Abstract
Background:
Hypervariability of HCV proteins is an important obstacle to design an efficient vaccine for HCV infection. Multi-epitope vaccines containing conserved epitopes of the virus could be a promising approach for protection against HCV.
Objectives:
Cellular and humoral immune responses against multi-epitope DNA and peptide vaccines were evaluated in BALB/c mice.
Materials and Methods:
In this experimental study, multi-epitope DNA- and peptide-based vaccines for HCV infection harboring immunodominant CD8+ T cell epitopes (HLA-A2 and H2-Dd) from Core (132-142), NS3 (1073-1081) and NS5B (2727-2735), a Th CD4+ epitope from NS3 (1248-1262) and a B-cell epitope from E2 (412-426) were designed. Multi-epitope DNA and peptide vaccines were tested in two regimens as heterologous DNA/peptide (group 1) and homologous peptide/peptide (group 2) prime/boost vaccine in BALB/c mice model. Electroporation was used for delivery of the DNA vaccine. Peptide vaccine was formulated with Montanide ISA 720 (M720) as adjuvant. Cytokine assay and antibody detection were performed to analyze the immune responses.
Results:
Mice immunized with multi-epitope peptide formulated with M720 developed higher HCV-specific levels of total IgG, IgG1 and IgG2a than those immunized with multi-epitope DNA vaccine. IFN-γ levels in group 2 were significantly higher than group 1 (i.e. 3 weeks after the last immunization; 37.61 ± 2.39 vs. 14.43 ± 0.43, P < 0.05). Moreover, group 2 had a higher IFN-γ/IL-4 ratio compared to group 1, suggesting a shift toward Th1 response. In addition, in the present study, induced immune responses were long lasting and stable after 9 weeks of the last immunization.
Conclusions:
Evaluation of multi-epitope DNA and peptide-vaccines confirmed their specific immunogenicity in BALB/c mice. However, lower Th1 immune responses in mice immunized with DNA vaccine suggests further investigations to improve the immunogenicity of the multi-epitope DNA vaccine through immune enhancers.
Keywords: Vaccine, Epitope, Electroporation, Prime-Boost
1. Background
Hepatitis C virus (HCV) has positive-strand RNA genome encoding three structural proteins of Core, E1 and E2 and seven nonstructural (NS) proteins including p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B. HCV infection is a major cause of liver disease and about 3% of the world’s population are infected with the virus (1). Over 75% of infected patients develop a chronic disease, which might ultimately progress to cirrhosis and hepatocellular carcinoma (2).
Despite the recent progress in the development of new drugs for treating chronic HCV infection, several important issues still remain. Therefore, an affordable preventive vaccine provides the best long term goal for controlling the global epidemic (3).
Inducing cross-neutralizing antibodies against HCV envelope proteins may be necessary to prevent attachment and entry of circulating virus into the hepatocytes (4). However, several studies indicated that cytotoxic T lymphocytes (CTL) have crucial roles in defense against HCV (5). There are evidences that cooperative responses of CD8+ and CD4+ T-lymphocytes against HCV antigens induce strong cellular immune responses and play critical roles in natural or therapeutic viral resolution (6, 7). Multi-epitope type vaccines containing conserved B- and T-cells epitopes seem to be promising approaches to combat against infectious agents with a highly variable antigenic context (8-10). Various studies have investigated the immunogenicity and potency of multi-epitope DNA- or peptide-based vaccines in HCV infection and have evidenced the capability of these vaccines to elicit strong cellular immune responses in mice models (11-14). Multi-epitope vaccines have advantageous over those encoding the whole antigens. They could induce immune responses against a particular set of conserved and critical epitopes and prevent deleterious function of the whole antigens. They also do not have immunosuppressive regions of whole proteins that interfere with the function of protective epitopes (15, 16).
Different strategies developed to increase the immunogenicity of multi-epitope DNA and peptide vaccines include using an appropriate adjuvant, improvement of delivery and/or different vaccination regimens. Montanide ISA 720 (M720; SEPPIC, France) is an efficient human-compatible adjuvant able to enhance Th1 immune responses during HCV protein vaccination (17, 18). In vivo electroporation (EP) is a potent strategy for DNA vaccine delivery, causing efficient uptake of DNA by cells and increasing expression of desirable gene (19). Application of different vaccination regimens such as priming with multi-epitope DNA and boosting with protein or peptide has been also shown to basically increase the immune response, particularly against viruses like HIV (20) and HCV (14, 21).
2. Objectives
In this study, multiple conserved HLA-A2 and H2-Dd restricted CD8+ T-cell epitopes from E2, core, NS3 and NS5B, a T-helper (Th) epitope from NS3 and a B-cell epitope from E2 antigens of HCV were selected and designed in a single multi-epitope construct. Our study aimed to provide a comparative evaluation of BALB/c mouse immune response versus the mentioned epitopes within multi-epitope DNA and peptide contexts. Electroporation for DNA injection, application of M720 adjuvant for peptide immunization and DNA prime-peptide boost immunization regimen were further used to enhance the vaccination efficacy. The stability of the immune responses was also determined in immunized mice.
3. Material and Methods
3.1. Selection of the Epitopes
Selection of CD8+ T-cell epitopes represented in the current HCV vaccine (Figure 1) was based on three different criteria:
Figure 1. Polytope DNA Construct Containing Six Different Epitopes of HCV in Different Blocks.
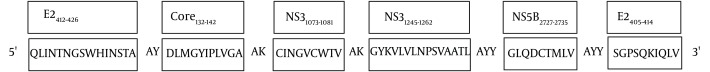
Their association with spontaneously clearance of the virus in patients with infection (6, 22-26).
Conservation of the sequence, especially between genotypes 1a and 1b, as the most frequent genotypes worldwide (27, 28).
Potential of the epitopes to induce immune responses in an HCV vaccine (22-26).
Due to a high prevalence of HLA-A2 alleles in Caucasian population (29), we initially focused on HLA-A2-restricted epitopes of HCV to design this model. Desirable epitopes were analyzed with two computer-based programs, SYFPEITI (http://www.syfpeithi.de) (30) and BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind) (31) to choose high binding HLA-A2 epitopes (Table 1). Accordingly, three immunodominant HLA-A2 restricted epitopes of core (Core132-142), NS3 (NS31073-1081) and NS5B (NS5B2727-2735), together with one H2-Dd mouse epitope of E2 (E2405-414) were selected. To enhance the immunogenicity of CD8+ T-cell responses, a Th CD4+ epitope (HLA-DR) from NS3 (NS31248-1262) was also chosen. Since clearance of HCV infection is associated with early appearance of broad neutralizing antibody responses, a neutralizing B-cell epitope from E2 (E2412-426) conserved between genotypes of HCV was considered for incorporation into the set of epitopes (25, 26, 32).
Table 1. Epitopes and Their Binding Scores Using Computer Based Programs.
| Protein | Amino Acid Position | Character (HLA) | Peptide Sequence | SYFPEITHI Score |
|---|---|---|---|---|
| NS3 | 1073-1081 | HLA-A2 | CINGVCWTV | 23 |
| NS5B | 2727-2735 | HLA-A2 | GLQDCTMLV | 24 |
| Core | 132-142 | HLA-A2 | DLMGYIPLVGA | 28 |
| E2 | 405-414 | H2-Dd | SGPSQKIQLV | - |
| NS3 | 1248-1262 | HLA-DR | GYKVLVLNPSVAATL | - |
| E2 | 412-426 | B cell epitope | QLINTNGSWHINSTA | - |
3.2. Construction of Multi-Epitope DNA Vaccine
An arrangement of the selected epitopes with suitable spacer combination was designed using two computer based modeling for peptide binding (http://imed.med.ucm.es/Tools/rankpep.html) and optimization of proteasomal processing (http://www.paproc.de/). Furthermore, to minimize creation of junctional epitopes, a suitable arrangement of epitopes was designed using two web-based algorithms of SYFPEITI and BIMAS.
According to mammalian Mus musculus codon frequency table (www .kasusa.org.jp/codon), the nucleotide sequence of the construct was further optimized to support mammalian codon usage. The Kozak sequence was added to the N-terminus of the multi-epitope construct to enhance its expression. The multi-epitope DNA (PT) was designed with suitable restriction enzyme sites (HindIII and BamHI) at the 5΄ and 3΄ ends, respectively. The PT construct was synthesized by Biamatik (Biomatik Corporation, Canada).
Constructed HCV DNA PT was digested with HindIII and BamHI restriction enzymes and inserted to the corresponding sites of expression vectors, pEGFP-N1 and pcDNA3.1 (+) (Invitrogen, Germany). The plasmid with the correct insert was detected by restriction analysis, PCR and sequencing.
pEGFP-N1 plasmid containing PT (pEGFP-PT) was purified by an alkaline lysis method (Qiagen, Plasmid Midi kit, Germany). The pcDNA-PT and pcDNA3.1 (+) plasmids were purified by ion exchange chromatography with EndoFree plasmid Purification Giga kit (Qiagen, Germany). DNA concentration was determined by analyzing the absorbance at 260 nm.
3.3. Peptides
For mice immunization and evaluation of immune responses, peptides of Core132-142 (DLMGYIPLVGA), NS31073-1081 (CINGVCWTV), NS5B2727-2735 (GLQDCTMLV), E2412-426 (QLINTNGSWHINSTA), NS31248-1262 (GYKVLVLNPSVAATL) and E2405-414 (SGPSQKIQLV) were synthesized by Biomatik with 90% purity (Biomatik Corporation, Canada).
All lyophilized peptides were dissolved in DMSO (Sigma, Germany) at a concentration of 10 μg/μL and stored at -20°C.
3.4. Expression of Multi-Epitope DNA in COS-7 Cell Line
Up to 5×104 COS-7 cells per well of a four-well plate (Griner, Germany) were incubated until the cells reached a confluence of 75%. Five micrograms of pEGFP-PT or pEGFP plasmids were mixed with LINPEI 25 KDa (10 μM; Polysciences, Europe) in HBS buffer (HEPES buffered saline) in a final volume of 200 μL and incubated at room temperature for 15 minutes. These PEI/DNA complexes were added to COS-7 cells in serum free media. Cells were incubated at 37°C in 5% CO2 for six hours, and then the medium was changed with complete MEM medium supplemented with 10% FCS. Transfection efficacy was determined with fluorescence microscopy (Nikon E200, USA) at different time intervals (24, 48, and 72 hours).
3.5. Mice
Six to eight weeks old female BALB/c mice were purchased from breeding stocks maintained at Pasteur Institute of Iran. All mice were maintained under pathogen-free conditions. All procedures were performed according to approval protocols and in accordance with recommendations for the proper use and care of laboratory animals. Five groups of 10 mice were considered for immunization.
3.6. Immunization
This was an experimental study. Immunization was performed subcutaneously (Sc) in the left footpad of five groups of susceptible BALB/c mice three times at three-week intervals. Mice of group 1 were immunized using two DNA injections with 50 μg of pcDNA-PT (in 50 μL of PBS) followed by a mix of all peptides (10 μg of each peptide per mouse) emulsified with 70% (v/v) M720. Mice of group 2 were immunized three times with formulated peptides with M720. PBS formulated with M720 was injected to group 3 mice for three times. Group 4 received empty vector (50 μg of pcDNA3.1) two times, and then PBS emulsified with 70% (v/v) M720. Group 5 received PBS three times. Groups 3, 4 and 5 were as controls. All DNA injections were performed using Electroporation (BTX Harvard apparatus, USA), during light anesthesia of mice with ketamine/xylazine solution. Vaccination schedule was shown in Table 2. Three weeks after the second and third immunizations, three mice per group were euthanized and spleens were collected. The rest of mice were killed after nine weeks of the third immunization to assess long-lasting immunity.
Table 2. Schedule of BALB/c Mice Immunization.
| Groups | 1st Immunization | 2nd Immunization | 3rd Immunization |
|---|---|---|---|
| 1 | pcDNA-PT | pcDNA-PT | Peptide + M720 |
| 2 | Peptide + M720 | Peptide + M720 | Peptide + M720 |
| 3 | PBS + M720 | PBS + M720 | PBS + M720 |
| 4 | pcDNA3.1 (+) | pcDNA3.1 (+) | PBS + M720 |
| 5 | PBS | PBS | PBS |
3.7. Cytokine Assay by ELISA
Preparation of single-cell suspension of murine splenocytes was performed according to a previously described protocol (33). Up to 2 × 106 cells /mL of pooled splenocyte from each group of mice were plated in 96-well plates in 200 μL of cell culture medium. Splenic lymphocytes were stimulated for three or five days in the presence of 10 μg/mL of synthetic peptides or 5 μg/mL of Concavalin A (Con A) as positive control. Unstimulated cells served as negative control. IFN-γ and IL-4 production levels were measured in supernatants of splenocyte cell cultures by sandwich-based ELISA using a DuoSet ELISA system (R&D, USA). The absorbance was measured at 450 nm (Anthos reader 2020, Labsystems-multiskan EX, Finland). All data was represented as mean ± SEM for each set of samples. Detection limits were 2 pg/mL and 7 pg/mL for IFN-γ and IL-4, respectively.
3.8. Antibody Assay
Pooled sera were prepared through retro-orbital bleeding the mice of each group prior to the first immunization, three weeks after the second and three and nine weeks after the third injection. ELISA was performed to determine specific IgG, IgG1 and IgG2a as described previously (33). Briefly, ELISA plate was coated with 100 μL of peptides (10 μg/mL of each peptide) diluted in 0.5M carbonate/bicarbonate buffer of pH 9.6. The plate was rinsed with PBS containing 0.05% Tween 20 (PBS-T), incubated with blocking buffer (5% skim milk in PBS) for two hours at 37°C. After washing with PBS-T, diluted sera (1:300) in PBS buffer containing 1% skim milk and 0.05% Tween-20 were added and incubated at 37°C for two hours. Plate was washed and incubated at 37°C for two hours with Horseradish Peroxidase (HRP) conjugated Goat Anti-mouse IgG, IgG1, IgG2a (1:4000; Southern Biotechnology Association, USA). Detection was performed with Tetramethylbenzidine (TMB) substrate and absorbance (OD) was measured at 450 nm.
3.9. Statistical analysis
To analysis and compare data, One-way analysis of variance (ANOVA) with post hoc HSD-Tukey test was performed using the Graph Pad Prism version 5 software. P value less than 0.05 was considered significant.
4. Results
4.1. Multi-Epitope DNA Vaccine
A DNA construct encoding four HCV immunodominant CD8+ T cell epitopes (HLA-A2 and H2-Dd restricted) from E2, Core, NS3 and NS5B, a Th CD4+ epitope from NS3 and a B-cell epitope from E2 was used as an HCV DNA vaccine candidate. Six epitopes were linked with appropriate spacer linker amino acid (Figure 1). DNA sequence with 252 bp in length was successfully cloned into the expression vector of pcDNA3.1 (+). PCR assay and restriction enzyme digestion approved correct insertion of PT. Accuracy of cloned sequence was further confirmed by sequencing analysis (data not shown).
4.2. In Vitro Expression of the Multi-Epitope Construct
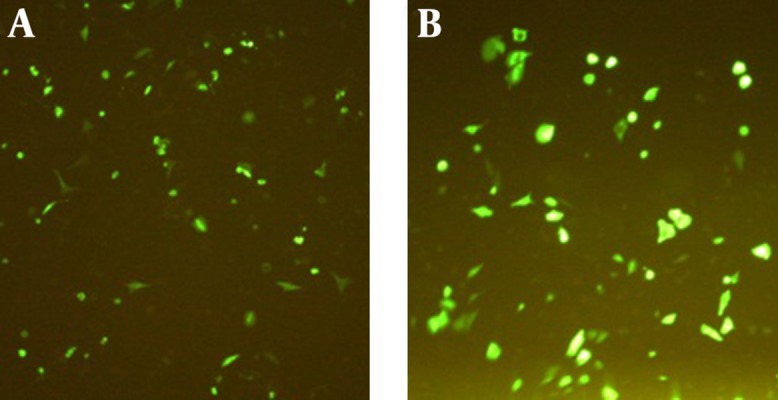
To confirm the PT DNA expression in vitro, PT was cloned at the N-terminus of green fluorescent protein (GFP) in pEGFP-N1 reporter plasmid (pEGFP-PT). GFP expression was evaluated by fluorescence microscopy at 24, 48 and 72 hours after transfection of the COS-7 cells with 5 μg pEGFP-PT and pEGFP-N1 (as positive control). As shown in Figure 2, PT was efficiently expressed in COS-7 cells, and the antigen expression level was comparable with GFP expression of the positive control. The best results were shown after 24 hours of transfection (Figure 2).
Figure 2. Transfected COS-7 Cells With pEGFP-PT (A) and pEGFP-N1 (B) Using PEI 25 kDa 24 Hours After Transfection.
4.3. Humoral Immune Responses
Blood samples of immunized and control groups were collected at regular intervals during the vaccination schedules. HCV-specific total IgG and its subclasses (IgG1 and IgG2a) were measured by ELISA.
We determined specific total IgG values three weeks after each immunization. As shown in Figure 3 A, marginal HCV-specific antibodies were detected after two times immunization with DNA (group 1); whereas, an increased antibody responses were observed in mice vaccinated two times with peptides (group 2). After the third immunization, the amount of total IgG in group 2 was significantly higher than those of group 1 and negative control groups (P < 0.05). At this time, in the mice of group 2, the level of IgG antibody significantly enhanced compared to those of the same group after the second immunization. In group 2, after the second boosting with peptides, IgG statistically increased up to a peak value and then fell into a slow decline. Interestingly, in group 1, nine weeks after the second boosting, antibody production increased statistically compared to those at three weeks after the second boosting. Considerable level of anti-HCV IgG1 was not detected in any of three time points in group 1 (Figure 3 B). However, in peptide-immunized mice in week nine after the last immunization, level of IgG1 increased significantly compared to six weeks before (P < 0.05).
Figure 3. IgG Profile of Antibody Responses in Immunized Mice.
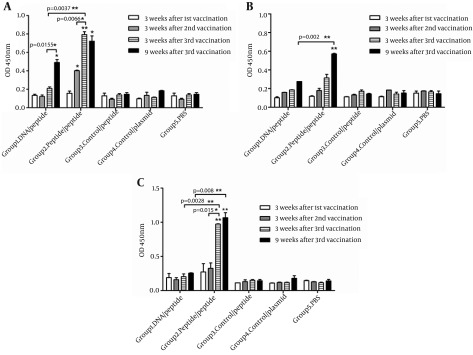
(A) IgG Total, (B) IgG1, (C) IgG2a. Comparison between the two indicated groups was significant when * P < 0.05 and ** P < 0.01. The asterisks shown directly above the bars, mark the statistically significant difference between vaccinated groups and the related control group of mice.
In addition, HCV-specific IgG2a generated by various vaccination regimens was measured. As shown in Figure 3 C, after the second immunization, IgG2a level in all vaccinated groups was low. After the third vaccination, group 1 generated only marginal IgG2a antibody titers above that of control groups. By contrast, at the same time, significantly higher level of IgG2a antibody was detected in group 2, compared to those of group 1 (P < 0.005) and background value obtained from control groups ( P < 0.0005) (Figure 3 C).
To assess long-lasting immunity, the amount of antibodies were measured nine weeks after the third vaccination. The results showed that in the tested groups, the antibody titer remained stable until nine weeks after the last immunization (Figure 3 C).
As indicated in Table 3, IgG2a/IgG1 ratio, as an indicator of Th1/Th2 polarization (34), was more than 1 value in mice of group 2 in all measured time points. It showed that HCV-peptide vaccination regimen efficiently shifted the immune response towards the Th1 phenotype.
Table 3. The Mean Ratio of IgG2a/IgG1 in Various Groups.
| Groups | Three Weeks After the 2nd Vaccination | Three Weeks After the 3rd Vaccination | Nine Weeks After the 3rd Vaccination |
|---|---|---|---|
| 1 | 1.0 | 1.1 | 0.9 |
| 2 | 1.8 | 3.0 | 1.9 |
| 3 | 1.0 | 0.9 | 1.0 |
| 4 | 0.6 | 0.8 | 1.2 |
| 5 | 0.7 | 0.7 | 1.0 |
4.4. Cellular Immune Responses
To compare the effects of two vaccination strategies on Th cell development, IFN-γ and IL-4 secretion from splenocytes of immunized mice recalled with peptides were measured by cytokine assay after three weeks of the second, and three and nine weeks after the third vaccinations. As demonstrated in Figure 4 A, at three weeks after the second vaccination, two immunized groups (Group 1, 2) developed higher levels of IFN-γ compared to control groups, and this enhancement was statistically significant in group 2 (P < 0.05).
Figure 4. IFN-γ (A) and IL-4 (B) Measurement at Regular Intervals During the Vaccination Schedules.
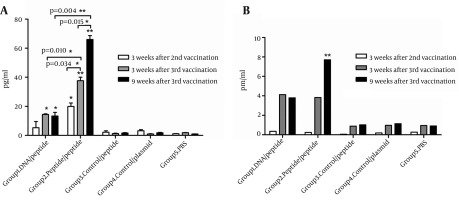
The comparison between the two indicated groups was significant when * P < 0.05 and ** P < 0.01. The asterisks shown directly above the bars mark the statistically significant difference between vaccinated groups and the related control group of mice.
At three weeks after the third immunization, group 2 mice produced higher amount of IFN-γ compared to group 1 (P < 0.05) and control animals (P < 0.005) significantly. At the same time, in group 1 mice, the amount of peptide specific IFN-γ responses increased significantly compared to the control groups (P < 0.05).
As indicated in Figure 4 B, at three-week time points after each immunization, the level of IL-4 was very low in all vaccinated groups (Figure 4 B). However, group 2 produced significantly higher level of IL-4 compared to group 1 (P < 0.05) and control (P < 0.005) mice after nine weeks of the last vaccination.
To determine long-lasting immunity, IFN-γ secretion was measured after nine weeks of the last vaccination. As shown in Figure 4 A, a high level of IFN-γ was found in groups 1 and 2 after nine weeks of the third immunization. Furthermore, in group 2, IFN-γ secretion was significantly higher at nine week than three weeks after the third immunization (P < 0.05). IFN-γ/IL-4 ratio in tested groups in all three-time points was higher than that of control groups (Table 4). Moreover, IFN-γ/IL-4 was approximately 3-folds higher in group 2 than that of group 1, indicating a stronger Th1 polarization due to HCV-peptide immunization regimen.
Table 4. The Mean Ratio of IFN- γ/ IL-4 in Various Groups.
| Groups | 3 Weeks After 3rd Vaccination | 9 Weeks After 3rd Vaccination |
|---|---|---|
| 1 | 3.5 | 3.3 |
| 2 | 9.8 | 8.6 |
| 3 | 1.4 | 1.6 |
| 4 | 1.0 | 1.5 |
| 5 | 2.0 | 1.1 |
5. Discussion
An ideal prophylactic or therapeutic vaccine against HCV infection should induce strong and broadly cross-reactive CD4+, CD8+ T-cell and neutralizing antibody responses (4). Variability of HCV antigens and immunosuppressive properties of its proteins has made multi-epitope type vaccines as a promising strategy in vaccine development against HCV (35). In this study, multi-epitope DNA-and peptide-vaccines harboring several selected immunodominant T- and B-cell epitopes from structural and non-structural antigens were designed (26, 36-39). Recent reports on multi-epitope vaccines have shown a need for improvement of their efficacy (14, 40). In this study, we tried to address this concern using in vivo electroporation as a DNA vaccine delivery method, DNA-prime/peptide boost regimen, and M720 as an efficient adjuvant for peptide vaccination. DNA vaccine delivery by in vivo electroporation has been reported to significantly enhance T-cell responses by increasing antigen specific T-cell frequencies as well as T cell function such as IFN-γ secretion (19).
Studies on cytokine profile of patients With HCV infection have indicated that Th1 cytokines are essential in viral clearance (24). Subclasses of antibodies (IgG1 and IgG2a) and IgG2a/IgG1 ratio serve as indirect indicators of Th1-type immune responses (34). Moreover, there is often a positive association between IgG2a levels and IFN-γ synthesis, which predicts tendency of immune responses for switching to Th1 type (20, 33). In the present study, DNA/peptide and peptide/peptide regimens were compared in parallel in BALB/c mice. To clarify immunological features behind our results, humoral immune responses derived from two vaccination regimens at three and nine weeks after immunization were analyzed. These two regimens had different effects on humoral immune responses. Marginal IgG1 and IgG2a were detected by DNA /peptide regimen (group 1). The two antibodies increased slowly after boosting with peptide in group 1. Peptide/peptide regimen (group 2) could induce the production of IgG2a after the second peptide injection. The level of this antibody rose significantly and continued to increase to a peak value at 3 and 9 weeks after the third immunization, respectively. Production of IgG1 started and significantly increased at 3 and 9 weeks after the third injection, respectively. Moreover, there was a higher IgG2a /IgG1 ratio in group 2 compared to group 1. These results suggested that efficient priming of Th1 cells with peptide immunogene and continuous stimulation of the immune system by slow release of peptides formulated with M720 caused an efficient antibody rise in group 2.
Since Th1-type cellular immune responses have a major role in spontaneous clearance of HCV infection, whether vaccines can stimulate a Th1-type immune response against HCV is the main feature of their evaluation. In this preliminary study, for cellular immune response, IFN-γ/IL-4 secretion assay was performed. Of course, some experiments such as in vivo/in vitro cytotoxicity assay for CD8+ T-cells are also considered in studies evaluating the efficacy of vaccines. Cytotoxicity assay for CD8+ T-cells shows their functional capability; however, this killing ability is usually in parallel with and sometimes dependent on the capacity of cells to produce IFN-γ (39). In fact, IFN-γ secretion is an important mechanism, which CD8+ T-cells use to stimulate the apoptosis pathway in their target cells. Moreover, an inverse association between CTL responsiveness and disease activity indicates that HCV is responsive to noncytolytic control. In this regard, several studies indicated that HCV is efficiently controllable noncytolytically when CD8+ T-cells recognize viral antigen and produce IFN-γ. In contrast, if activated CD8+ T-cells recognize the antigen but fail to produce IFN-γ, they cannot eradicate the infection. That is why IFN-γ detection via either intracellular staining or ELISPOT or ELISA assay is considered as a routine method for functional analysis of CD8+ T-cell responses (41). Therefore, in this study for further determination of the phenotype (Th1 vs. Th2) of the immune responses induced by multi-epitope DNA or peptide vaccines, splenocytes from immunized mice were stimulated with pool of peptides in vitro after three and nine weeks of the third vaccination. Our data showed a significant difference in IFN-γ levels in favor of immunized mice (multi-epitope DNA and peptide vaccines) compared to the control groups and significantly higher for peptide vaccine. Induction of higher Th1 immune responses by DNA/peptide immunization regimen compared to control groups indicated the efficiency of DNA construct to activate the Th1 response and peptide boosting in maintenance of its level. However, lower T-cell immunity responses in this group compared to those primed and boosted with peptide showed a need for more optimizations of multi-epitope-DNA construct in addition to DNA/peptide regimen and using in vivo electroporation as a delivery method. Our finding is in contrast to a previous study reported by Memarnejadian et al. (14) who observed the highest Th1-type responses in mice immunized by DNA/peptide regimen. This difference could be due to application of an HBsAg moiety fused to their multi-epitope construct and use of CpG in addition to M720 as adjuvant formulation.
Moreover, our findings showed lower T-cell priming by DNA- priming compared to peptide priming strategy. This low response can be most likely explained by low level antigen expression generally observed with DNA vaccination (42). However, priming T cells and generating memory Th1 responses by peptide immunization could be efficiently restimulated by peptide boosting. It is possible that enhanced immune responses observed for peptide/peptide vaccination strategy were the consequences of nature and function of the selected epitopes to induce Th1 type responses, ability of M720 adjuvant to slow release of antigens and the presence of CD4+ epitope in the designed peptide vaccine. CD8+ T-cells need for CD4+ cells for clearance of HCV infection has been reported by several researches (2, 18). Some studies on HCV peptide vaccines containing peptide derived from conserved T-cell epitopic regions of structural and nonstructural proteins have been recently published. Immunogenicity of these vaccines in phase I studies requires further optimization (43). Moreover, immunization with multi-epitope peptide construct led to a higher IFN-γ /IL-4 secretion ratio in a recall test, supporting a shift toward a Th1-type cellular response.
Interestingly, nine weeks after the last immunization, IgG1, IgG2a and IFN-γ levels remained stable in group1; whereas, the two antibodies and IFN-γ secretion increased in group 2. This long-lasting immunity could be due to:
Induction of long-lived Th1-type cellular responses through immunization with peptide formulated in M720.
Maintenance of the pool of these cells.
Mechanisms underlying this maintenance are long-term antigen expression or release of antigens continuously, which may be related to the adjuvant. In this study, peptides were formulated with M720, which is reported to induce strong immune responses with secretion of high amounts of IFN-γ, IgG2a and long- lived CD8+ T cells. Similar to our findings, a combination of HCV Core protein with M720+CpG was shown to elicit strong humoral and cellular responses with long-lasting polyfunctional Th1 and CD8+ T-cell responses in mouse model (18).
In summary, multi-epitope DNA and peptide vaccines containing Th, B-cell and four immunodominant CD8+ T cell epitopes from structural and non-structural antigens were designed to induce Th1 responses in BALB/c mice. T-cell immune responses in H2-Dd mice immunized with peptide vaccine were significantly higher compared to the multi-epitope DNA vaccine. The present findings showed the potency of M720 adjuvant and peptide-prime/peptide–boosting regimen as efficient approaches to improve multi-epitope peptide vaccines. It is also suggested to manipulate multi-epitope DNA construct to enhance its efficacy. In this regard, fusion of a potent adjuvant to the multi-epitope DNA construct can be a promising strategy to improve the vaccine efficacy in future studies. Moreover, the present study was primarily designed to ensure the immunogenicity of the multi-epitope constructs and to find the best immunization protocol on BALB/c model. After modification of DNA construct, further investigations on the potency of this modified and optimized vaccination protocol would be on HLA-A2 transgenic mouse models, which assure the application of more comprehensive and detailed immune response analysis against HLA-A2 restricted epitopes of the vaccine.
Acknowledgments
We would like to thank SEPPIC, France for providing Montanide ISA 720 required for this research. We are thankful to Dr. Mehdi Yosefi at Department of Immunology, School of Medical School, Tabriz University of Medical Sciences, Tabriz, Iran, for his technical assistance.
Footnotes
Authors’ Contributions:Conceived and design the experiment: SR, KS, AM. Performed the experiments: LPS, TT, KS, FA. Analyzed the data: LPS, SR. Contributed reagents/ materials/ analysis tools: KS, SR, SMA, RR, TMA. Wrote the paper: KS, LPS.
Funding/Support:This study was funded and supported by grant No. 13270 from Tehran University of Medical Sciences (TUMS); and grant No. 90-803 from Baqiyatallah University of Medical Sciences.
References
- 1.Roohvand F, Kossari N. Advances in hepatitis C virus vaccines, Part one: Advances in basic knowledge for hepatitis C virus vaccine design. Expert Opin Ther Pat. 2011;21(12):1811–30. doi: 10.1517/13543776.2011.630662. [DOI] [PubMed] [Google Scholar]
- 2.Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 3.Ippolito G, Capobianchi MR, Lanini S, Antonelli G. Is hepatitis C virus eradication around the corner only 25 years after its discovery? Int J Antimicrob Agents. 2014:[Epub ahead of Print]. doi: 10.1016/j.ijantimicag.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Torresi J, Johnson D, Wedemeyer H. Progress in the development of preventive and therapeutic vaccines for hepatitis C virus. J Hepatol. 2011;54(6):1273–85. doi: 10.1016/j.jhep.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 5.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5(3):215–29. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 6.Smyk-Pearson S, Tester IA, Lezotte D, Sasaki AW, Lewinsohn DM, Rosen HR. Differential antigenic hierarchy associated with spontaneous recovery from hepatitis C virus infection: implications for vaccine design. J Infect Dis. 2006;194(4):454–63. doi: 10.1086/505714. [DOI] [PubMed] [Google Scholar]
- 7.Vertuani S, Bazzaro M, Gualandi G, Micheletti F, Marastoni M, Fortini C, et al. Effect of interferon-alpha therapy on epitope-specific cytotoxic T lymphocyte responses in hepatitis C virus-infected individuals. Eur J Immunol. 2002;32(1):144–54. doi: 10.1002/1521-4141(200201)32:1<144::AID-IMMU144>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Suhrbier A. Polytope vaccines for the codelivery of multiple CD8 T-cell epitopes. Expert Rev Vaccines. 2002;1(2):207–13. doi: 10.1586/14760584.1.2.207. [DOI] [PubMed] [Google Scholar]
- 9.Iglesias MC, Mollier K, Beignon AS, Souque P, Adotevi O, Lemonnier F, et al. Lentiviral vectors encoding HIV-1 polyepitopes induce broad CTL responses in vivo. Mol Ther. 2007;15(6):1203–10. doi: 10.1038/sj.mt.6300135. [DOI] [PubMed] [Google Scholar]
- 10.Martin P, Simon B, Lone YC, Chatel L, Barry R, Inchauspe G, et al. A vector-based minigene vaccine approach results in strong induction of T-cell responses specific of hepatitis C virus. Vaccine. 2008;26(20):2471–81. doi: 10.1016/j.vaccine.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Huang XJ, Lu X, Lei YF, Yang J, Yao M, Lan HY, et al. Cellular immunogenicity of a multi-epitope peptide vaccine candidate based on hepatitis C virus NS5A, NS4B and core proteins in HHD-2 mice. J Virol Methods. 2013;189(1):47–52. doi: 10.1016/j.jviromet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Kuang Y, Liu Y, Li W, Jiang Z, Xiao L, et al. Immunogenicity of multiple-epitope antigen gene of HCV carried by novel biodegradable polymers. Comp Immunol Microbiol Infect Dis. 2011;34(1):65–72. doi: 10.1016/j.cimid.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Arashkia A, Roohvand F, Memarnejadian A, Aghasadeghi MR, Rafati S. Construction of HCV-polytope vaccine candidates harbouring immune-enhancer sequences and primary evaluation of their immunogenicity in BALB/c mice. Virus Genes. 2010;40(1):44–52. doi: 10.1007/s11262-009-0417-3. [DOI] [PubMed] [Google Scholar]
- 14.Memarnejadian A, Roohvand F. Fusion of HBsAg and prime/boosting augment Th1 and CTL responses to HCV polytope DNA vaccine. Cell Immunol. 2010;261(2):93–8. doi: 10.1016/j.cellimm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Subbramanian RA, Kuroda MJ, Charini WA, Barouch DH, Costantino C, Santra S, et al. Magnitude and diversity of cytotoxic-T-lymphocyte responses elicited by multiepitope DNA vaccination in rhesus monkeys. J Virol. 2003;77(18):10113–8. doi: 10.1128/JVI.77.18.10113-10118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tine JA, Firat H, Payne A, Russo G, Davis SW, Tartaglia J, et al. Enhanced multiepitope-based vaccines elicit CD8+ cytotoxic T cells against both immunodominant and cryptic epitopes. Vaccine. 2005;23(8):1085–91. doi: 10.1016/j.vaccine.2003.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Qiu Q, Wang RY, Jiao X, Jin B, Sugauchi F, Grandinetti T, et al. Induction of multispecific Th-1 type immune response against HCV in mice by protein immunization using CpG and Montanide ISA 720 as adjuvants. Vaccine. 2008;26(43):5527–34. doi: 10.1016/j.vaccine.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roohvand F, Aghasadeghi MR, Sadat SM, Budkowska A, Khabiri AR. HCV core protein immunization with Montanide/CpG elicits strong Th1/Th2 and long-lived CTL responses. Biochem Biophys Res Commun. 2007;354(3):641–9. doi: 10.1016/j.bbrc.2006.12.232. [DOI] [PubMed] [Google Scholar]
- 19.Weiland O, Ahlen G, Diepolder H, Jung MC, Levander S, Fons M, et al. Therapeutic DNA vaccination using in vivo electroporation followed by standard of care therapy in patients with genotype 1 chronic hepatitis C. Mol Ther. 2013;21(9):1796–805. doi: 10.1038/mt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy JS, Co M, Green S, Longtine K, Longtine J, O'Neill MA, et al. The safety and tolerability of an HIV-1 DNA prime-protein boost vaccine (DP6-001) in healthy adult volunteers. Vaccine. 2008;26(35):4420–4. doi: 10.1016/j.vaccine.2008.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duenas-Carrera S. DNA vaccination against hepatitis C. Curr Opin Mol Ther. 2004;6(2):146–50. [PubMed] [Google Scholar]
- 22.Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, et al. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71(8):6011–9. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirai M, Okada H, Nishioka M, Akatsuka T, Wychowski C, Houghten R, et al. An epitope in hepatitis C virus core region recognized by cytotoxic T cells in mice and humans. J Virol. 1994;68(5):3334–42. doi: 10.1128/jvi.68.5.3334-3342.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward S, Lauer G, Isba R, Walker B, Klenerman P. Cellular immune responses against hepatitis C virus: the evidence base 2002. Clin Exp Immunol. 2002;128(2):195–203. doi: 10.1046/j.1365-2249.2002.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, et al. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci U S A. 2007;104(20):8449–54. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, et al. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci U S A. 2009;106(18):7537–41. doi: 10.1073/pnas.0902749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zali MR, Mayumi M, Raoufi M, Nowroozi A. Hepatitis C virus genotypes in the Islamic Republic of Iran: a preliminary study. East Mediterr Health J. 2000;6(2-3):372–7. [PubMed] [Google Scholar]
- 28.Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13(2):223–35. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen KY, Liu J, Ren EC. Structural and functional distinctiveness of HLA-A2 allelic variants. Immunol Res. 2012;53(1-3):182–90. doi: 10.1007/s12026-012-8295-5. [DOI] [PubMed] [Google Scholar]
- 30.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 31.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152(1):163–75. [PubMed] [Google Scholar]
- 32.Ikram A, Anjum S, Tahir M. In Silico Identification and Conservation Analysis of B-cell and T-Cell Epitopes of Hepatitis C Virus 3a Genotype Enveloped Glycoprotein 2 From Pakistan: A Step Towards Heterologous Vaccine Design. Hepat Mon. 2014;14(6):e22215. doi: 10.5812/hepatmon.9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdian N, Gholami E, Zahedifard F, Safaee N, Rafati S. Evaluation of DNA/DNA and prime-boost vaccination using LPG3 against Leishmania major infection in susceptible BALB/c mice and its antigenic properties in human leishmaniasis. Exp Parasitol. 2011;127(3):627–36. doi: 10.1016/j.exppara.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Petrushina I, Tran M, Sadzikava N, Ghochikyan A, Vasilevko V, Agadjanyan MG, et al. Importance of IgG2c isotype in the immune response to beta-amyloid in amyloid precursor protein/transgenic mice. Neurosci Lett. 2003;338(1):5–8. doi: 10.1016/s0304-3940(02)01357-5. [DOI] [PubMed] [Google Scholar]
- 35.Yao ZQ, Eisen-Vandervelde A, Waggoner SN, Cale EM, Hahn YS. Direct binding of hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004;78(12):6409–19. doi: 10.1128/JVI.78.12.6409-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohr HF, Schmitz D, Arenz M, Weyer S, Gerken G, Meyer zum Buschenfelde KH. The viral clearance in interferon-treated chronic hepatitis C is associated with increased cytotoxic T cell frequencies. J Hepatol. 1999;31(3):407–15. doi: 10.1016/s0168-8278(99)80030-0. [DOI] [PubMed] [Google Scholar]
- 37.Park SH, Yang SH, Lee CG, Youn JW, Chang J, Sung YC. Efficient induction of T helper 1 CD4+ T-cell responses to hepatitis C virus core and E2 by a DNA prime-adenovirus boost. Vaccine. 2003;21(31):4555–64. doi: 10.1016/s0264-410x(03)00499-7. [DOI] [PubMed] [Google Scholar]
- 38.Chang KM, Thimme R, Melpolder JJ, Oldach D, Pemberton J, Moorhead-Loudis J, et al. Differential CD4(+) and CD8(+) T-cell responsiveness in hepatitis C virus infection. Hepatology. 2001;33(1):267–76. doi: 10.1053/jhep.2001.21162. [DOI] [PubMed] [Google Scholar]
- 39.Zhu F, Eckels DD. Functionally distinct helper T-cell epitopes of HCV and their role in modulation of NS3-specific, CD8+/tetramer positive CTL. Hum Immunol. 2002;63(9):710–8. doi: 10.1016/s0198-8859(02)00430-5. [DOI] [PubMed] [Google Scholar]
- 40.Shi L, Liu S, Fan GX, Sheng L, Ren HX, Yuan YK. Effective induction of type 1 cytotoxic T cell responses in mice with DNA vaccine encoding two hepatitis C virus cytotoxic T lymphocyte epitopes. Viral Immunol. 2006;19(4):702–11. doi: 10.1089/vim.2006.19.702. [DOI] [PubMed] [Google Scholar]
- 41.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194(10):1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. 2000;18:927–74. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 43.Roohvand F, Kossari N. Advances in hepatitis C virus vaccines, part two: advances in hepatitis C virus vaccine formulations and modalities. Expert Opin Ther Pat. 2012;22(4):391–415. doi: 10.1517/13543776.2012.673589. [DOI] [PubMed] [Google Scholar]


