Abstract
Eosinophilic esophagitis (EoE) is an allergic disorder characterized by infiltration of the esophagus with eosinophils. We had previously reported association of the TSLP/WDR36 locus with EoE. Here we report genome-wide significant associations at four additional loci; c11orf30 and STAT6, which have been previously associated with both atopic and autoimmune disease, and two EoE-specific loci, ANKRD27 that regulates the trafficking of melanogenic enzymes to epidermal melanocytes and CAPN14, that encodes a calpain whose expression is highly enriched in the esophagus. The identification of five EoE loci, not only expands our etiological understanding of the disease but may also represent new therapeutic targets to treat the most debilitating aspect of EoE, esophageal inflammation and remodeling.
Introduction
Eosinophilic esophagitis (EoE) is an inflammatory disorder of the esophagus histologically characterized by accumulation of eosinophils in the esophageal epithelium. Clinical symptoms of EoE include dysphagia, failure to thrive, vomiting and epigastric or chest pain. A diagnosis of EoE is made following endoscopy and biopsy upon finding isolated eosinophils in the esophagus having ruled out gastroesophageal reflux1. Multiple reports indicate a gender bias, with males predominantly affected2. The rate of co-existing atopic disease in other organs is high, with up to 70% of subjects presenting with asthma or atopic dermatitis2. EoE is considered a food allergy–related disorder based on the high rate of food allergen sensitization and a higher rate of food anaphylaxis in cases compared with the general population1, 3. Furthermore, the majority of EoE cases undergo disease remission following introduction of an elemental formula diet that lacks allergens. Experimental modeling of EoE in mice has demonstrated a key role for adaptive immunity and Th2-cell cytokines (especially IL-5 and IL-13) in the disease process and a strong connection between allergic sensitization and inflammation in the respiratory tract and skin4. The stringent diagnostic criteria for EoE, that include biopsy proven eosinophilic infiltration of the esophagus, result in a phenotypically homogenous case series that is well powered for GWAS and a potentially powerful model to study the genetics of food allergy and atopy in general.
Increasing evidence suggests a strong genetic component to EoE5. In a pediatric study, nearly 10% of parents of EoE patients had a history of esophageal strictures and ~8% had biopsy proven EoE5, 6. However, there has only been one replicated locus identified to date. Using a GWAS approach, we have previously reported genome wide association of multiple variants at the thymic stromal lymphopoetin (TSLP) locus in a cohort of EoE patients7. Here we report the results of an expanded GWAS totaling 936 cases and 4312 controls in an imputed dataset that included ~2.3M variants, identifying four novel EoE associated loci.
Results
EoE GWAS
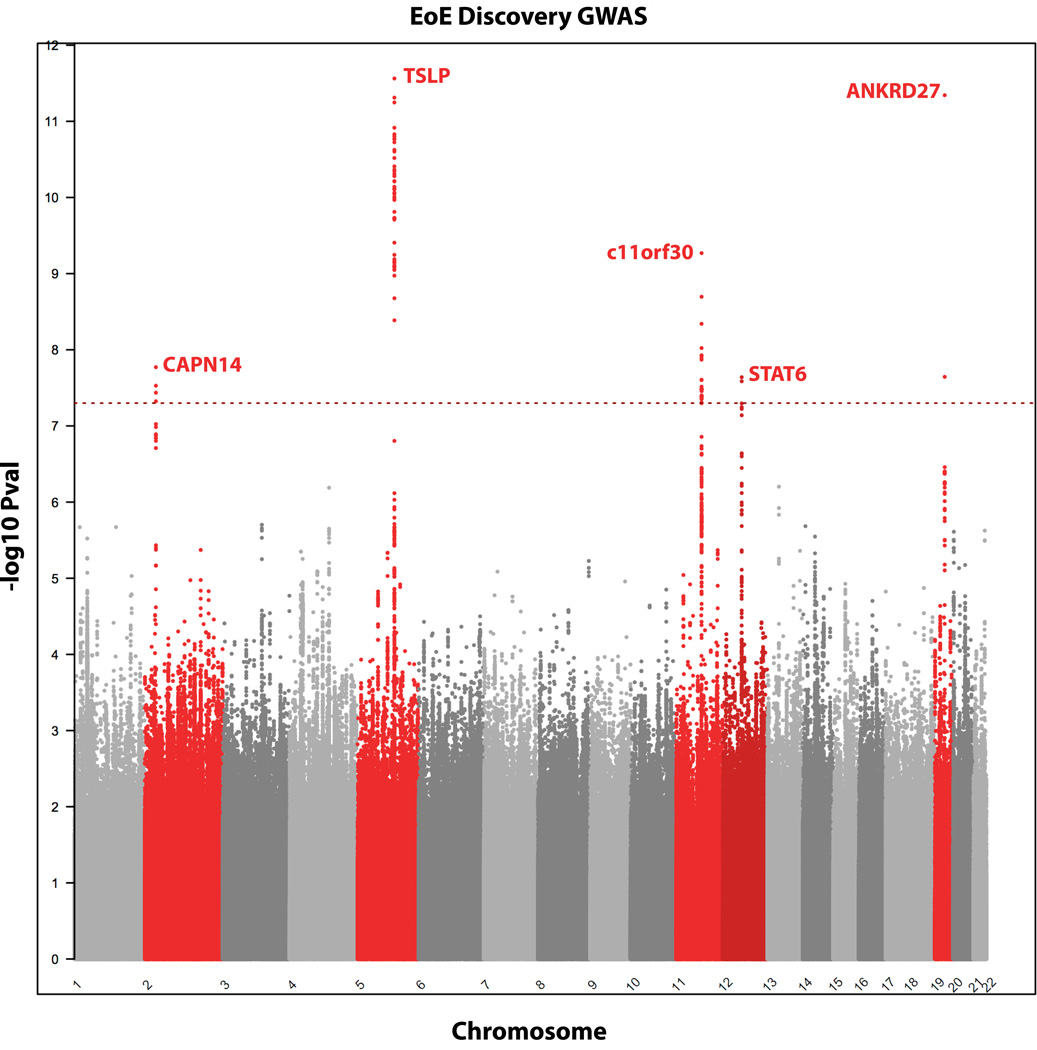
The dataset was split into discovery and replication sets based on the Illumina arrays on which the samples were genotyped (HH550/HH610 or OmniExpress). Following GWAS of the discovery cohort (n= 603 cases and 3637 controls) by logistic regression of the binary EoE phenotype adjusting for sex and the first 10 eigenvectors of the principal component analysis, five loci remained genome wide significant (cutoff P ≤ 5×10−8) following multiple testing correction (Figure 1). The same variants at the TSLP, c110rf30 and CAPN14 loci were also associated with EoE in the replication cohort (n= 333 cases 675 controls). The genome-wide significant variants mapped to the previously reported TSLP locus7 (top SNP discovery cohort rs1438673; P 1.74×10−12, OR 0.62; P replication 3.84×10−3, OR replication 0.792; P combined 1.5×10−13, OR 0.67; Supplementary Table 1, Supplementary Figure 2) a novel locus on chr11q13.5 that contains the c11orf30 gene (top SNP rs55646091; P discovery 5.83×10−10, OR 2.21; P replication 4.33×10−3, OR replication 1.584; P combined 7.67×10−11, OR 2.41; Supplementary Table 1), and a novel locus on chr2p23.1 that spans the CAPN14 gene (top SNP rs74732520; P discovery 1.69×10−8, OR 1.78; P replication 5.86×10−3, OR replication 1.56; P combined 4.16×10−9, OR 1.91; Supplementary Table 1, Supplementary Figure 4). Two further novel loci surpassed genome-wide significance in the discovery cohort that we were not sufficiently powered to replicate, a locus on chr12q13.3 that spans the STAT6 gene (top SNP rs167769, P discovery 2.29×10−8, OR 1.49; Supplementary Figure 5) and a locus on chr19q13.11 spanning the ANKRD27 gene (top SNP rs3815700, P discovery 4.54×10−12, OR 1.65; Supplementary Figure 6). Meta-analysis of the discovery and replication cohorts did not identify any additional genome-wide significant loci, however, a sixth intergenic locus upstream of NOVA1 at chr14q12 showed a trend towards association (top SNP rs8008716, P combined 6.9×10−8, OR 1.71; P discovery 2.07×10−6, OR 1.45; P replication 2.2×10−3, OR 1.57). To determine if the c11orf30 and STAT6 signals were driven by the high rates of EoE comorbidities we carried out conditional analyses at the two loci, including asthma, atopic dermatitis and allergic rhinitis status as a covariate for the c11orf30 locus and sensitization as a covariate at the STAT6 locus in a subset of 265 cases for which we had individual level comorbidity data. Residual association with EoE was detected at both loci following the conditional analyses (Supplementary Table 2).
Figure 1. Manhattan plot of the EoE discovery GWAS.
N= 603 cases and 3637 controls. -log10 Pvals on the y-axis plotted against ascending physical position on the x-axis. The dotted red line represents the genome-wide significance threshold Pval ≥ 5×10−8
The LD patterns between the associated variants at the c11orf30 locus indicated the presence of independent effects (Supplementary Figure 3). Conditional analyses in the discovery cohort on the top SNP, rs55646091, confirmed the existence of an independent effect, tagged by the rs11236791 variant, at the locus (Supplementary Table 3).
Esophageal biopsy transcriptome sequencing
RNAseq of primary epithelial cells derived from esophageal biopsy of 9 EoE patients and 3 controls confirmed expression of TSLP, c11orf30, CAPN14, STAT6, ANKRD27 and NOVA1 in esophageal epithelial cells. We detected expression of 12,407 genes out of an estimated 21,0008. Examining differential expression between cases and controls CAPN14 expression was almost 4 fold increased in EoE cases compared to controls (cases FPKM 9.82807, control FPKM 0.630785; log2(fold change) 3.96169; P 5×10−5; Supplementary Table 4). The remaining four genes showed subtle, albeit not statistically significant expression level changes. Examining other genes at the association loci, expression of both WDR36 and GALNT14 was detected but without any significant differences in cases and controls. LRRC32 was not expressed at appreciable levels (Supplementary Table 4).
Pathway analysis of the differentially expressed genes in cases and controls from the transcriptome sequencing experiment indicated an enrichment of cell cycle-related GO-terms amongst genes whose expression was decreased in cases vs controls and an enrichment of epidermis and epithelial cell development and differentiation GO-terms in the list of gene whose expression increased in cases vs controls (Supplementary Table 5).
Discussion
Since our initial report of association of TSLP variants with EoE in under 200 patients, TSLP has been associated with allergic sensitization9, 10, asthma11, 12 and allergic rhinitis13 in GWAS that required thousands of cases to achieve significance. Variants at the c11orf30 locus have been associated with seasonal allergic rhinitis13, ulcerative colitis14, Crohn's disease15, atopic dermatitis16, 17, asthma18 and allergic sensitization10, albeit with much lower odds ratios (range 1.09 in asthma to 1.22 in atopic dermatitis). Asthma, atopic dermatitis and allergic rhinitis are common comorbidities of EoE we therefore carried out a conditional analysis on asthma, atopic dermatitis and allergic rhinitis status in the EoE cases demonstrating that the observed c11orf30 association with EoE was independent of comorbidity status. The c11orf30 gene encodes, EMSY, a transcriptional regulator that was initially identified as a BRCA-2-associated protein that is amplified in human mammary adenocarcinomas19. More recently, EMSY has been identified as a central component in a novel Akt-dependent mechanism by which IFN and other growth factors regulate the expression of interferon-stimulated genes (ISGs)20. STAT6 is a key player in the IL4 pathway. STAT6 when activated by IL-4, through it's receptor IL-4R, controls the expression of GATA3, the Th2 master regulatory transcription factor, as well as the Il4 locus control region21. STAT6 has been associated with serum IgE levels22 and allergic sensitization 10, through GWAS. Conditional analysis at the STAT6 locus on sensitization status indicated the observed association with EoE was independent of sensitization. In addition to TSLP and the c11orf30 and STAT6 loci which have previously been associated with allergic / inflammatory conditions by GWAS we identified two loci that appear to be EoE specific. The chr19 locus which spans three genes, ANKRD27, PDCD5 and RGS9BP and a locus at chr2p23.1 that spans the CAPN14 gene. CAPN14 has recently been reported to be associated with EoE following a meta analysis of 736 samples23. The same study also reported associations at two additional loci at XKR6 and an intergenic region on 15q13, neither of these loci showed any evidence of association in our study (XKR6 rs2898261 P 0.663; 15q13 rs8041227 P 0.5686).
Of the three genes at the chr19 locus, ANKRD27 (also referred to as Varp), appears to be the most likely candidate, it has been shown to regulate the trafficking of melanogenic enzymes to epidermal melanocytes24, interestingly, discoloration of the esophagus has recently been reported in 90% of EoE patients25. ANKRD27 has also recently been shown to act as a kinetic inhibitor of SNARE complex formations involving VAMP726, which is involved in apical transport in epithelial cells27 and wound healing28.
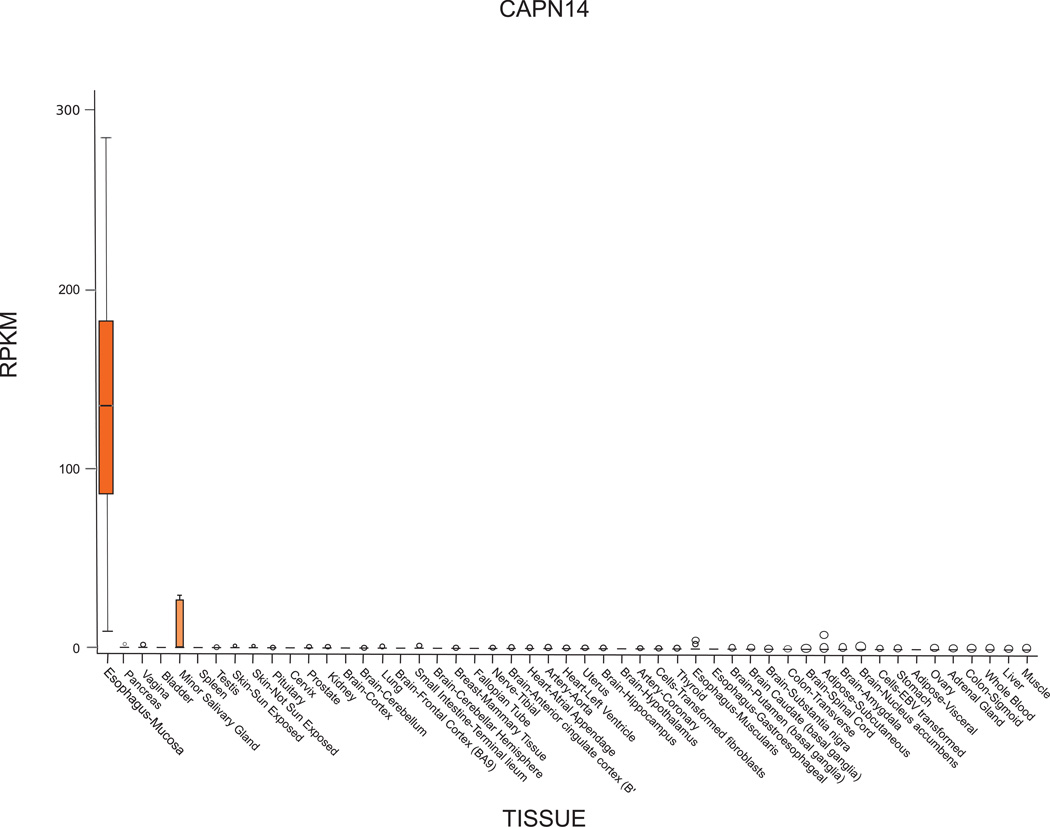
CAPN14 is a member of the calpain family. Calpains are a family of intracellular Ca2+-regulated cysteine proteases that have been shown to function in diverse biological processes including the cell cycle, platelet aggregation, and myoblast fusion through proteolytic cleavage of their substrates. Calpains include both ubiquitous and tissue-specific members29, CAPN14 shows highly specific expression, initial publications did not detect expression in any tissues tested30, however, the test panels used appear to have not included esophagus. Data from the GTEx project31 and The Human Protein Atlas32 both indicate that CAPN14 expression is limited to the esophageal mucosa (Figure 2). Phylogenetically, CAPN14 is most closely related to calpain 13 and both are divergent from the remainder of the protein family. A recent evolutionary study of the calpain family indicates that CAPN14 has undergone persistent functional divergence during evolution33.
Figure 2. CAPN14 is highly expressed in the esophagus.
transcriptome sequencing data from the GTEx project indicates CAPN14 is predominantly expressed in the esophageal mucosa (n= 106). Each boxplot represent a measure of CAPN14 gene expression, plotted on the y-axis for a given tissue, plotted on the x-axis. The measure of expression used is reads per kilobase million (RPKM). The boxplot whiskers represent the data range across the 106 replicates, with the notch in the box representing the median expression value.
The tissue specificity of calpains can result in tissue-specific disease phenotypes34, mutations in CAPN3, a muscle-specific large subunit35, result in limb-girdle muscular dystrophy, type 2A (LGMD2A)36. The expression of both CAPN8 and CAPN9 is predominantly restricted to the gastric surface mucus (pit) cells in the stomach. Neither gene has yet been implicated in human disease, however, mouse knock out models are susceptible to ethanol induced gastric mucosal injury, implicating both in gastric mucosal defense from external stressors37.
Not only does CAPN14 appear to be expressed exclusively in the esophagus, our results also indicate CAPN14 is overexpressed in EoE esophageal epithelial cells compared with controls, consistent with a gain of function. Similar results have also recently been published showing upregulation of CAPN14 in primary epithelial cells from EoE biopsies and organotypic cultures after IL-13 simulation23. CAPN14 has previously been implicated in allergy and inflammation, it has been shown to be unregulated by IL-4 stimulation38. In a recent study of an asthma mouse model, inhibition of calpain by calpeptin resulted in a marked improvement of the asthma phenotype, reversing airway hyper-responsiveness, reducing airway inflammation, bronchoalveolar lavage (BAL) fluid eosinophilia, sub-epithelial fibrosis and the inflammatory cytokine profile, including IL-4, IL-5, IL-13, transforming growth factor (TGF)-β1 and ova-specific immunoglobulin E39. Inhibition of CAPN14 activity may therefore constitute a potential therapy for the most debilitating aspect of EoE, esophageal inflammation and remodeling.
Methods
Samples
The EoE discovery cohort consisted of 603 clinically confirmed EoE patients of European ancestry and 3637 matched controls. 529 samples were collected from 5 US sites, including CHOP, UCSD, Northwestern, Stanford and UCSM, the mean age of these cases was 8.75 years. A further 74 samples were collected from AMC, mean age was 39.9. The replication cohort consisted of 333 cases and 675 controls of European ancestry. The mean age of the replication cohort cases was 8.4 SD years. All cases were biopsy proven with an eosinophils/hpf (400X) count of ≥24 on proton pump inhibitor (PPI) therapy for at least 8 weeks. The majority of EoE subjects in both discovery and replication cohorts were male making up 73% in the discovery cohort and 75% in the replication cohort. Moreover, 70% of the discovery cohort and 72% of the replication cohort had asthma, allergic rhinitis or atopic dermatitis. The study was approved by the Institutional Review Board (IRB) of the Children's Hospital of Philadelphia (CHOP). Written informed consent for participation in the study was obtained from all participants and their parents or guardians.
Genotyping
The discovery samples were genotyped on either the Illumina HumanHap550, HH610 and the replication samples were genotyped on the Illumina HumanOmni Express-12v1 arrays at the Center for Applied Genomics at CHOP.
Standard quality control parameters were applied to the dataset, samples with chip-wide genotyping failure rate < 5% were excluded. SNPs with minor allele frequencies of < 1%, genotyping failure rates of greater that 2% and Hardy-Weinberg P-Values less that 1×10−6 were excluded from further analysis.
Genetic ancestry was determined by computing principal components on the dataset using smartpca, a part of the EIGENSTRAT package, on 100,000 random autosomal SNPs in linkage equilibrium. Samples were clustered into 4 Continental ancestry groups (Caucasian, African including admixed African-American, Asian and native American / admixed Hispanic) by K-means clustering using the kmeans package in R.
Population stratification
smartPCA eigenvectors were included as covariates in a logistic regression to control for population stratification as required. To determine the genomic inflation for each case control set, we carried out an association analysis on the genotype data using plink prior to imputation. If genomic inflation exceeded 1.03, principal components were included as covariates in the post-imputation GWAS.
Duplicate samples and cryptic relatedness
pairwise IBD values were generated for all samples using the plink genome command. IBD was performed independently on the samples of Caucasians ancestry and African ancestry. A random sample from any pair with a PI_HAT value exceeding 0.3 was excluded from further analysis.
Imputation
Imputation of untyped markers (~39M) was carried out using IMPUTE2 after prephasing with Shapeit. Each chromosome was prephased separately. To prevent chip-based batch effects due to differences in variant densities, each chip type was prephased and imputed separately. Reference phased cosmopolitan haplotypes and recombination rates were obtained from the 1000 genomes project (1000 Genomes Phase I integrated variant set b37 March 2012 release). Imputation was carried out in 5Mb intervals using an effective population size of 20000 as recommended. As a measure of the overall imputation accuracy we compared the concordance between the imputed and known genotypes in the subset of SNPs for which genotyping data was available. At a call threshold of 0.9, over 99% of the imputed genotypes were called and over 96% of those were concordant with the known genotypes.
Post-imputation association analysis
Statistical tests for association were carried out using the SNPTESTv2 package. Single marker analyses for the genome-wide data were carried out using linear regression taking genotype uncertainty introduced by the imputation into account. Call threshold was set at 0.9. SNPs with an info score below 0.8 were excluded from further analysis; the score is a measure of the observed information for the estimate of the allele frequencies at each imputed SNP which is obtained by splitting the data into two components, observed and missing, the observed data likelihood is then integrated over the missing data. Combined P-values across the individual data sets were generated using both fixed-effect and random-effect meta-analyses as implemented in the metal package for the fixed effects and the RE2 model in the METASOFT package for the random effects.
Transcriptome sequencing
mRNA libraries were constructed from primary esophageal epithelial cells derived from 9 cases (55% male and 44% female; mean age 11.6) and 3 controls (33% male and 66% female; mean age 12.1) using the Illumina TruSeq RNA Sample Preparation Kit v2, according to the manufacturer's instructions with 12 unique indexed adapters. Libraries were sequenced on an Illumina HiSeq 2000, generating 7.5 Gb 100bp paired-end reads per sample. Transcripts were assembled, transcript abundances estimated and tested for differential expression between cases and controls using the cufflinks package.
Pathway analysis
Differentially expressed genes from the transcriptome sequencing experiment were separated into two lists of up or down regulated genes in the cases vs controls. Inclusion criteria included a statistically significant differential expression test (P range 5×10−5 - 0.0019) and a minimum two log2 fold-change. Enrichment of KEGG pathways, Gene Ontology (GO) terms and Functional categories (SP_PIR_KEYWORDS) was analyzed using DAVID (http://david.abcc.ncifcrf.gov/).
Supplementary Material
Acknowledgements
We thank all the participants of this study. We will also thank Christina Gussfason, Kathryn Ruymann, Alain Benitez and Rosetta Chiavacci is helping with the collection of samples; Chris Liacouras, Michele Shuker, Terri Brown-Whitehorn, Ritu Verma, and Laura Gober for identification of patients for the study. Genotyping was funded by an Institute Development Fund to The Center for Applied Genomics, Food Phight Fund for Eosinophilic Esophagitis and an Adele S. and Daniel S. Kubert Estate gift to the Center for Applied Genomics. Additional funding was provided by a U01 award from the National Institute of Health (HG006830-02). J Spergel was supported by an award from the US Department of Defense (A-16809.2) and the Stuart Starr Chair of Pediatrics. GT Furuta was supported by a grant from National Institutes of Health (K24DK100303). S Aceves was supported by an R01 award from the National Institute of Health (AI 092135).
Footnotes
Author contribution statement:
PMAS analyzed the data; MLW provided cell lines; HH, PMAS designed the experiments and wrote the paper; AC, SA, NG, KN, AJB, GTF, JMS provided samples and carried out phenotyping. All authors reviewed the manuscript.
Conflict of Interest: the authors have no conflicts of interest to declare
References
- 1.Furuta GT, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Spergel JM, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48:30–36. doi: 10.1097/MPG.0b013e3181788282. [DOI] [PubMed] [Google Scholar]
- 3.Spergel JM. Eosinophilic esophagitis in adults and children: evidence for a food allergy component in many patients. Curr Opin Allergy Clin Immunol. 2007;7:274–278. doi: 10.1097/ACI.0b013e32813aee4a. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard C, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. The New England journal of medicine. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 6.Johansson SG, et al. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004;113:832–836. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 7.Rothenberg ME, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nature genetics. 2010;42:289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 9.Hinds DA, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnelykke K, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–906. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirota T, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torgerson DG, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramasamy A, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esparza-Gordillo J, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 17.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira MA, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes-Davies L, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 20.Ezell SA, et al. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc Natl Acad Sci U S A. 2012;109:E613–E621. doi: 10.1073/pnas.1115029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual review of immunology. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 22.Granada M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012;129:840–845. e821. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kottyan LC, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Ohbayashi N, Maruta Y, Kanno E, Itoh T, Fukuda M. Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol Biol Cell. 2009;20:2900–2908. doi: 10.1091/mbc.E08-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K, et al. Narrow-band imaging magnifying endoscopy in adult patients with eosinophilic esophagitis/esophageal eosinophilia and lymphocytic esophagitis. Gastrointest Endosc. 2013;78:659–664. doi: 10.1016/j.gie.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 26.Schafer IB, et al. The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nature structural & molecular biology. 2012;19:1300–1309. doi: 10.1038/nsmb.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaineau M, Danglot L, Galli T. Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 2009;583:3817–3826. doi: 10.1016/j.febslet.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Rao SK, Huynh C, Proux-Gillardeaux V, Galli T, Andrews NW. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J Biol Chem. 2004;279:20471–20479. doi: 10.1074/jbc.M400798200. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53(Suppl 1):S12–S18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- 30.Dear TN, Boehm T. Identification and characterization of two novel calpain large subunit genes. Gene. 2001;274:245–252. doi: 10.1016/s0378-1119(01)00599-6. [DOI] [PubMed] [Google Scholar]
- 31.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 33.Macqueen DJ, Wilcox AH. Characterization of the definitive classical calpain family of vertebrates using phylogenetic, evolutionary and expression analyses. Open biology. 2014;4:130219. doi: 10.1098/rsob.130219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorimachi H, Hata S, Ono Y. Calpain chronicle--an enzyme family under multidisciplinary characterization. Proceedings of the Japan Academy Series B, Physical and biological sciences. 2011;87:287–327. doi: 10.2183/pjab.87.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorimachi H, et al. Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J Biol Chem. 1989;264:20106–20111. [PubMed] [Google Scholar]
- 36.Richard I, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 37.Hata S, et al. Calpain 8/nCL-2 and calpain 9/nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense. PLoS Genet. 2010;6:e1001040. doi: 10.1371/journal.pgen.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueta M, Sotozono C, Kinoshita S. Expression of interleukin-4 receptor alpha in human corneal epithelial cells. Japanese journal of ophthalmology. 2011;55:405–410. doi: 10.1007/s10384-011-0030-6. [DOI] [PubMed] [Google Scholar]
- 39.Aich J, Mabalirajan U, Ahmad T, Agrawal A, Ghosh B. Loss-of-function of inositol polyphosphate-4-phosphatase reversibly increases the severity of allergic airway inflammation. Nature communications. 2012;3:877. doi: 10.1038/ncomms1880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.