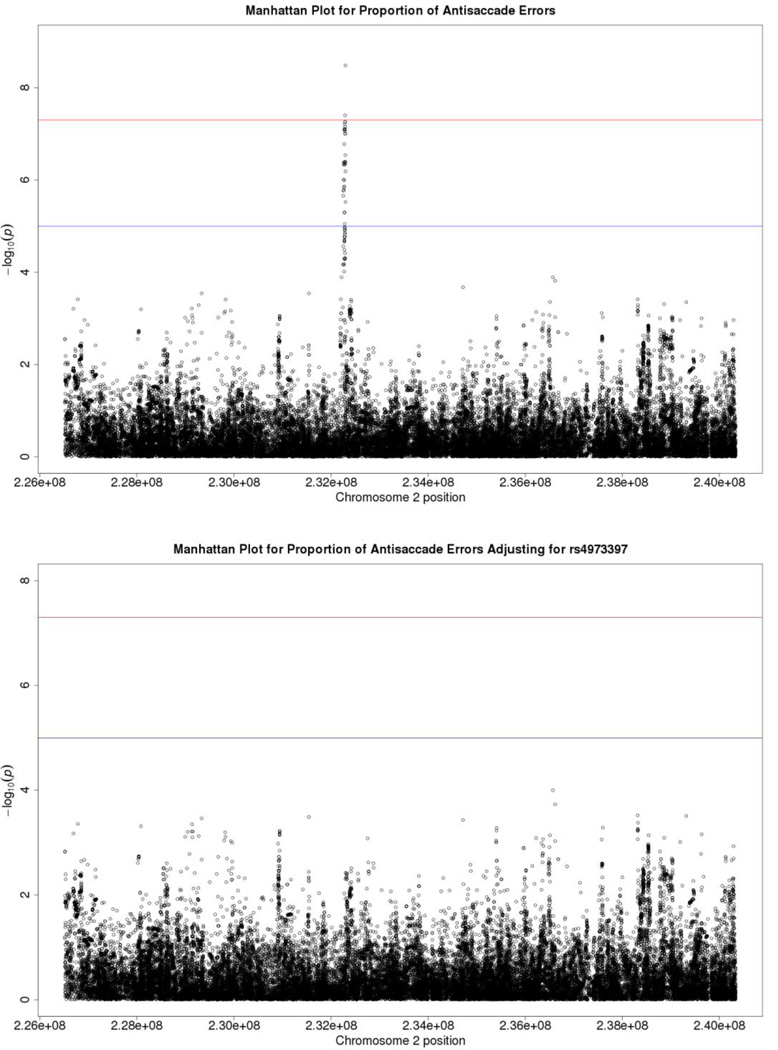
Figure 3.
Manhattan plots from GWAS of imputed SNPs on Chromosome 2 before and after covarying out rs4973397. The top panel depicts p values before covarying out rs4973397, while the bottom panel depicts p values after doing so. Manhattan plots also depict the distribution of −log10(p values) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p values. The horizontal upper line indicates the genome-wide significance level (5E-08). The horizontal lower line indicates E-05, which is sometimes used to indicate “suggestive” significance.
Table 4 lists SNPs with p values less than 1 × 10−4, corresponding to the data presented in Figure 1. In addition to rs4973397, which is located in the untranslated region of LOC729898, rs12998237 (in the untranslated region of gene B3GNT7) came very close to statistical significance. More strikingly, though, as Figure 2 shows, a number of SNPs among those with the smallest p values were on Chromosome 2, and Table 4 confirms that 6 of the top 10 SNPs were on Chromosome 2. From the base pair positions of these SNPs on Chromosome 2, it is evident that all are located in the same region, suggesting that these SNPs may be acting in concert. To further explore this possibility, we added rs4973397 as a covariate to RFGLS and reran our analyses for the other SNPs on Chromosome 2—effectively testing whether any of the markers in the same region on Chromosome 2 had an effect that was statistically independent of rs4973397. If the other markers were reduced to statistical nonsignificance after removing the effect of rs4973397, this would suggest that all these markers on Chromosome 2 were acting together. Results indicated that this was indeed the case in that the p values of all nearby markers were reduced to nonsignificance (as can be seen in Table 5\t5\) when accounting for the contribution of rs4973397. Only one marker, rs1840108, was affected little by the adjustment; it was 50 Mb away from rs4973397. The finding that all p values of SNPs in this region of Chromosome 2 become nonsignificant after adjustment suggests that there is only one effect in the region.
Table 4.
SNPs Associated with Antisaccade Error Rate with p Values Less than 10−4
| SNP | Chr | Position | Allele1 | Alelle2 | Beta | SE | t | p | Gene | Class |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4973397 | 2 | 232291471 | G | A | 0.031 | 0.006 | 5.425 | 6.10E-08 | ||
| rs12998237 | 2 | 232276651 | G | A | 0.031 | 0.006 | 5.405 | 6.83E-08 | ||
| rs2290130 | 2 | 232263127 | A | G | 0.029 | 0.006 | 5.114 | 3.28E-07 | B3GNT7 | multiple |
| rs13030174 | 2 | 232271284 | C | A | 0.028 | 0.006 | 4.867 | 1.17E-06 | ||
| rs1868455 | 2 | 232278400 | G | A | 0.023 | 0.005 | 4.597 | 4.40E-06 | ||
| rs13240504 | 7 | 126089319 | A | G | 0.032 | 0.007 | 4.568 | 5.06E-06 | GRM8 | intron |
| rs1840108 | 2 | 181366251 | A | C | 0.040 | 0.009 | 4.544 | 5.66E-06 | ||
| rs2028162 | 11 | 16247475 | G | A | −0.025 | 0.006 | −4.526 | 6.17E-06 | SOX6 | intron |
| rs13235082 | 7 | 126099842 | A | C | 0.032 | 0.007 | 4.493 | 7.20E-06 | GRM8 | intron |
| rs763564 | 12 | 91024916 | A | G | 0.031 | 0.007 | 4.438 | 9.29E-06 | ||
| rs3857348 | 5 | 10297847 | A | G | −0.022 | 0.005 | −4.371 | 1.27E-05 | CMBL | intron |
| rs4621162 | 2 | 232284911 | G | A | 0.022 | 0.005 | 4.361 | 1.32E-05 | ||
| rs4368910 | 7 | 98439700 | A | G | −0.022 | 0.005 | −4.319 | 1.60E-05 | ||
| rs2520497 | 12 | 91022310 | G | A | 0.033 | 0.008 | 4.303 | 1.72E-05 | ||
| rs1913754 | 10 | 82423687 | G | A | 0.023 | 0.005 | 4.261 | 2.07E-05 | ||
| rs9310429 | 3 | 13711684 | G | A | 0.033 | 0.008 | 4.247 | 2.21E-05 | LINC00620 | intron |
| rs11135869 | 8 | 25335354 | G | A | 0.022 | 0.005 | 4.196 | 2.77E-05 | CDCA2 | intron |
| rs583297 | 6 | 153349813 | G | A | −0.021 | 0.005 | −4.196 | 2.77E-05 | RGS17 | intron |
| rs10254771 | 7 | 125960953 | A | G | 0.029 | 0.007 | 4.134 | 3.64E-05 | ||
| rs7136250 | 12 | 131417075 | G | A | −0.021 | 0.005 | −4.121 | 3.84E-05 | ||
| rs6901674 | 6 | 78946797 | A | G | 0.110 | 0.027 | 4.110 | 4.03E-05 | ||
| rs12907899 | 15 | 34151519 | G | A | 0.020 | 0.005 | 4.091 | 4.37E-05 | RYR3 | intron |
| rs6074043 | 20 | 44781380 | A | G | 0.024 | 0.006 | 4.090 | 4.40E-05 | ||
| rs9644041 | 8 | 25321281 | G | A | 0.021 | 0.005 | 4.087 | 4.45E-05 | CDCA2 | intron |
| rs1836789 | 19 | 56489456 | G | A | −0.022 | 0.005 | −4.084 | 4.50E-05 | NLRP8 | intron |
| rs1422812 | 5 | 169069114 | G | A | 0.027 | 0.007 | 4.077 | 4.64E-05 | DOCK2 | intron |
| rs2369271 | 2 | 232289244 | G | A | 0.022 | 0.005 | 4.066 | 4.86E-05 | ||
| rs4770248 | 13 | 22794240 | A | G | 0.022 | 0.005 | 4.049 | 5.23E-05 | LINC00540 | intron |
| rs1061375 | 3 | 13679203 | A | G | 0.022 | 0.005 | 4.038 | 5.49E-05 | FBLN2 | synon codon |
| rs4236538 | 7 | 98434486 | A | G | −0.020 | 0.005 | −4.033 | 5.60E-05 | ||
| rs701265 | 3 | 152554357 | G | A | 0.028 | 0.007 | 4.011 | 6.14E-05 | P2RY1 | synon codon |
| rs6786253 | 3 | 152575924 | G | A | 0.028 | 0.007 | 4.003 | 6.37E-05 | ||
| rs2903455 | 4 | 79095407 | G | A | 0.028 | 0.007 | 4.001 | 6.42E-05 | FRAS1 | intron |
| rs6440825 | 3 | 152572930 | G | A | 0.028 | 0.007 | 3.995 | 6.57E-05 | ||
| rs1415707 | 13 | 98496595 | A | G | −0.020 | 0.005 | −3.993 | 6.64E-05 | ||
| rs6733349 | 2 | 232268312 | G | A | 0.021 | 0.005 | 3.979 | 7.02E-05 | ||
| rs2180477 | 20 | 14759169 | A | G | 0.051 | 0.013 | 3.967 | 7.38E-05 | MACROD2 | intron |
| rs1776438 | 6 | 37553497 | A | G | 0.023 | 0.006 | 3.963 | 7.52E-05 | ||
| rs2922446 | 8 | 134664999 | C | A | −0.020 | 0.005 | −3.961 | 7.57E-05 | ||
| rs10007572 | 4 | 183691547 | G | A | 0.021 | 0.005 | 3.950 | 7.95E-05 | TENM3 | intron |
| rs1595373 | 11 | 16266740 | G | A | −0.024 | 0.006 | −3.946 | 8.07E-05 | SOX6 | intron |
| rs13241603 | 7 | 125901145 | A | C | 0.027 | 0.007 | 3.939 | 8.31E-05 | ||
| rs10282703 | 7 | 125907965 | A | G | 0.027 | 0.007 | 3.939 | 8.31E-05 | ||
| rs744270 | 6 | 89299046 | A | G | 0.041 | 0.010 | 3.916 | 9.14E-05 | ||
| rs17046362 | 12 | 79483041 | A | G | −0.098 | 0.025 | −3.911 | 9.34E-05 | SYT1 | intron |
| rs17005224 | 12 | 79506862 | A | G | −0.098 | 0.025 | −3.911 | 9.34E-05 | SYT1 | intron |
| rs6881702 | 5 | 59277134 | G | A | 0.022 | 0.006 | 3.907 | 9.48E-05 | PDE4D | intron |
| rs4542191 | 1 | 18813445 | G | A | 0.027 | 0.007 | 3.902 | 9.69E-05 | ||
| rs10766314 | 11 | 16285515 | G | A | −0.024 | 0.006 | −3.902 | 9.69E-05 | SOX6 | intron |
| rs3985603 | 11 | 16283644 | A | C | −0.024 | 0.006 | −3.900 | 9.75E-05 | SOX6 | intron |
| rs486390 | 7 | 152783129 | A | G | 0.039 | 0.010 | 3.900 | 9.78E-05 |
Note. SNP = single nucleotide polymorphism; Chr = chromosome on which each SNP is located; Position = location in assembly GRCh37 (hg19). The two alleles at each locus are listed, with the minor allele listed first. Alleles are aligned to the forward strand of assembly GRCh37 (hg19). Beta = RFGLS regression coefficient; SE = standard error; df range = 4,449–4,452. Gene = nearest gene based on the NCBI database. If none is listed, then the SNP is located in an intergenic region. Class = SNP’s NCBI function class; intron = a noncoding variant.
Table 5.
Analyses Testing the Effects of Covarying Out rs4973397 on Other SNPs in Same Region on Chromosome 2 that Produced p Values Less than 10−7 in GWAS
| SNP | Chr | Position | Original p value | Adjusted p value |
|---|---|---|---|---|
| rs1840108 | 2 | 181366251 | 5.6677E-06 | 0.00001 |
| rs2290130 | 2 | 232263127 | 3.287E-07 | 0.52847 |
| rs13030174 | 2 | 232271284 | 1.17E-06 | 0.94767 |
| rs12998237 | 2 | 232276651 | 6.83E-08 | 0.39861 |
| rs1868455 | 2 | 232278400 | 4.40E-06 | 0.16977 |
| rs4973397 | 2 | 232291471 | 6.10E-08 | – |
Chr = chromosome; SNP = single nucleotide polymorphism; Position = base pair position in assembly GRCh37 (hg19); Original p value = p value from initial GWAS; Adjusted p value = p value after covarying out rs4973397 in GWAS.
The top SNP (rs4973397) in this analysis on Chromosome 2 had a p value of 6.1 × 10−8, barely short of genome-wide significance. It is possible that there are SNPs in this region of Chromosome 2 that are not on the Illumina chip that could rise to genome-wide significance if they were to be tested. To evaluate this possibility, we imputed SNPs in LD with SNPs in this apparent genomic “hot spot” to see if they rose to genome-wide significance. We repeated the RFGLS analysis on a collection of about 50,000 imputed SNPS targeting this region of Chromosome 2. The smallest p value among the imputed markers was 3.28 × 10−9 from SNP rs1868457, which is located only 9,530 bases telomeric from rs4973397 (see Table 6\t6\), and is statistically significant at a genome-wide level. As we did with the nonimputed markers, we adjusted for rs4973397 again for SNPs on Chromosome 2 with p values less than 10−7; doing so made all small p values large with the imputed markers as they did with the Illumina genotypes, indicating that there is only one source of genetic influence in the region (see Table 6). The marker with the smallest p value, rs1868457, was imputed with an r2 of .982, and it has a minor allele frequency of .311. This suggests a high degree of confidence in the result for the imputed SNP in that it is correlated highly with the observed SNP, and its allelic variant is common as well.
Table 6.
Analyses Testing the Effects of Covarying Out rs4973397 on Imputed SNPs in Same Region on Chromosome 2 with p Values Less than 10−7 in GWAS
| SNP | Chr | Position | Original p value | Adjusted p value |
|---|---|---|---|---|
| \rs12694890 | 2 | 232276553 | 8.21E-08 | 0.40291 |
| rs12998237 | 2 | 232276651 | 8.20E-08 | 0.40255 |
| rs12999051 | 2 | 232276992 | 7.86E-08 | 0.39270 |
| rs12993290 | 2 | 232279687 | 6.15E-08 | 0.45461 |
| rs4973392 | 2 | 232286552 | 6.10E-08 | - |
| rs12622248 | 2 | 232289127 | 7.80E-08 | 0.74416 |
| rs6721978 | 2 | 232290157 | 9.75E-08 | 0.88992 |
| rs12614599 | 2 | 232290640 | 3.96E-08 | 0.31522 |
| rs12614600 | 2 | 232290672 | 6.99E-08 | 0.87376 |
| rs4973396 | 2 | 232291320 | 5.47E-08 | 0.32961 |
| rs4973397 | 2 | 232291471 | 5.49E-08 | 0.22324 |
| rs16825944 | 2 | 232291866 | 9.96E-08 | 0.91805 |
| rs1868457 | 2 | 232296082 | 3.28E-09 | 0.01189 |
Chr = Chromosome; SNP = single nucleotide polymorphism; Position = base pair position in build GRCh37 (hg19); Original p value = p value from GWAS of imputed SNP data; Adjusted p value = p value after covarying out rs4973397 of imputed SNPs.
The results of this imputation analysis are depicted in Figure 3. The two Manhattan plots were drawn to amplify the x-axis region around the hot spot to facilitate visual inspection of the imputation results. The upper panel shows the effects of imputed markers without adjusting for rs4973397, and illustrates the several imputed SNPs that achieve genome-wide significance. The lower panel depicts p values for the imputed SNPs after accounting for the effect of rs4973397, and highlights the drop in p values expected if the SNPs in this region are functioning together as a unit.
SNP Effects: Candidate SNPs
Table S1 lists p values for each of the 1,180 endophenotype-general candidate SNPs from the current GWAS. None was statistically significant at 4.24 × 10−5. However, the top scoring SNP, rs1625579 with p = .00025, was identified as the strongest finding associated with schizophrenia in a large study that included over 50,000 participants (Ripke et al., 2011).
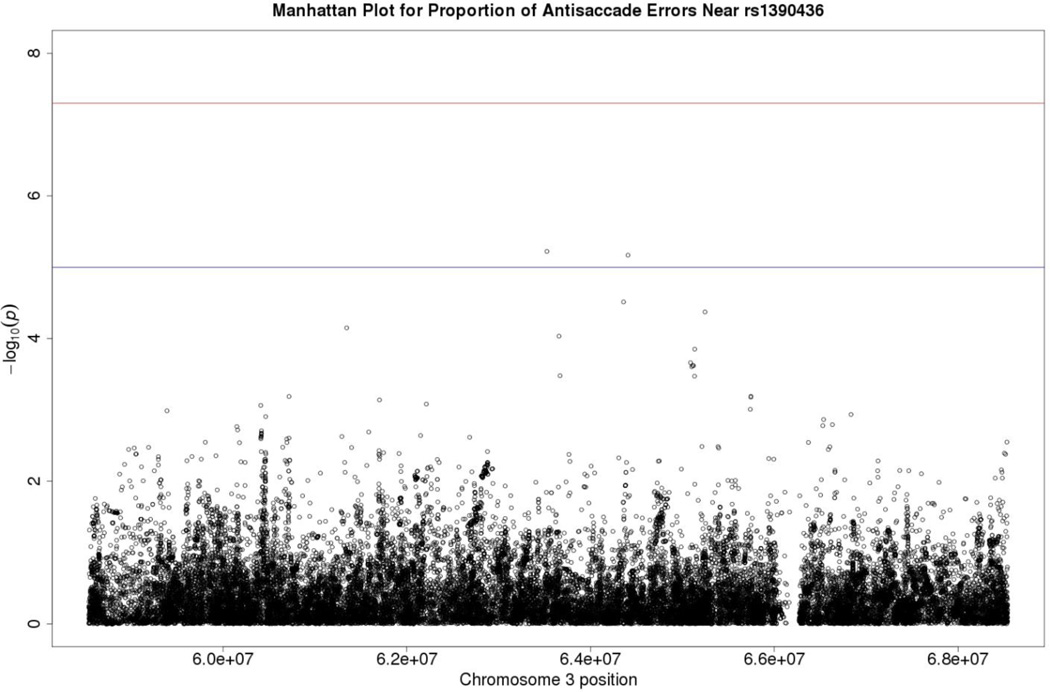
In an additional attempt to build on the existing literature, we extended the linkage analyses undertaken by Greenwood et al. (2013) in relation to schizophrenia using our general population sample. These authors reported a significant LOD (logarithm (base 10) of odds) score of 3.96 for an antisaccade task with the peak at about 87 to 88 centimorgans (cM) on Chromosome 3, at 3p14. The SNP closest to the peak was rs1390436 at 87.14 cM, which corresponds to base pair 63,563,882 on the NCBI build 37 map.2\fn2\ We followed up the Greenwood et al. finding by testing association to all of our GWAS SNPs that were within 5 Mb of rs1390436. In addition to association with our genotyped markers, we tested association with dosages imputed using the 1000 Genomes reference sample of European Caucasians. Altogether, we tested 38,897 markers for association with antisaccade errors in that 5 Mb region. The two smallest p values were from markers rs17069000 (p = 6.00e-06 at base 63,525,288) and rs78803532 (p = 6.77e-06 at base 64,407,690), neither of which is below the Bonferroni criterion of 1.29e-06 (.05/38,897) for the 10 Mb region (see Table 7\t7\ and Manhattan plot in Figure 4\f4\). Both markers have low minor allele frequencies (.018 for rs17069000 and .007 for rs78803532), warranting caution when interpreting these possible leads because the effects appear to be based on a small number of people. Furthermore, while the imputation r2 for rs78803532 was .96, for rs17069000, it was only .45.
Table 7.
Results for Imputed SNPs in Regions Used in Linkage Analyses in Greenwood et al. (2013)
| Position | SNP | Chr | Allele1 | Allele2 | R2 | Beta | SE | Test | p value |
|---|---|---|---|---|---|---|---|---|---|
| 61346176 | rs73104810 | 3 | G | A | 0.73 | .075 | .019 | 3.977 | 7.08E-05 |
| 63525288 | rs17069000 | 3 | C | A | 0.45 | −.125 | .028 | −4.532 | 6.00E-06 |
| 63657227 | rs144806580 | 3 | C | T | 0.78 | −.088 | .023 | −3.913 | 9.25E-05 |
| 64359281 | rs76018258 | 3 | C | T | 0.94 | −.142 | .034 | −4.173 | 3.07E-05 |
| 64407690 | rs78803532 | 3 | A | G | 0.96 | −.142 | .032 | −4.506 | 6.77E-06 |
| 65246133 | rs187682812 | 3 | T | C | 0.51 | −.350 | .085 | −4.098 | 4.24E-05 |
Note. Chr = chromosome on which each given SNP is located; SNP = single nucleotide polymorphism; Position = base pair location in build GRCh37(hg19); Beta = regression coefficient; SE = standard error; p values = p values for SNP being tested; Allele1 and Allele2 = major and minor alleles, respectively; R2 = imputation R2, a measure of imputation accuracy in our dataset.
Figure 4.
Manhattan plot for linkage analyses for Chromosome 3 based on Greenwood et al. (2013). Manhattan plots also depict the distribution of −log10(p values) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p values. The horizontal upper line indicates the genome-wide significance level (5E-08). The horizontal lower line indicates E-05, which is sometimes used to indicate “suggestive” significance.
Gene Effects: Genome-Wide Scan
In the genome-wide tests conducted with VEGAS, two genes proved statistically significant at 2.84 × 10−6 (see Table 8\t8\). The two genes (B3GNT7 and NCL) with the smallest p values contained rs4973397 (i.e., the same SNP with the smallest p value from RFGLS). These results are not entirely surprising given that the p values for individual SNPs from RFGLS are used to calculate gene-based p values in VEGAS.
Table 8.
Genome-Wide Gene Test VEGAS Results with p < 2.84 × 10−6
| SNP | Chr | nSNPs | Test | Gene p value | Best SNP | SNP p value |
|---|---|---|---|---|---|---|
| B3GNT7 | 2 | 18 | 207.440 | 2.80E-07 | rs4973397 | 6.10E-08 |
| NCL | 2 | 19 | 170.887 | 1.00E-07 | rs4973397 | 6.10E+00 |
Note. Chr = chromosome; SNP = single nucleotide polymorphism; nSNPs = number of SNPs in gene; Test = test statistic.
Gene Effects: Candidate Genes
Next, we examined our list of 204 gene candidates. While no single gene was statistically significant at 2.45 × 10−4, the gene with the smallest p value in this set of 204 was GRM8 (p = 0.0025; see Table S2), a protein-coding gene in the glutamatergic system located on Chromosome 7. Finally, we evaluated the list of 92 genes that had been specifically linked to endophenotypes for schizophrenia (including antisaccade performance) by COGS (Greenwood et al., 2011).3\fn3\ No gene in this set crossed the threshold for statistical significance set at 5.32 × 10−4 (see Table S3). In addition to this, we examined 10 candidate genes that have been reported to be related to the antisaccade response (see Table S4). None of the genes on this list was significant at p < .005.
Discussion
The current study was the first to undertake a GWAS of the antisaccade response—a key psychophysiological psychiatric liability index that has been frequently cited as an endophenotype for schizophrenia. We built on prior work that has used conventional biometric models and association analyses to uncover the behavioral and molecular genetic bases of the antisaccade response and affiliated disorders (Greenwood et al., 2011, 2012, 2013; Malone & Iacono, 2002). In the current study, we extended this line of inquiry by investigating the amount of phenotypic variance in antisaccade error accounted for by the combined effect of all SNPs used in GWAS (through GCTA) and by undertaking a GWAS for antisaccade error rate. We also attempted to capitalize on the a priori likelihood of uncovering an effect by investigating the significance of SNPs and genes implicated in neurotransmitter and other brain systems potentially relevant to the antisaccade response.
As a first step, we examined the biometric heritability of the antisaccade phenotype in the GWAS sample. We found that approximately 50% of the variance in this endophenotype can be attributed to additive genetic effects—a result that is in accordance with previous reports (Greenwood et al., 2007; Malone & Iacono, 2002). Next, we examined the degree to which this heritability could be captured by the GWAS SNPs using GCTA. Our results suggested that most of the biometric heritability of antisaccade error could be accounted for by the GWAS SNPs. GCTA-based SNP heritability for other endophenotypes, including those for the other psychophysiological indices examined in these special issue papers, often fall far short of capturing the genetic variance estimated from biometric methods, a phenomenon referred to as the “missing heritability” problem. Results from this study, however, suggest that this missing heritability is less of a problem for antisaccade error rate, with most of the genetic influence on this endophenotype arising from the additive effects of common genetic variants.
Next, to investigate whether any single (or a few) SNPs influenced the antisaccade response more than others, we utilized GWAS analyses where we regressed the antisaccade error rate against 527,829 SNPs. Results indicated that no SNP crossed the threshold for statistical significance. Intriguingly, however, an apparent Chromosome 2 hot spot emerged that, when further fleshed out using imputation, yielded genome-wide significant results (for rs1868457, and almost significant results for rs4973397). The significance of this finding remains uncertain, however, because this region has not been linked with executive function, eye tracking, or related disorders such as schizophrenia in prior work. We also carried out a genome-wide analysis using VEGAS software and found two potential genes of interest: B3GNT7 and NCL (in which rs4973397 was contained; rs1868457 did not appear to be located near any gene). In terms of function, B3GNT7 and NCL are protein-coding genes, but it is not evident that they are expressed in the brain. NCL or nucleolin has been implicated in ribosomal synthesis and maturation. Neither gene appears to have any obvious link to antisaccade function or psychiatric disorder.
Since GWAS analyses are atheoretical, to reduce the likelihood that we had not missed a finer-grained signal in our data, we examined p values for a list of 1,180 candidate SNPs that had been formulated from prior work. Again, none reached statistical significance. However, the SNP with the smallest p value (rs1625579, p = 0.00025 in the current study) has been noted as of interest in relation to schizophrenia (Ripke et al., 2011; Sullivan, Daly, & O'Donovan, 2012), thus tentatively tying antisaccade genetics to prior findings for schizophrenia. This SNP is located within intron 3 of the nonprotein coding gene AK094607, which contains the primary transcript for MIR137, (microRNA 137) a known regulator of neuronal development (Ripke et al., 2011).
To examine whether genes specifically related to various neurotransmitter systems and substances such as alcohol, nicotine, or drugs were related to the antisaccade response as well, we evaluated a list of 204 NeuroSNP candidate genes. Again, although none was statistically significant when we used a Bonferroni-corrected threshold, one gene from this list of 204—GRM8—had a nominally significant p value (p = .0025). Results from the GWAS analyses also indicated that one SNP from GRM8 on Chromosome 7 had a small p value (rs13240504, 5.06 × 10−6; see top 10 SNPs in Table 4). Because these correlated gene and SNP findings fall below our significance threshold, they can be viewed as suggestive at best. However, because GRM8 has been identified as of potential interest in the past, we take this opportunity to characterize its relevance. GRM8 codes for proteins in the glutamatergic system; glutamate itself is the primary excitatory neurotransmitter involved in several brain functions. Prior research has implicated GRM8 in a variety of disorders including schizophrenia (Takaki et al., 2004), alcohol dependence (Chen et al., 2009), vulnerability to nicotine dependence (Vink et al., 2009), and depression (Terracciano et al., 2010). GRM8 is not a part of the list of genes compiled by COGS (Greenwood et al., 2011), though other glutamate receptors are a part of their list.
Given the evidence supporting antisaccade task performance as an endophenotype for schizophrenia, of particular relevance was how our findings compared to those of COGS (Greenwood et al., 2011, 2012, 2013). None of the 92 candidate schizophrenia-relevant genes identified by COGS emerged as significant in our analyses. Focusing on their candidate gene findings that were specific to the antisaccade response, we also failed to find a result that surpassed our Bonferroni threshold of p = .005. The top scoring gene in this set, CACNG2 (calcium channel, voltage-dependent, gamma subunit 2), is expressed in the cerebral cortex and has been associated with bipolar disorder in addition to schizophrenia (Jan et al., 2014; Y.-L. Liu et al., 2008). Finally, we followed up the linkage study of Greenwood et al. (2013) that found a peak near 3p14 by examining GWAS SNPs in that region for their strength of association with antisaccade error. Although there were SNPs in this region that achieved p values less than 10−5 (see Figure 4), these effects were not significant.
Limitations
Antisaccade error has shown a robust association with schizophrenia, a severe mental disorder that occurs in about 1.1% of the population age 18 and older (http://www.nimh.nih.gov/health/publications/the-numbers-count-mental-disorders-in-america/index.shtml#Schizophrenia). For the MTFS, a general population study of twin children and their parents, the prevalence of schizophrenia is likely lower than 1%. Hence, it is possible that our results may have been different had our sample included more psychosis-prone individuals. The nonsignificant findings we obtained trying to follow up leads generated by COGs cannot be seen as disconfirming or as replication failures because it is not known what strength of effect to expect in our general population sample. The COGS findings derive from family data that include a large number of individuals with severe phenotypes, and thus come from a sample where genetic variants might be expected to show stronger association with an endophenotype than in a community sample with few such individuals.
Of possible concern was our exclusion of 16 individuals whose error rate was 100%, an exclusion that could have further reduced our likelihood of finding significant effects. These individuals were excluded because, in addition to producing all errors, they did not demonstrate that they understood the task by self-correcting their error on even a single trial. Instead, they generated only prosaccades, just as they did on the task that immediately preceded the antisaccade procedure. At 4,469 participants, our sample was large, and these individuals constitute less than .4% of this total; their inclusion would thus be unlikely to have much effect. Of note, we did include 25 subjects who had 100% error rates but who had self-corrected at least one error, showing that they understood and were following instructions. Hence, it was not the case that the extreme end of the distribution of error performance was eliminated from the sample.
Conclusion
Our findings support the conclusion that antisaccade error, like other complex phenotypes, is largely a polygenic trait stemming from the additive effects of many genetic variants each contributing small effects. However, the results we show here for antisaccade error are somewhat different from those found for the other endophenotypes examined in this special issue in that the GCTA analyses produced considerably higher estimates of SNP heritability for antisaccade performance than they did for the other MTFS endophenotypes, albeit still with large standard errors (Malone, Burwell et al., 2014; Malone, Vaidyanathan et al., 2014; Vaidyanathan, Isen et al., 2014; Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014). In fact, the biometric and GCTA estimates were quite similar, suggesting that much of the genetic influence on antisaccade error stems from the influence of common genetic variants. Nevertheless, given recent work indicating that rare variants may contribute importantly to schizophrenia, it is possible that rare variants contribute as well to this schizophrenia endophenotype. This possibility is further explored in the Vrieze et al. (Vrieze, Malone, Pankratz et al., 2014; Vrieze, Malone, Vaidyanathan et al., 2014) companion articles that are part of this special issue.
We obtained a genome-wide significant effect involving two genes on Chromosome 2—B3GNT7 and NCL. Replication is required to verify the relevance of this potential discovery to antisaccade error. Of note, compared to many psychophysiological measures, collecting antisaccade data is relatively easy, and the task takes only a minute or so to complete. For this reason, and given the promise antisaccade error has shown here and in other investigations as a possible endophenotype, we strongly endorse the collection of antisaccade performance in investigations that also include the collection of DNA from participants, thus enabling the acquisition of pooled samples large enough for meta-analyses examining the genetic basis of antisaccade error.
Supplementary Material
Acknowledgments
This research was supported by NIH grants DA 024417, DA 05147, DA 036216, AA 09367, and DA 13240.
Footnotes
COGS has a list of 94 genes, two of which are on the sex chromosomes and not a part of our VEGAS tests. Thus, we have a total of 92 genes.
E-mail communication with Dr. Greenwood, March 10, 2014.
This list of 92 genes shared 42 genes with a list of 204 genes. However, whether we Bonferroni-corrected p values for 92 or 48 (i.e., 92 – 44 = 48), no gene crossed the statistical significance threshold.
Supporting Information
Additional supporting information may be found in the online version of this article:
Figure S1: Distribution of antisaccade error rate residuals.
Table S1: SNP associations for endophenotype-general candidate SNPs.
Table S2: Results of VEGAS gene-based tests of endophenotype-general candidate genes.
Table S3: Results of VEGAS gene-based tests of COGS endophenotype candidate genes.
Table S4: Results of VEGAS gene-based tests of antisaccade-specific candidate genes.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- \ref\1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Greenwood TA, Swerdlow NR, Light GA, Schork NJ Investigators of the Consortium on the Genetics of Schizophrenia. Advances in endophenotyping schizophrenia. World Psychiatry. 2008;7:11–18. doi: 10.1002/j.2051-5545.2008.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins ME, Curtis CE, Iacono WG, Grove WM. Antisaccade performance is impaired in medically and psychiatrically healthy biological relatives of schizophrenia patients. Schizophrenia Research. 2004;71:167–178. doi: 10.1016/j.schres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Calkins ME, Iacono WG, Curtis CE. Smooth pursuit and antisaccade performance evidence trait stability in schizophrenia patients and their relatives. International Journal of Psychophysiology. 2003;49:139–146. doi: 10.1016/s0167-8760(03)00101-6. [DOI] [PubMed] [Google Scholar]
- Cameron IGM, Pari G, Alahyane N, Brien DC, Coe BC, Stroman PW, Munoz DP. Impaired executive function signals in motor brain regions in Parkinson's disease. NeuroImage. 2012;60:1156–1170. doi: 10.1016/j.neuroimage.2012.01.057. [DOI] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. American Journal of Medical Genetics B: Neuropsychiatric Genetics. 2009;150B:359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Eggenberger E. Neuro-ophthalmology of movement disorders. Current Opinion in Ophthalmology. 2012;23:491–496. doi: 10.1097/ICU.0b013e328358ba14. [DOI] [PubMed] [Google Scholar]
- Clementz BA. Psychophysiological measures of (dis)inhibition as liability indicators for schizophrenia. Psychophysiology. 1998;35:648–668. [PubMed] [Google Scholar]
- Curtis CE, Calkins ME, Grove WM, Feil KJ, Iacono WG. Saccadic disinhibition in patients with acute and remitted schizophrenia and their first-degree biological relatives. American Journal of Psychiatry. 2001;158:100–106. doi: 10.1176/appi.ajp.158.1.100. [DOI] [PubMed] [Google Scholar]
- Feifel D, Farber RH, Clementz BA, Perry W, Anllo-Vento L. Inhibitory deficits in ocular motor behavior in adults with attention-deficit/hyperactivity disorder. Biological Psychiatry. 2004;56:333–339. doi: 10.1016/j.biopsych.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Gooding DC, Tallent KA. The association between antisaccade task and working memory task performance in schizophrenia and bipolar disorder. Journal of Nervous and Mental Disease. 2001;189:8–16. doi: 10.1097/00005053-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Gooding DC, Tallent KA. Spatial working memory performance in patients with schizoaffective psychosis versus schizophrenia: A tale of two disorders? Schizophrenia Research. 2002;53:209–218. doi: 10.1016/s0920-9964(01)00258-4. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, Schork NJ. Initial heritability analyses of endophenotypic measures for schizophrenia: The consortium on the genetics of schizophrenia. Archives of General Psychiatry. 2007;64:1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Hardiman G. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2011;168:930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Light GA, Swerdlow NR, Radant AD, Braff DL. Association analysis of 94 candidate genes and schizophrenia-related endophenotypes. PloS One. 2012;7:e29630. doi: 10.1371/journal.pone.0029630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ, Lazzeroni LC. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeych ME, Folan MM, Luna B, Tarter RE. Impaired oculomotor response inhibition in children of alcoholics: The role of attention deficit hyperactivity disorder. Drug and Alcohol Dependence. 2006;82:11–17. doi: 10.1016/j.drugalcdep.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Haraldsson HM, Ettinger U, Magnusdottir BB, Ingason A, Hutton SB, Sigmundsson T, Petursson H. Neuregulin-1 genotypes and eye movements in schizophrenia. European Archives of Psychiatry and Clinical Neuroscience. 2010;260:77–85. doi: 10.1007/s00406-009-0032-2. [DOI] [PubMed] [Google Scholar]
- Haraldsson HM, Ettinger U, Magnusdottir BB, Sigmundsson T, Sigurdsson E, Ingason A, Petursson H. Catechol-O-methyltransferase val158met polymorphism and antisaccade eye movements in schizophrenia. Schizophrenia Bulletin. 2010;36:157–164. doi: 10.1093/schbul/sbn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth J, Mirsky J, Heuer HW, Matlin A, Jafari A, Garbutt S, Miller BL. Multicenter validation of a bedside antisaccade task as a measure of executive function. Neurology. 2012;78:1824–1831. doi: 10.1212/WNL.0b013e318258f785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer HW, Mirsky JB, Kong EL, Dickerson BC, Miller BL, Kramer JH, Boxer AL. Antisaccade task reflects cortical involvement in mild cognitive impairment. Neurology. 2013;81:1235–1243. doi: 10.1212/WNL.0b013e3182a6cbfe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SB, Ettinger U. The antisaccade task as a research tool in psychopathology: A critical review. Psychophysiology. 2006;43:302–313. doi: 10.1111/j.1469-8986.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- Iacono WG. Psychophysiologic markers of psychopathology: A review. Canadian Psychology. 1985;26:96–112. [Google Scholar]
- Iacono WG, Malone SM. Developmental endophenotypes: Indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Development Perspectives. 2011;5:239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome-wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology. 2014 doi: 10.1111/psyp.12343. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, McGue M. Minnesota Twin Family Study. Twin Research and Human Genetics. 2002;5:482–487. doi: 10.1375/136905202320906327. [DOI] [PubMed] [Google Scholar]
- Insel TR, Cuthbert BN. Endophenotypes: Bridging genomic complexity and disorder heterogeneity. Biological Psychiatry. 2009;66:988–989. doi: 10.1016/j.biopsych.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Insel TR, Cuthbert BN, Garvey M, Heinssen R, Pine DS, Quinn K, Wang P. Research Domain Criteria (RDoC): Toward a new classification framework for research on mental disorders. American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Jan W-C, Yang S-Y, Chuang L-C, Lu R-B, Lu M-K, Sunny Sun H, Kuo P-H. Exploring the associations between genetic variants in genes encoding for subunits of calcium channel and subtypes of bipolar disorder. Journal of Affective Disorders. 2014;157:80–86. doi: 10.1016/j.jad.2013.12.044. [DOI] [PubMed] [Google Scholar]
- Kattoulas E, Stefanis NC, Avramopoulos D, Stefanis CN, Evdokimidis I, Smyrnis N. Schizophrenia-related RGS4 gene variations specifically disrupt prefrontal control of saccadic eye movements. Psychological Medicine. 2012;42:757–767. doi: 10.1017/S003329171100167X. [DOI] [PubMed] [Google Scholar]
- Kelly DJ, Walker R, Norbury CF. Deficits in volitional oculomotor control align with language status in autism spectrum disorders. Developmental Science. 2013;16:56–66. doi: 10.1111/j.1467-7687.2012.01188.x. [DOI] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota Twin Family Study: Increasing the yield of twin families at high risk for externalizing psychopathology. Twin Research and Human Genetics. 2009;12:489–501. doi: 10.1375/twin.12.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono WG, McGue M. A rapid generalized least squares model for a genome-wide quantitative trait association analysis in families. Human Heredity. 2011;71:67–82. doi: 10.1159/000324839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light GA, Swerdlow NR, Rissling AJ, Radant A, Sugar CA, Sprock J, Braff DL. Characterization of neurophysiologic and neurocognitive biomarkers for use in genomic and clinical outcome studies of schizophrenia. PloS One. 2012;7:e39434. doi: 10.1371/journal.pone.0039434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-L, Fann CS-J, Liu C-M, Chen WJ, Wu J-Y, Hung S-I, Hwang T-J. RASD2, MYH9, and CACNG2 genes at chromosome 22q12 associated with the subgroup of schizophrenia with non-deficit in sustained attention and executive function. Biological Psychiatry. 2008;64:789–796. doi: 10.1016/j.biopsych.2008.04.035. [DOI] [PubMed] [Google Scholar]
- Luna B, Doll SK, Hegedus SJ, Minshew NJ, Sweeney JA. Maturation of executive function in autism. Biological Psychiatry. 2007;61:474–481. doi: 10.1016/j.biopsych.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of resting EEG activity: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12344. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Iacono WG. Error rate on the antisaccade task: Heritability and developmental change in performance among preadolescent and late-adolescent female twin youth. Psychophysiology. 2002;39:664–673. [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of P3 event-related brain potential amplitude: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12345. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A, Friedman NP. The nature and organization of individual differences in executive functions: Four general conclusions. Current Directions in Psychological Science. 2012;21:8–14. doi: 10.1177/0963721411429458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DP, Armstrong IT, Hampton KA, Moore KD. Altered control of visual fixation and saccadic eye movements in attention-deficit hyperactivity disorder. Journal of Neurophysiology. 2003;90:503–514. doi: 10.1152/jn.00192.2003. [DOI] [PubMed] [Google Scholar]
- Munoz DP, Everling S. Look away: The anti-saccade task and the voluntary control of eye movement. Nature Reviews Neuroscience. 2004;5:218–228. doi: 10.1038/nrn1345. [DOI] [PubMed] [Google Scholar]
- O’Driscoll GA, Dépatie L, Holahan A-LV, Savion-Lemieux T, Barr RG, Jolicoeur C, Douglas VI. Executive functions and methylphenidate response in subtypes of attention-deficit/hyperactivity disorder. Biological Psychiatry. 2005;57:1452–1460. doi: 10.1016/j.biopsych.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Schmechtig A, Flomen RH, Kumari V, Collier D, Makoff A, Ettinger U. CHRFAM7A copy number and 2-bp deletion polymorphisms and antisaccade performance. International Journal of Neuropsychopharmacology. 2009;12:267–273. doi: 10.1017/S1461145708009784. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Reilly JL, Frankovich K, Hill SY, Gershon ES, Keefe RSE, Keshavan MS, Sweeney JA. Elevated antisaccade error rate as an intermediate phenotype for psychosis across diagnostic categories. Schizophrenia Bulletin. 2013 doi: 10.1093/schbul/sbt132. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski JK, Borkowska A, Czerski PM, Hauser J. Eye movement disturbances in schizophrenia and a polymorphism of catechol-O-methyltransferase gene. Psychiatry Research. 2002;113:49–57. doi: 10.1016/s0165-1781(02)00245-7. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Kendler K, Craddock NJ, Lee PH, Neale BM, Nurnberger JI, Sklar P. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nature Reviews Genetics. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki H, Kikuta R, Shibata H, Ninomiya H, Tashiro N, Fukumaki Y. Positive associations of polymorphisms in the metabotropic glutamate receptor type 8 gene (GRM8) with schizophrenia. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;128:6–14. doi: 10.1002/ajmg.b.20108. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, Ferrucci L. Genome-wide association scan of trait depression. Biological Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: A genomewide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12346. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: A genome-wide association study. Psychophysiology. 2014 doi: 10.1111/psyp.12348. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Smit AB, De Geus EJ, Sullivan PF, Willemsen G, Hottenga JJ, Peltonen L. Genome-wide association study of smoking initiation and current smoking. American Journal of Human Genetics. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12349. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Vaidyanathan U, Kwong A, Kang HM, Zhan X, Iacono WG. In search of rare variants: Preliminary results from whole genome sequencing of 1325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12350. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide association studies and genomic prediction. Vol. 1019. New York: Humana Press; 2013. Genome-wide complex trait analysis (GCTA): Methods, data analyses, and interpretations; pp. 215–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.