Abstract
Background
Mutations in the T-box gene Brachyury have well known effects on invagination of the endomesodermal layer during gastrulation, but the gene also plays a role in the determination of left/right axis determination that is less well studied. Previous work has implicated node morphology in this effect. We use the TWis allele of Brachyury to investigate the molecular and morphological effects of the T locus on axis determination in the mouse.
Results
Similar to embryos mutant for the T allele, TWis/TWis embryos have a high incidence of ventral and/or reversed heart looping. In addition, heterotaxia between the direction of heart looping and the direction of embryo turning is common. Scanning electron microscopy reveals defects in node morphology including irregularity, smaller size and a decreased number of cilia, although the cilia appear morphologically normal. Molecular analysis shows a loss of perinodal expression of genes involved in Nodal signaling, namely Cer2, Gdf1 and Nodal itself. There is also loss of Dll1 expression, a key component of the Notch signaling pathway, in the presomitic mesoderm.
Conclusions
Morphological abnormalities of the node as well as disruptions of the molecular cascade of left/right axis determination characterize TWis/TWis mutants. Decreased Notch signaling may account for both the morphological defects and the absence of expression of genes in the Nodal signaling pathway.
Keywords: Brachyury, TWis, Left/right asymmetry, embryonic node, Nodal, Gdf1, Dll1, Cer2, Left/right asymmetry, embryonic node, Nodal, Gdf1, Dll1, Cer2
INTRODUCTION
Brachyury (T) was the first T-box gene identified in mice (Dobrovolskaïa-Zavadskaïa, 1927). It is first expressed in the epiblast in the pre-primitive streak stage embryo and, as the embryo develops, is expressed later in the primitive streak and embryonic node. Mice heterozygous for a T null allele are viable and fertile but have shortened tails; homozygous embryos lack posterior somites after the seventh pair, have a convoluted neural tube, no distinguishable notochord and die at midgestation (Grüneberg, 1958; Beddington et al., 1992). T homozygous mutant embryos show heart morphology and looping defects, randomized situs and lack expression of Nodal in the perinodal region and left lateral plate mesoderm (LPM), indicating a disruption in the process of left/right axis determination. There is also abnormal expression of Lefty1 and Lefty2, two TGFβ-related genes that normally show asymmetric expression (King et al., 1998). In addition, homozygous T mutants have morphological defects in node formation, leading to the idea that the inductive signal for left/right axis formation from the node is abnormal in T/T embryos (Beddington et al., 1992; King et al., 1998). However, at the time these studies were done, the nodal flow hypothesis for left/right axis determination (Hamada et al., 2002) had not been formulated and node cilia, which are essential for setting up the cascade of asymmetric gene expression leading to normal situs, were not examined in T/T mutants, nor were a number of molecular players now known to be involved in the Nodal signaling pathway leading to left/right axis determination.
TWis is a spontaneous mutation of T that comprises an insertion of a transposon-like element into the 3’ end of the seventh exon. The insertion creates a mutated splice site that results in the loss of wild type T mRNA. TWis produces transcripts that bypass the mutated exon 7 splice site but contain the T-box binding domain. Mutant proteins are hypothesized to compete with wild type T for DNA binding sites thus producing a dominant negative phenotype that is similar to the null allele but more extreme (Shedlovsky et al., 1986; Herrmann et al., 1990; Goldin and Papaioannou, 2003). TWis heterozygous mice have a more severe tail truncation than T heterozygotes; TWis homozygous mutant embryos, like T homozygotes lack a notochord, but have no discernable somites (Shedlovsky et al., 1988). In TWis homozygous mutants, precursors of the notochord are present, as seen by double labeling with T antibody and a probe for Shh. However, precursor cells do not survive and no distinguishable notochord is formed. The lack of a notochord leads to neural tube defects and TWis/TWis mutants die at midgestation (Shedlovsky et al., 1988; Conlon et al., 1995). Heart development and left/right patterning have not been previously reported for TWis/TWis mutants. In this study, we investigate the left/right asymmetry phenotype of the TWis/TWis mutant to determine the extent of disruption of heart looping and morphology, as well as the morphology of the node and nodal cilia. We also investigate the expression of left/right specific genes in TWis/TWis mutants as well as components of the Wnt and Notch signaling pathways. In agreement with previous studies of the T locus, our results show disruptions in left/right axis determination and node morphology and extend previous observations to show heterotaxia of heart looping and embryo turning. We also detail the effect on node morphology, including node cilia, and document disruptions in additional molecular markers of the signaling cascade involved in the determination of left/right asymmetry.
RESULTS
Abnormalities in heart looping and embryo turning in TWis/TWis mutants
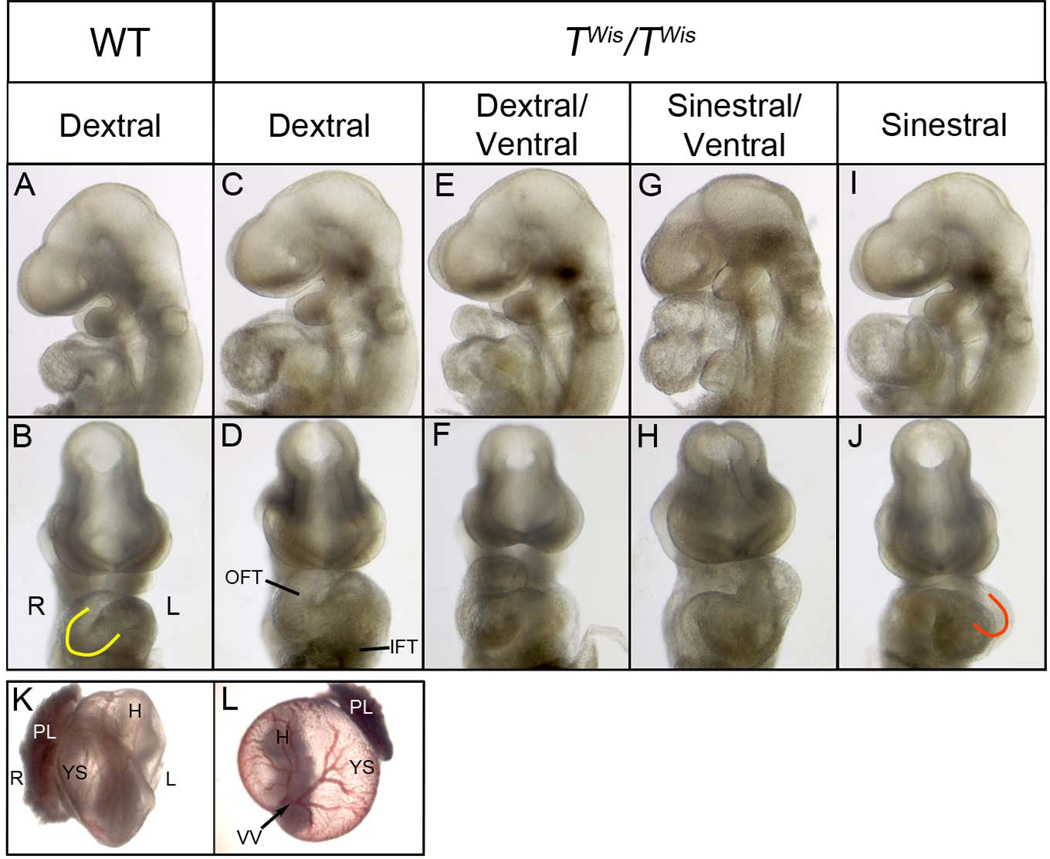
The dextral looping of the heart is one of the earliest morphological signs of bilateral asymmetry. To investigate left/right asymmetry in TWis homozygous mutants prior to their death at embryonic day (E) 11.5, embryos were collected from TWis heterozygous matings between E9.5 and E10.5 and the direction of heart looping and embryo turning was recorded. TWis/TWis mutant embryos are morphologically distinguishable by the severe axial truncation that results in loss of posterior structures including the allantois. Analysis of heart looping in wild type controls, including both +/+ and TWis/+ embryos, showed that 98% of embryos (97/99) undergo dextral heart looping, resulting in the normal configuration in which the inflow and outflow tract are on the left and right sides of the embryo, respectively, and the ventricles are perpendicular to the rostral/caudal axis forming a ‘C’ shaped loop when viewed from the ventral aspect (Table 1; Figure 1A, B). Among TWis/TWis embryos, 6% (2/32) showed normal dextral looping (Table1; Figure 1C, D); 13% (4/32) had dextral/ventral looped hearts (Table1; Figure 1E, F), in which there is correct placement of the inflow and outflow tracts while the right and left ventricles are aligned on the rostral-caudal rather than the left-right axis; 9% (3/32) showed hearts that were ventrally looped, in which the ventricles as well as the inflow and outflow tracts aligned on the rostral-caudal axis (Table 1); 22% (7/32) showed sinestral/ventral looped hearts, where the right-left position of the inflow and outflow tract was reversed (Table 1; Figure 1G, H); and 50% (16/32) had sinestral looped or ‘J’ shaped hearts (Table 1; Figure 1I, J), indicating a complete reversal of heart looping.
Table 1.
Number of wild type and TWis/TWis mutant embryos with different heart looping and embryo turning phenotypes at E9.5–E10.5.
| Wild type1 (%) | TWis/TWis (%) | ||
|---|---|---|---|
| Direction of Heart Looping | Dextral | 97 (98) | 2 (6) |
| Dextral/ Ventral | 0 | 4 (13) | |
| Ventral | 0 | 3 (9) | |
| Sinestral/Ventral | 0 | 7 (22) | |
| Sinestral | 2 (2) | 16 (50) | |
| Total | 99 | 32 | |
| Direction of Embryo Turning/Situs | Normal/Normal | 90 (97) | 4 (17) |
| Reversed/Inversus | 1 (1) | 10 (43) | |
| Reversed/Heterotaxia | 2 (2) | 9 (39) | |
| Total | 93 | 23 |
Wildtype includes both +/+ and TWis/+ embryos.
Figure 1. Heart looping and embryo turning in wild type and TWis/TWis embryos at E9.5.
A–J: Left and frontal views of head and heart of E9.5 embryos showing a wild type embryo with dextral looped heart (A, B) and TWis/TWis mutant embryos with a dextral looped heart (C, D), dextral/ventral looped heart (E, F), sinestral/ventral looped heart (G, H), and sinestral looped heart (I, J). Yellow ‘C’ in panel B indicates dextral heart looping and ‘J’ in panel J indicates sinestral heart looping. K, L: Frontal view of embryos within the yolk sac with the placenta attached. The wild type embryo (K) has turned toward its right side; vitelline vessels exit on the embryo’s left side and the placenta is on the right side. In the TWis/TWis mutant embryo (L) the vitelline vessels exit the embryo on the right side and placenta is on the left. H, head; IFT, inflow tract; L, left; OFT, outflow tract; PL, placenta; R, right; VV, vitelline vessels; YS, yolk sac.
Embryo turning is another early indicator of left/right asymmetry. Embryos normally rotate in an anticlockwise direction toward their right side to assume a fetal position within the membranes with the umbilical vessels, placenta and tail on the right side of the embryo and the vitelline vessels entering the yolk sac from the left side. Among embryos that were scored for both heart looping and embryo turning, 97% (90/93) of wild type embryos, including TWis/+ and +/+ embryos, had undergone normal embryo turning and heart looping resulting in normal situs (Table 1; Figure 1K). Among TWis/TWis mutants, 52% (12/23) showed reversal of embryo turning in which the placenta was located on the left side of the embryo and vitelline vessels were positioned on the right side (Figure 1L). However, the direction of embryo turning and heart looping were normal in only 17% (4/23) of TWis/TWis mutants whereas 43% (10/23) of mutants showed situs inversus, in which both the direction of heart looping and embryo turning were reversed, and 39% (9/23) showed heterotaxia, in which direction of heart looping was not concordant with the direction of embryo turning (Table 1). TWis/TWis mutants therefore have a high incidence of situs inversus and heterotaxia compared with wild type embryos.
Characterization of the node and midline of TWis/TWis mutants at E8.0
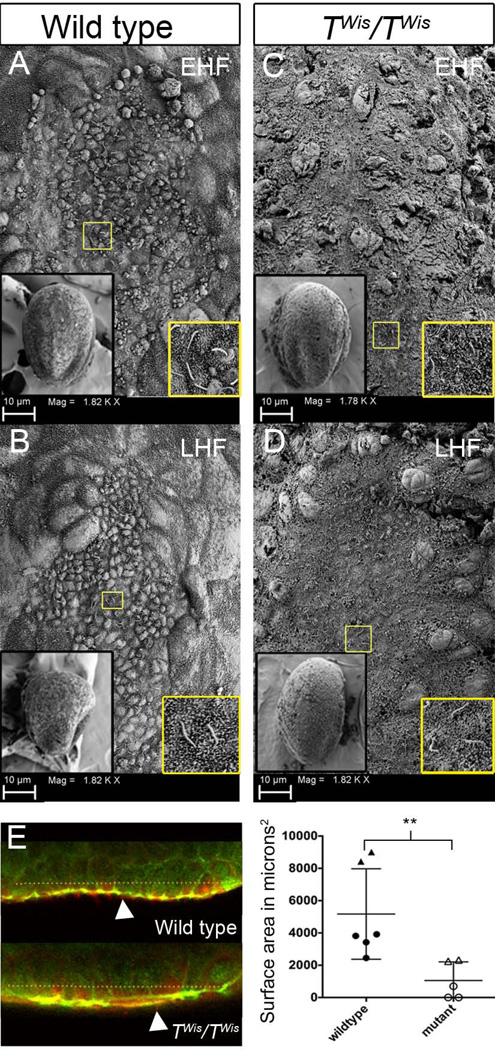
To investigate node morphology of TWis/TWis mutants at the time left/right axis determination is taking place, we used scanning electron microscopy (SEM) and immunofluorescence with confocal microscopy at E8.0. By SEM, wild type embryonic nodes (n=6) appear oval shaped at the early head fold (EHF) stage (Figure 2A) and shield shaped at the late head fold (LHF) stage (Figure 2B). The nodes have a distinct border between the smaller, cobblestone-like node cells and the larger surrounding endoderm cells. Monocilia are present on the node cells (Figure 2A, B insets). In TWis/TWis mutant embryos, three out of four nodes lacked a well defined border and individual cells of all mutant nodes were less regular in size and less distinct, lacking the cobblestone appearance of the nodes of wild type embryos. Normal-appearing cilia were present (Figure 2C, D insets) as confirmed by confocal microscopy and immunofluorescence with antibodies against acetylated tubulin, a marker of node cilia (Figure 2E). However, wild type embryos contained a significantly larger number of nodal cilia on average (138 ± 22) than TWis/TWis mutants (35 ± 8) (Mann Whitney U test; p=0.009).
Figure 2. Node morphology and cilia of wild type and TWis/TWis embryos at E8.0.
A–D: Scanning electron microscopy of the embryonic node in wild type (A, B) and TWis/TWis mutants (C, D) at early head fold (EHF) and late head fold (LHF) stages. The left inset of each panel corresponds to a ventral view of the embryo at 300× magnification. The right inset of each panel is an image of cilia found in the corresponding node taken at 3300×. Yellow box indicates magnified area in the right inset. E. Optical slices of wild type and TWis/TWis mutant nodes stained for acetylated tubulin (red) and ZO1 (green). Dotted line indicates the xy length of the bilaminar node taken to estimate the surface area of the node. White arrowheads indicate acetylated tubulin positive cilia. Scatter plot shows the distribution of node surface areas measured from confocal z stacks. Circles indicate EHF stage embryos and triangles are LHF stages. Double asterisks represent a statistically significant difference between the two populations (Mann-Whitney U test; p <0.01).
The size of the node was estimated both from direct measurements of the SEM images and from the analysis of confocal z-stacks. From the SEM images, the surface area of wild type nodes was significantly greater (4001 ± 324 µm2; n=6) than TWis/TWis mutants (1307 ± 440 µm2; n=4) (Mann Whitney U test; p <0.01) and similarly, from analysis of confocal z-stacks, the surface area of wild type embryos (n=6) was significantly greater than TWis/TWis mutants (n=5) (Mann Whitney U test; p <0.01, Figure 2E). A bilaminar epithelial node could not be identified in two of three EHF stage TWis/TWis mutants, which were scored as zero for surface area (Figure 2E).
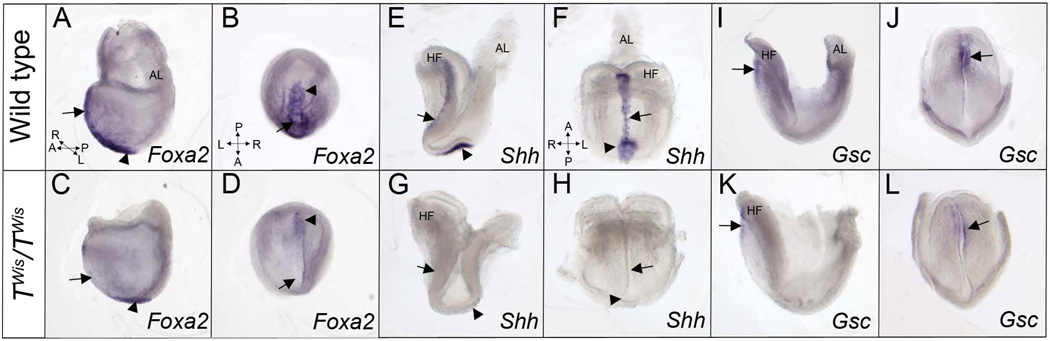
We also examined the molecular differentiation of the node and its derivative midline structures at E8.0. Foxa2 marks the prospective floor plate and the node in wild type embryos (Figure 3A, B) (Kinder et al., 2001). Expression was not detected in the floor plate in TWis/TWis mutants (n=4), but was present in the node at the distal tip of the embryo at a lower expression level than wild type controls (Figure 3C, D). Shh, which is expressed in the prechordal plate, notochordal plate and embryonic node in wild type embryos (Figure 3E, F) (Jeong and Epstein, 2003) was not observed in three TWis/TWis mutants (Figure 3G, H), whereas a fourth showed Shh expression in the prechordal plate, but not in the rest of the midline or the node (data not shown). Gsc is expressed in the anterior foregut endoderm, prechordal plate and midline neuroectoderm, in both wild type and TWis/TWis mutants (n=4) (Chu et al., 2005). However Gsc is expressed at a lower level in TWis/TWis mutants (n=4) when compared to wild type controls (Figure 3I–L).
Figure 3. Molecular differentiation of the node and anterior midline in wild type and TWis/TWis embryos at E8.0.
A–D: Left lateral and ventral views of wild type (A, B) and TWis/TWis mutant (C, D) embryos showing expression of Foxa2 by in situ hybridization. Wild type embryos express Foxa2 in the midline and node while TWis/TWis mutant embryos show expression in the node but not in the midline. E–H: Left lateral and frontal views of wild type (E, F) and TWis/TWis mutant embryos (G, H) showing expression of Shh in the node and midline of wild type embryos but no expression in the mutant embryos. I–L: Left lateral and frontal views of wild type embryos (I, J) and TWis/TWis mutant embryos (K, L) showing Gsc expression in the anterior foregut endoderm and prechordal plate in wild type embryos and a similar pattern but lower expression level in mutant embryos. Arrows point to the anterior midline and arrowheads point to the node. Compass in A refers to A, C, E, G, I, K; compass in B refers to B, D, J, L; compass in F refers to F, H. AL, allantois; HF, headfolds.
Disruption of the Nodal signaling pathway in TWis/TWis mutants
In wild type embryos, Nodal is first expressed in the perinodal region at E8.0 and by the 3-somite stage shows higher expression on the left side as well as expression in the left LPM (n=12) (Figure 4A, B) (Brennan et al., 2002). Like T/T embryos, TWis/TWis embryos show a complete absence of Nodal expression at comparable stages (n=5; Figure 4C, D). Gdf1, a Nodal agonist, is expressed bilaterally in the perinodal region at E8.0, as well as bilaterally in the LPM in wild type embryos (n=8; Figure 4E, F) (Rankin et al., 2000). In TWis/TWis mutant embryos, Gdf1 expression is absent from the perinodal region; however, bilateral LPM expression is maintained (n=5; Figure 4G, H). Cer2, a Nodal antagonist, shows bilateral perinodal expression in wild type embryos at E8.0 with higher expression on the right side (n=8; Figure 4I, J) (Nakamura et al., 2012). In TWis/TWis mutant embryos there was no expression of Cer2 (n=6; Figure 4K, L). Pitx2, a downstream target of Nodal, is asymmetrically expressed in the left LPM and regulates the direction of heart looping. In wild type embryos at E8.5, Pitx2 expression is observed in the left inflow tract of the heart, as well as the head folds (n=11; Figure 4M, N) (Lin et al., 1999). In TWis/TWis mutants, Pitx2 expression is present in the head folds but absent from the inflow tract of the heart (n=5; Figure 4O, P).
Figure 4. Disruption of Nodal signaling pathway in TWis/TWis embryos at E8.0.
A–L: Left lateral and ventral views of wild type and TWis/TWis mutant embryos showing the expression of Nodal in the perinodal region and left LPM in wild type embryos (A, B) but no expression in TWis/TWis mutants (C, D); Gdf1 bilateral expression in the perinodal region and LPM of wild type embryos (E, F) but only bilateral LPM expression in TWis/TWis mutants; and Cer2 bilateral perinodal expression in wild type embryos (I, J) but no expression in TWis/TWis mutants (K, L). Arrowheads indicate LPM expression and arrows indicate perinodal expression. Compass in A refers to A, E, C, G, I, and K; compass in B refers to B, F, D, H, J, and L. M–P: Frontal and left lateral views of Pitx2 expression in the head folds and left inflow tract in wild type embryos (M, N) but only in the head folds of TWis/TWis mutant embryos (O, P). Pitx2 expression on the right side of TWis/TWis mutant is in the yolk sac remnants still attached to the embryo. AL, allantois; H, heart; HF, head folds; LIT, left inflow tract.
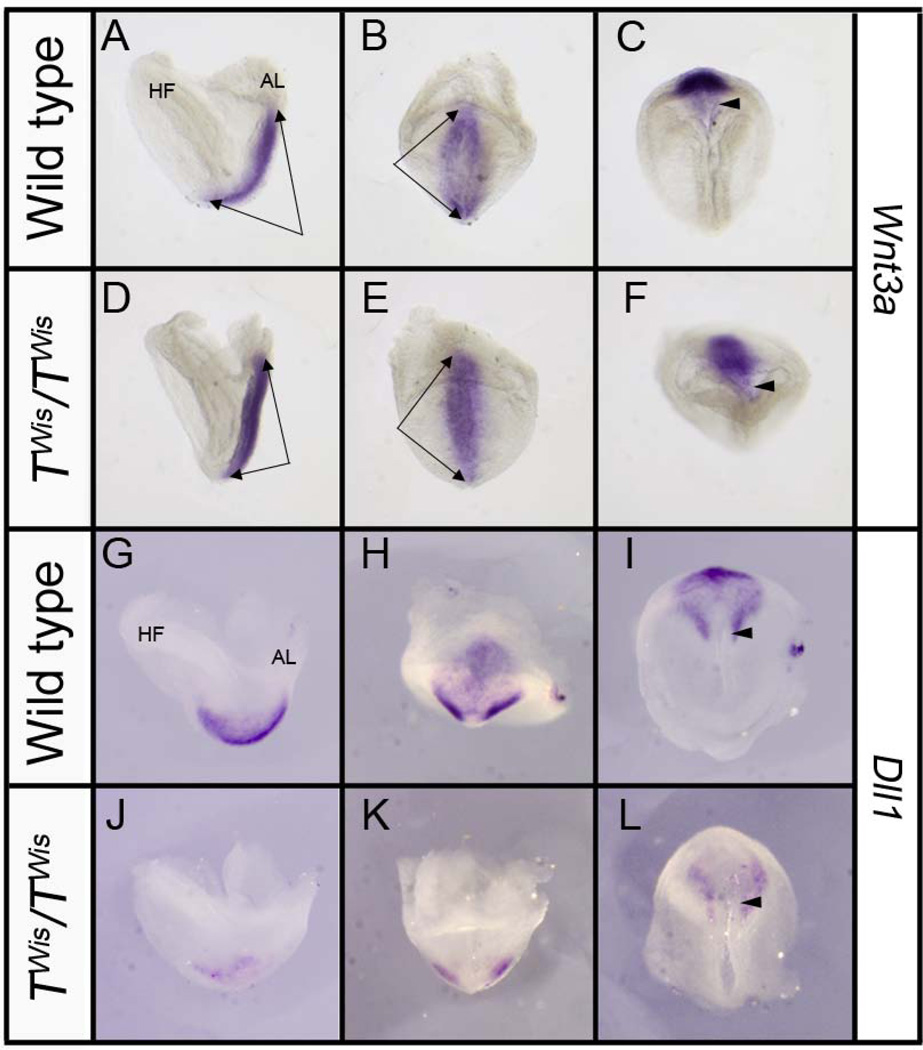
The canonical Wnt signaling pathway is known to regulate perinodal expression of Nodal by regulating Notch signaling (Nakaya et al., 2005) and mutations in Notch signaling components lead to a loss of perinodal Nodal expression (Krebs et al., 2003). In wild type embryos at E8.0, Wnt3a is expressed throughout the primitive streak (n=5, Figure 5A–C) (Nakaya et al., 2005) and stage-matched TWis/TWis mutants showed no difference compared to their wild type littermates (n=5; Figure 5D–F). Dll1 is an essential ligand in the Notch pathway and in wild type embryos at E8.0 is expressed in the primitive streak and the presomitic mesoderm (n=5; Figure 5G–I) (Krebs et al., 2003). Stage matched TWis/TWis mutant embryos show an absence of Dll1 expression in most of the presomitic mesoderm while maintaining a low level of bilateral Dll1 expression at the distal tip of the embryo (n=5; Figure 5J–L).
Figure 5. Expression of Wnt3a and Dll1 in wild type and TWis/TWis embryos at E8.0.
A–F: Left lateral (A, D), posterior (B, E), and ventral (C, F) views of the distal tip of wild type and TWis/TWis embryos showing similar Wnt3a expression throughout the entire primitive streak (double arrows); arrowheads point to the node in ventral views. G–L: Left lateral (G, J), posterior H, K), and ventral (I, L) views of Dll1 expression in the presomitic mesoderm and perinodal region in wild type embryos and greatly reduced expression in TWis/TWis mutants, particularly in the presomitic mesoderm. AL, allantois; HF, headfold. Arrowheads point to the node in ventral views.
Discussion
The involvement of Brachyury in the invagination of the endomesodermal layer has been functionally conserved throughout metazoan evolution and its role in the specification and differentiation of the notochord is a feature of chordate development (Satoh et al., 2013). The mutant phenotypes associated with T mutant alleles in mice are well known to affect posterior mesoderm formation resulting in loss of somites, allantois defects and shortened or absent tails, as well as affecting the survival of the midline notochord (Showell et al., 2004; Naiche et al., 2005; Inman and Downs, 2006). Less is known about another feature of T mutants, the disruption of the left/right body axis determination, which has been seen in T mutants in mice, as well as in mutants of the orthologous gene ntl in zebrafish and misexpression studies of the orthologous gene XBra in Xenopus (King et al., 1998; Kitaguchi et al., 2002; Amack and Yost, 2004).
In this study, the TWis allele of Brachyury was used to further explore the nature and extent of left/right asymmetry defects in the mouse. Similar to the T allele, the TWis allele has no effect on axis determination in heterozygotes, but causes a disruption of asymmetry when homozygous. The range of phenotypes in heart morphogenesis of homozygous mutants is similar for the two alleles including a high incidence of ventral looping, although in terms of situs, there was a higher incidence of reversed heart looping in TWis/TWis mutants (72%) compared with T/T mutants (50%) (King et al., 1998). This difference could be due to strain differences or to chance in the smaller sample size in the earlier study. The direction of embryo turning, or axial rotation, was also found to be reversed in a majority of TWis/TWis mutant embryos. However, the direction of turning was not concordant with the direction of heart looping in 39% of the embryos. The process of axial rotation is initiated at about the same time as heart looping. The two morphogenetic movements proceed simultaneously and are tightly correlated in normal embryos. Although it is still a poorly understood process, axial rotation is thought to be driven by asymmetric cell proliferation in the lateral body wall and gut that is somehow linked to visceral asymmetries (Miller and White, 1998). However, there are a number of situations in axis-determination mutants or in experimental perturbations of embryos in which the direction of axial rotation and heart looping are not in the same direction (Hadjantonakis et al., 2008; Gardner, 2010). Thus, the direction of embryo turning is separable from left/right axis determination at the node.
How does this abnormal left/right patterning come about? The initial breaking of bilateral symmetry in the mouse is brought about by the unidirectional nodal flow created by the beating of motile cilia in the pit of the node (Nonaka et al., 1998). Cilia in the crown cells surrounding the node sense the direction of flow which initiates a cascade of asymmetric Ca2+ signaling and gene expression (Yoshiba et al., 2012). The signaling molecule Nodal is expressed more strongly in the left perinodal region while its antagonist, Cer2, which responds directly to the nodal flow, is stronger on the right. Perinodal expression of the Nodal agonist, Gdf1, is essential for the transfer of the perinodal Nodal signal to the left LPM where its target, Pitx2, directs asymmetric heart development (Tanaka et al., 2007).
In embryos with mutations in T, there are major morphological abnormalities of the node and midline and the abnormally shaped nodes of T/T embryos were thought to be a contributing factor to laterality defects (King et al., 1998). We have extended these light microscopy observations of the node using confocal and scanning electron microscopy and have shown that TWis/TWis mutants have variable node defects ranging from lack of a defined, bilaminar node with a distinct boundary to a smaller node with a flatter surface appearance. Nodal cilia are present and appear normal but are much less numerous in TWis/TWis mutant nodes, although the lower number of cilia is unlikely to be a factor because as few as two motile cilia have been shown to set up the asymmetric gene expression cascade (Shinohara et al., 2012).
At the early primitive streak stage both T and Foxa2 are expressed in precursors of the mouse organizer. Foxa2 homozygous mutants fail to form a node or notochord although mutants do express T in a limited region of the primitive streak (Beddington et al., 1992; Ang and Rossant, 1994; Kinder et al., 2001). Foxa2 also regulates Shh expression in the notochord and floor plate (Jeong and Epstein, 2003). In TWis/TWis mutants, Shh expression is completely lost, in agreement with previous work (Conlon et al., 1995), and Foxa2 expression is reduced in the node and absent from the prospective floor plate. This could indicate that T regulates Foxa2 or that these tissues are compromised in TWis/TWis mutants and there is simply less node and notochord tissue. A previous study suggested that precursor cells of the node are unaffected in TWis homozygous mutants as Gsc expression is normal at the early streak stage (E6.25–E6.75) (Conlon et al., 1995). Gsc null mutants have no node defects (Rivera-Perez et al., 1995), thus it is unlikely to be a target of T and the lower levels of Gsc we observed in TWis/TWis mutants at E8.0 most likely reflects the poor development of the node.
Our results are in agreement with studies in zebrafish where ntl is expressed in the dorsal forerunner cells, the precursors of the ciliated cells of Kupffer’s vesicle, which is a structure analogous to the node in the mouse. Knockdown of ntl specifically in the dorsal forerunner cells shows that ntl functions cell autonomously in these cells. In the absence of ntl, ciliated cells are present but disorganized and Kupffer’s vesicle fails to form, resulting in disruption of left/right axis determination (Amack and Yost, 2004; Amack et al., 2007).
T/T embryos have no perinodal expression of Nodal and no expression of either Nodal or Lefty2 in the left LPM (King et al., 1998). In this study we have expanded this observation to investigate factors both upstream and downstream of perinodal Nodal signaling. We show that in addition to the lack of Nodal expression and a resulting loss of Pitx2 expression in the left LPM and inflow tract of the heart, TWis/TWis embryos also show disruption in the expression of genes that regulate Nodal in the perinodal region, namely Cer2 and Gdf1. Consequently we investigated the expression of genes involved in cell signaling pathways known to regulate perinodally-expressed genes. Primitive streak expression of the canonical Wnt signaling pathway gene, Wnt3a, was unaffected in TWis homozygous mutants in agreement with previous work showing that Wnt3a acts upstream of T (Hofmann et al., 2004). Expression of Dll1, a ligand of the Notch signaling pathway, was absent from the primitive streak, posterior end of the embryo and node in TWis/TWis mutants. This observation is consistent with the regulation of Dll1 by canonical Wnt signaling in synergy with T and Tbx6 (Hofmann et al., 2004). Dll1 homozygous mutant embryos show no expression of Nodal in the perinodal region and have node and notochord defects and improper pattering of the floor plate. Node and midline defects in Dll1 mutant embryos are hypothesized to be the cause of the observed left/right asymmetry phenotype (Przemeck et al., 2003). Furthermore, the perinodal expression of Gdf1 and Cer2 is thought to be under control of Notch signaling (Kitajima et al., 2013), potentially providing a unifying cause for the morphological and molecular defects observed in TWis/TWis mutants.
King et al. (King et al., 1998) concluded that the most parsimonious explanation for the left/right axis defects of T/T mutants – bilateral Lefty1 expression in the midline and lack of Lefty2 expression in the left LPM – was an abnormal signal emanating from the node, possibly caused by abnormal node morphology. Our results support this idea by documenting specific morphological defects in the node including a lower number of node cilia. Mutation of the homeobox gene Noto, which acts downstream of both T and Foxa2 (Abdelkhalek et al., 2004), causes a very similar node and cilia phenotype (Beckers et al., 2007). The transcription factor Zic3, a known target of T that controls left-right asymmetry in Xenopus (Kitaguchi et al., 2002), is another candidate target for contributing to TWis/TWis node defects as Zic3 has a role in regulating node morphogenesis specifically in the T-expressing tissue of the primitive streak (Jiang et al., 2013; Sutherland et al., 2013). In addition, we further illuminate the phenotype by showing defects in Notch signaling upstream of perinodal signaling that could underlie both the morphological and molecular defects caused by the T locus mutations.
EXPERIMENTAL PROCEDURES
Mice and embryo collection
The TWis mutant allele (Shedlovsky et al., 1988) was kept on a mixed genetic background including A/J, C57,129 and ICR (Taconic). TWis heterozygous mice, which are viable and fertile, were mated together to obtain +/+, TWis/+, and TWis/TWis embryos between E8.0–E9.5 (E0.5 denotes noon of the day a vaginal plug was observed). Embryos were removed from the uterus and dissected out of the decidua leaving the yolk sac and placenta intact. Placement of the tail, placenta and vitelline vessels was recorded. The yolk sac was then removed and heart morphology and the direction of heart looping was recorded. At E8.0–E8.5, TWis/TWis mutant embryos were identified based on the lack of an allantois. At E9.5–E10.5 TWis/TWis mutant embryos were identified based on axial truncation defects. TWis/+ and +/+ embryos were indistinguishable at this early age and were considered together as wild type controls. Wild type embryos were staged by somite number and TWis/TWis mutant embryos were staged by morphology of the head. All procedures were approved by the Columbia University Medical Center Institutional Animal Care and Use Committee.
In situ hybridization
Embryos were dissected in phosphate buffered saline (PBS) containing 0.2% bovine serum albumin (BSA)(Sigma), fixed overnight in 4% paraformaldehyde (PFA) in PBS at 4°C, dehydrated in 100% methanol and stored at −20°C. Whole mount in situ hybridization using antisense RNA probes was performed essentially as described previously (Wilkinson, 1992).
Immunofluorescence and confocal microscopy
Embryos were dissected in PBS and fixed in fresh prechilled (4°C) PBS with 0.5% PFA overnight at 4°C. Embryos were blocked in cold 1% BSA and 10% goat serum (heat inactivated and filtered, 22µM), 0.5 Triton X-100 (Sigma) in PBS from 45 min to overnight at 4°C. Primary antibody incubation was carried out in 1% BSA, 2% goat serum and 0.5% Triton X-100 in PBS overnight at 4°C. Embryos were incubated in secondary antibody in 1% BSA, 2% goat serum, and 0.5% Triton X-100 in PBS overnight at 4°C. Embryos were mounted in Vectashield (Vector Labs) between two coverslips in a well cut out from several layers of electrical tape. ZO1 (Zymed) was used at a concentration of 1:200 and mouse anti-acetylated tubulin (Invitrogen) was used at a concentration of 1:400.
Embryos were imaged using a Nikon A1RMP two-photon microscope. Optical z sections of mounted embryos were taken at 0.5µm intervals using a Nikon 25x/1.1NA water immersion objective. Optical sections were compiled into z-stacks for individual embryos. NIS-elements AR 4.0 software was used for acquisition and analysis of z-stacks. Surface area analysis was done using ImageJ (http://rsbweb.nih.gov/ij/). The surface area was estimated by first identifying the node as a bilaminar epithelial cell structure and then multiplying the xy length of the bilayer in each optical section by the interval step of the z-stack and summing all the resulting areas.
Scanning Electron Microscopy
Embryos were dissected in prechilled PBS with 0.05% Triton X-100 (Fisher) and fixed in 2.5% PFA, 2.5% gluteraldehyde in 0.075M sodium cacodylate buffer, pH 7.4 (Scanning Electron Microscopy Sciences) rocking overnight at 4°C. Embryos are placed in 100% ethanol for critical point drying using a Denton JCP-1 critical point dryer. Critical point dried embryos were placed on a piece of carbon tape and plated with Gold/Palladium in a Denton Vacuum 1V Sputter/Dryer. Embryos are then imaged on a Zeiss Field Emission Scanning Electron microscope Supra 25.
Bullet points.
Brachyury mouse mutants have left/right axis defects including heterotaxia and heart looping defects
The node is abnormal and has fewer cilia
Nodal signaling pathway components are misregulated
The Notch ligand Dll1 is downregulated in the primitive streak
Acknowledgements
We thank Alexandra Castillejos and Elana Ernstoff for technical assistance, Nina Lampen at the Sloan Kettering Scanning Electron Microscopy Facility, and the Herbert J. Irving Comprehensive Cancer Center Confocal and Specialized Microscopy Core.
Grant sponsor: This work was supported by grant R01HD056248 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (V.E.P.) and a Ruth L. Kirschstein National Predoctoral Fellowship F3HD065441 (D.C.) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- Abdelkhalek HB, Beckers A, Schuster-Gossler K, Pavlova MN, Burkhardt H, Lickert H, Rossant J, Reinhardt R, Schalkwyk LC, Muller I, Herrmann BG, Ceolin M, Rivera-Pomar R, Gossler A. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004;18:1725–1736. doi: 10.1101/gad.303504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer's vesicle in zebrafish. Developmental Biology. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Current Biology. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ang S, Rossant J. HNF-3b is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Beckers A, Alten L, Viebahn C, Andre P, Gossler A. The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left-right patterning. Proceedings of the National Academy of Sciences. 2007;104:15765–15770. doi: 10.1073/pnas.0704344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington RSP, Rashbass P, Wilson V. Brachyury-a gene affecting mouse gastrulation and early organogenesis. Development Supplement. 1992:157–165. [PubMed] [Google Scholar]
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes & Development. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Ding J, Jeays-Ward K, Price SM, Placzek M, Shen MM. Non-cell-autonomous role for Cripto in axial midline formation during vertebrate embryogenesis. Development. 2005;132:5539–5551. doi: 10.1242/dev.02157. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Wright CVE, Robertson EJ. Effects of the TWis mutation on notochord formation and mesodermal patterning. Mechanisms of Development. 1995;49:201–209. doi: 10.1016/0925-4773(94)00318-h. [DOI] [PubMed] [Google Scholar]
- Dobrovolskaïa-Zavadskaïa N. Sur la mortification spontanée de la queue che la souris nouveau-née et sur l'existence d'un caractère (facteur) héréditaire "non viable". C. R. Seanc. Soc. Biol. 1927;97:114–116. [Google Scholar]
- Gardner RL. Normal bias in the direction of fetal rotation depends on blastomere composition during early cleavage in the mouse. PLoS One. 2010;5:e9610. doi: 10.1371/journal.pone.0009610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin SN, Papaioannou VE. Unusual misregulation of RNA splicing caused by insertion of a transposable element into the T (Brachyury) locus. BMC Genomics. 2003;4:14. doi: 10.1186/1471-2164-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüneberg H. Genetical studies on the skeleton of the mouse XXIII. The development of brachyury and Anury. Journal of Embryology and experimental Morphology. 1958;6:424–443. [PubMed] [Google Scholar]
- Hadjantonakis A-K, Pisano E, Papaioannou VE. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PloS ONE. 2008;3:e2511. doi: 10.1371/journal.pone.0002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nature Reviews Genetics. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labiet S, Poustka A, King TR, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hofmann M, Schuster-Gossler K, Watabe-Rudolph M, Aulehla A, Herrmann BG, Gossler A. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes & Development. 2004;18:2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Brachyury is required for elongation and vasculogenesis in the murine allantois. Development. 2006;133:2947–2959. doi: 10.1242/dev.02454. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Epstein D. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development (Cambridge, England) 2003;130:3891–3902. doi: 10.1242/dev.00590. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Zhu L, Hu L, Slesnick TC, Pautler RG, Justice MJ, Belmont JW. Zic3 is required in the extra-cardiac perinodal region of the lateral plate mesoderm for left-right patterning and heart development. Hum Mol Genet. 2013;22:879–889. doi: 10.1093/hmg/dds494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PPL. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- King T, Beddington RSP, Brown NA. The role of the brachyury gene in heart development and left-right specification in the mouse. Mechanisms of Development. 1998;79:29–37. doi: 10.1016/s0925-4773(98)00166-x. [DOI] [PubMed] [Google Scholar]
- Kitaguchi T, Mizugishi K, Hatayama M, Aruga J, Mikoshiba K. Xenopus Brachyury regulates mesodermal expression of Zic3, a gene controlling left-right asymmetry. Development Growth and Differentiation. 2002;44:55–61. doi: 10.1046/j.1440-169x.2002.00624.x. [DOI] [PubMed] [Google Scholar]
- Kitajima K, Oki S, Ohkawa Y, Sumi T, Meno C. Wnt signaling regulates left-right axis formation in the node of mouse embryos. Dev Biol. 2013;380:222–232. doi: 10.1016/j.ydbio.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O'Brien TP, Hamada H, Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes and Development. 2003;17:1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisúa-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Miller SA, White RD. Right-left asymmetry of cell proliferation predominates in mouse embryos undergoing clockwise axial rotation. Anatomical Record. 1998;250:103–108. doi: 10.1002/(SICI)1097-0185(199801)250:1<103::AID-AR10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annual Review of Genetics. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Saito D, Kawasumi A, Shinohara K, Asai Y, Takaoka K, Dong F, Takamatsu A, Belo JA, Mochizuki A, Hamada H. Fluid flow and interlinked feedback loops establish left-right asymmetric decay of Cerl2 mRNA. Nature communications. 2012;3:1322. doi: 10.1038/ncomms2319. [DOI] [PubMed] [Google Scholar]
- Nakaya MA, Biris K, Tsukiyama T, Jaime S, Rawls JA, Yamaguchi TP. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development. 2005;132:5425–5436. doi: 10.1242/dev.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Przemeck GKH, Heinzmann U, Beckers J, Hrabe de Angelis M. Node and midline defects are associated with left-right development in Delta1 mutant embryos. Development. 2003;130:3–13. doi: 10.1242/dev.00176. [DOI] [PubMed] [Google Scholar]
- Rankin CT, Bunton T, Lawler AM, Lee S-J. Regulation of lert-right patterning in mice by growth/differentiation factor-1. Nature Genetics. 2000;24:262–265. doi: 10.1038/73472. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- Satoh N, Tagawa K, Takahashi H. How was the notochord born? Evol Dev. 2013;14:56–75. doi: 10.1111/j.1525-142X.2011.00522.x. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, Guenet J-L, Johnson LL, Dove WF. Induction of recessive lethal mutations in the T/t-H2 region of the mouse genome by a point mutagen. Genetic Research. 1986;47:135–142. doi: 10.1017/s0016672300022977. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, King TR, Dove WF. Saturation germ line mutagenesis of the murine t region including a lethal allele at the quaking locus. Proceedings of the National Academy of Sciences, USA. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K, Kawasumi A, Takamatsu A, Yoshiba S, Botilde Y, Motoyama N, Reith W, Durand B, Shiratori H, Hamada H. Two rotating cilia in the node cavity are sufficient to break left-right symmetry in the mouse embryo. Nat Commun. 2012;3:622. doi: 10.1038/ncomms1624. [DOI] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Developmental Dynamics. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MJ, Wang S, Quinn ME, Haaning A, Ware SM. Zic3 is required in the migrating primitive streak for node morphogenesis and left-right patterning. Hum Mol Genet. 2013;22:1913–1923. doi: 10.1093/hmg/ddt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka C, Sakuma R, Nakamura T, Hamada H, Siajoh Y. Long-range action of Nodal requirees interaction with GDF-1. Genes and Development. 2007;21:3272–3282. doi: 10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. Whole mount in situ hybridization of vertebrate embryos. Oxford: IRL Press; 1992. [Google Scholar]
- Yoshiba S, Shiratori H, Kuo IY, Kawasumi A, Shinohara K, Nonaka S, Asai Y, Sasaki G, Belo JA, Sasaki H, Nakai J, Dworniczak B, Ehrlich BE, Pennekamp P, Hamada H. Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science. 2012;338:226–231. doi: 10.1126/science.1222538. [DOI] [PMC free article] [PubMed] [Google Scholar]