Abstract
The umbilical cord (UC) matrix is a source of multipotent mesenchymal stem cells (MSCs) that have adipogenic potential and thus can be a model to study adipogenesis. However, existing variability in adipocytic differentiation outcomes may be due to discrepancies in methods utilized for adipogenic differentiation. Additionally, functional characterization of UCMSCs as adipocytes has not been described. We tested the potential of three well-established adipogenic cocktails containing IBMX, dexamethasone, and insulin (MDI) plus indomethacin (MDI-I) or rosiglitazone (MDI-R) to stimulate adipocyte differentiation in UCMSCs. MDI, MDI-I, and MDI-R treatment significantly increased PPARγ and C/EBPα mRNA and induced lipid droplet formation. However, MDI-I had the greatest impact on mRNA expressions of PPARγ, C/EBPα, FABP4, GPD1, PLIN1, PLIN2, and ADIPOQ and lipid accumulation, whereas MDI showed the least. Interestingly, there were no treatment group differences in the amount of PPARγ protein. However, MDI-I treated cells had significantly more C/EBPα protein compared to MDI or MDI-R, suggesting that indomethacin-dependent increased C/EBPα may contribute to the adipogenesis-inducing potency of MDI-I. Additionally, BMP4 treatment of UC-MSCs did not enhance responsiveness to MDI-induced differentiation. Finally to characterize adipocyte function, differentiated UCMSCs were stimulated with insulin and downstream signaling was assessed. Differentiated UCMSCs were responsive to insulin at two weeks but showed decreased sensitivity by five weeks following differentiation, suggesting that long-term differentiation may induce insulin resistance. Together, these data indicate that UCMSCs undergo adipogenesis when differentiated in MDI, MDI-I, MDI-R, however the presence of indomethacin greatly enhances their adipogenic potential beyond that of rosiglitazone. Furthermore, our results suggest that insulin signaling pathways of differentiated UCMSCs are functionally similar to adipocytes.
Keywords: Adipogenesis, mesenchymal stem cells, umbilical cord, PPAR-gamma, insulin signaling
INTRODUCTION
The umbilical cord (UC) matrix, also known as Wharton’s Jelly, is a potential source of mesenchymal stem cells (MSCs) for clinical and therapeutic applications (1;2). UC matrix cells have been characterized to have cell-surface markers typical of MSCs (positive for CD10, CD13, CD29, CD44, CD73, CD90 and CD105 and negative for CD14, CD33, CD56, CD31, CD34 and CD45) (3–5) and can differentiate into a number of cell types including: dermal fibroblasts (6), osteoblasts (7;8), chondrocytes (7;9), adipocytes (7), myocytes (10), hepatocytes (11;12) and neural cells (13;14). Furthermore, compared to embryonic stem cells, UCMSCs are a non-controversial resource of multipotent stem cells that can be obtained in large quantities (4;11), and have been shown to retain stem-like qualities over long-term culture (15). In addition to their importance in tissue engineering, transplanted UCMSCs have recently been shown to provide therapeutic benefits in injured renal (16) and hepatic (17) tissue through the release of exosomes. UCMSCs also provide an easily accessible source of human stem cells that can be utilized to study the mechanisms regulating lineage differentiation. Finally, UCMSCs are likely to provide a unique resource to study to impact of developmental programming in the offspring.
The process of cellular differentiation into adipocytes (viz. adipogenesis) has been extensively studied due to the central role of adipose tissue in disorders such as obesity, type 2 diabetes and numerous other related metabolic disorders. Obesity develops from a chronic positive energy imbalance, where energy intake exceeds energy expenditure. Throughout development, adipose tissue expansion is regulated by processes of hypertrophy and hyperplasia. Extensive hypertrophy, as often seen with obesity, can induce a metabolically inflexible and hormone-resistant state, driving the metabolic perturbations that accompany obesity (18;19). On the other hand, adipose tissue hyperplasia has been shown to help maintain a healthy and metabolically responsive phenotype (19). The production of new adipocytes through adipogenesis proceeds through a series of finely tuned steps, beginning with commitment of cells to the adipocyte lineage and culminating in terminal differentiation and acquisition of adipocytic phenotype characterized by lipid accumulation (20;21). To date, many of the mechanisms regulating adipocyte function and differentiation have been widely examined using the mouse fibroblast-derived preadipocyte cell line, 3T3-L1 (22–24). Other studies have been carried out using bipotential or multipotential cell lines and primary mesenchymal cells isolated from the bone-marrow. UCMSCs also provide a relatively accessible human in vitro system to study adipogenesis and as a source of fetal stem cells, can be used to test the effects the in utero environment on differentiation potential. Several differentiation protocols have been utilized to induce adipogenesis in UCMSCs (reviewed in (25)), which can result in various phenotypic outcomes and levels of differentiation, indicating a need for studies that compare the outcomes of various differentiation protocols. Most importantly, characterization of the functionality of adipocytes differentiated from UCMSCs is yet to been described. Finally, although markers of terminal adipocyte differentiation have been characterized to some degree in these cells (7;26–29), adipocyte commitment has not been explored.
In vitro differentiation of UCMSCs into adipocytes can be achieved by inducing adipogenic transcription factors, peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT-enhancer binding proteins (C/EBPα and β) (30), with a differentiation cocktail termed MDI. MDI consists of a phosphodiesterase inhibitor (M: 3-isobutyl-1-methylxanthine, IBMX), a glucocorticoid (D: dexamethasone), and insulin (I). In most cases, MDI is supplemented with a cyclooxygenase inhibitor such as indomethacin or a PPARγ agonist like rosiglitazone. Both indomethacin and rosiglitazone have been shown to induce adipogenesis through the stimulation of PPARγ (31–33), via slightly different mechanisms. Indomethacin drives PPARγ expression, whereas rosiglitazone, a potent PPARγ ligand, increases its transcriptional activity (31). Furthermore, both chemicals have been shown to induce gene expression changes that are independent of PPARγ activation (31;34) that may lead to different levels of differentiation.
There were three main goals in this study to further characterize the adipogenic phenotype of UCMSCs. The first objective was to determine the adipogenic response of UCMSCs to three differentiation conditions: MDI, MDI-indomethacin (MDI-I) and MDI-rosiglitazone (MDI-R) via analysis of cellular morphology, gene expression, and protein levels. Secondly, we set out to determine if treatment with bone morphogenetic protein 4 (BMP4), a potent inducer of adipocyte commitment in the murine mesenchymal stem cell line C3H10T1/2 (35), will commit UCMSCs to an adipocyte lineage, thereby enhancing the differentiation response to MDI, MDI-I and/or MDI-R. Finally, we aimed to functionally characterize UCMSCs differentiated into adipocytes by testing their responsiveness to insulin challenge. We hypothesized that MDI-I will induce the greatest adipogenic response in UCMSCs, and that exposure to BMP4 will commit all cells to an adipocyte lineage, therefore eliminating the differential effects of the three types of differentiation media.
MATERIALS AND METHODS
Isolation of Umbilical Cord Mesenchymal Stem Cells
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Umbilical cords (UC) were collected at the University of Arkansas for Medical Sciences (UAMS), after obtaining written informed consent from mothers during the first trimester of pregnancy. The protocol was approved by the Institutional Review Board at the University of Arkansas for Medical Sciences (NCT01131117). Three inch pieces of UCs from two patients were washed three times with PBS containing 1% antibiotic-antimycotic (ABAM) (Life Technologies, Carlsbad, CA) and stored at 4°C until processing (within 2–3 h of collection). Cells from the UC matrix (UCMSCs) were isolated from the two UCs, pooled, and expanded using media as described elsewhere (36). In brief, UCs were divided into 1 inch pieces, sliced open longitudinally using a sterile surgical blade to expose the umbilical cord matrix, vessels were removed, and tissue was scored horizontally many times to help liberate the cells. Next, the tissue underwent a series of enzymatic digestions (300 U/ml collagenase + 1 mg/ml hyaluronidase (45 min), and 0.25% trypsin-EDTA (15 min)) followed by 10 minutes of scraping the tissue with a forceps in 1x PBS to remove cells. The solution was placed on a 100 µm cell strainer; cells were collected, and centrifuged at 1000 rpm for 5 minutes. UCMSCs were counted and plated in growth media (Supplemental Table 1) in a single well of a 6-well plate. Cells were expanded until the third passage after which they were fluorescently labeled with antibodies against CD13, CD29, CD44, CD90, CD105, CD31, CD34, and CD45 (BioLegend, San Diego, CA) and analyzed via FACS. UCMSCs were then plated for adipogenesis experiments described below.
Cell Culture and Adipocyte Differentiation
UCMSCs were maintained in growth media until reaching 80% confluence. Cells were plated for experimentation at 1×104 cells/cm2, one day prior to treatment with BMP4 (R&D Systems, Minneapolis, MN) or vehicle, after which media containing BMP4 (50, 100, 150 ng/ml) or vehicle, which was replaced every other day for 4 d. UCMSCs were either collected at this point or treated with adipogenic media: DMEM containing 10% FBS, 1% antibiotic-antimycotic, 1 µM dexamethasone, 500 µM IBMX, 1 µM insulin (MDI) with the addition of either 60 µM indomethacin (MDI-I), or 10 µM rosiglitazone (Cayman Chemicals, Boston, MA) (MDI-R). Cells were cultured for up to 5 weeks with the appropriate adipogenic media replaced every third day. An insulin challenge was performed in some cases on differentiated cells. For these experiments, cells were serum starved for 16 h followed by treatment with 100 nM insulin for 10-min. The insulin was removed, cells washed with PBS, and collected for protein analysis as described below.
mRNA Isolation and qRT-PCR
Total RNA was isolated using RNeasy Mini kit including on-column DNase digestion (Qiagen, Valencia, CA) from n = 6 wells per treatment group. RNA integrity was assessed using Experion RNA StdSens analysis kit (BioRad). Total RNA (1 µg) was reverse transcribed using IScript cDNA synthesis kit (BioRad), and subsequent real-time PCR analysis was performed using an ABI Prism 7500 FAST sequence detection system (Applied Biosystems, Foster City, CA). Gene specific primers were designed using Primer Express Software (Applied Biosystems). The relative amounts of mRNA were quantified using a standard curve and normalized to the expression of SRP14 mRNA. Primer sequences are depicted in Supplemental Table 2.
Oil Red O Staining and Microscopy
UCMSCs (n = 3 per treatment group) were fixed for 15 minutes in 3% paraformaldehyde, permeabilized with 0.1% Triton X-100 for 5 minutes, and blocked with PBS containing 5% BSA for 30 minutes at room temperature. To visualize lipid accumulation over the differentiation time course, cells were stained with a lipid soluble dye (Oil-Red O in 60% isopropanol) for 45 minutes, followed by 3 washes with PBS. Actin filaments were stained using Alexa fluor-488 phalloidin (Life Technologies) per manufacturer’s instructions. All fluorescent labeling dyes were examined using an Axio Vert 200 fluorescent microscope (Carl Zeiss, Oberkochen, Germany). Axiovision software (Carl Zeiss) was used to semi-quantitatively estimate the amount of triglyceride (TG) storage by measuring the percentage of Oil-red-O staining per 40x field and normalizing to nuclear content (count of DAPI-stained nuclei/field). Lipid droplet size was also semi-quantitatively estimated using Axiovision software, by dividing the total area stained with Oil-red O in each field by the number of individual particles. Four 40x fields were analyzed per sample (n = 3 per treatment group).
Protein Isolation and Immunoblotting
Total cell lysates from n = 3–5 wells per treatment group were prepared in RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1.0% NP-40, 1.0% deoxycholic acid, 0.1% SDS, 2 mM EDTA) containing 1 mM PMSF and protease inhibitor cocktail. Proteins were resolved by SDS-PAGE and immunoblotting was carried out using standard procedures (37). Membranes were incubated with primary antibodies against PPARγ (#sc-7196), phosphorylated ERK1/2 (P-ERK1/2) (#sc-81492) (both from Santa Cruz Biotechnology Inc., Dallas, Texas), C/EBPα (#3087), C/EBPβ (#229C), AKT (#9272), phosphorylated AKT (P-AKT) (#5106S), JNK (#9258), phosphorylated JNK (P-JNK) (#4668), ERK1/2 (#46955), and α-tubulin (#9099S) (Cell Signaling, Danvers, MA) for 16 h at 4°C. HRP-conjugated secondary antibodies against rabbit and mouse IgG (Santa Cruz Biotechnology, Inc.) were used for protein detection. Quantitation of immunoblots was performed using Quantity One software (Biorad).
Statistical Analysis
Real-time RT-PCR data are expressed as mean fold change from control ± SEM and all other data are expressed as means ± SEM. None of the data is transformed. Repeated measures two-way ANOVA followed by Tukey’s or Sidak’s multiple comparisons test was used to compare the three differentiation treatments across multiple time points. For BMP stimulation of UCMSCs, one-way ANOVA followed by all-pair wise comparison by the Student-Neuman-Keuls method was performed. For all statistical tests, P ≤ 0.05 was considered to be statistically significant. Statistical analyses were performed using GraphPad Prism 6 (v 6.02, La Jolla, CA).
RESULTS
UC matrix cells express mesenchymal stem cell surface markers
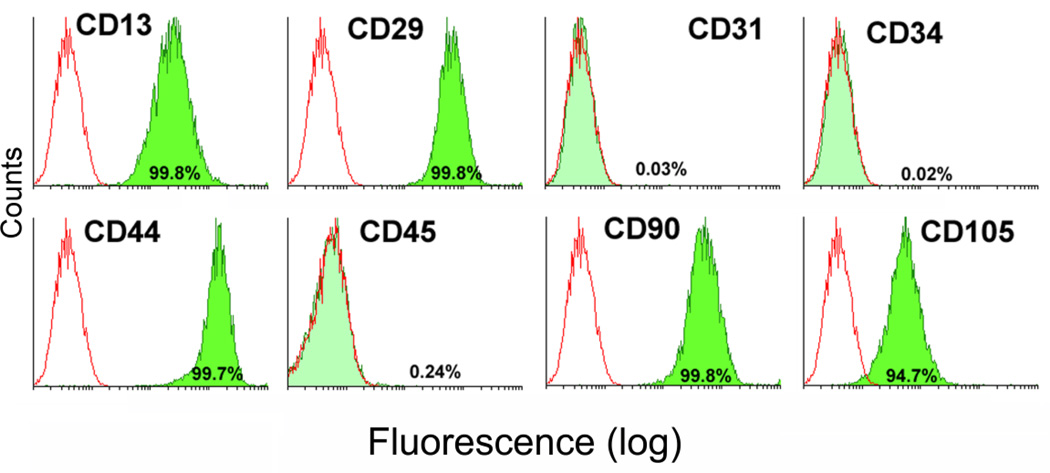
Flow cytometric analysis of cell surface antigens revealed that expanded UC cells homogenously expressed CD13, CD29, CD44, CD90 and CD105, whereas only 0.3% of cells expressed CD31, CD34, and CD45 (Figure 1), confirming a mesenchymal stem cell phenotype.
Figure 1. Analysis of stem cell surface markers on UCMSCs.
Expanded umbilical cord matrix cells (UCMSCs) that were collected from two individuals and pooled were labeled with antibodies against cell surface antigens (CD13, CD29, CD44, CD90 CD31, CD34, CD45, and CD105) and analyzed via flow cytometry. Open histograms represent the background signal; the green histogram indicates a positive signal for the indicated antibody. The percentage listed depicts the percentage of cells that showed a positive signal above background for the indicated antibody.
MDI-I induces the greatest adipogenic response by 5 weeks
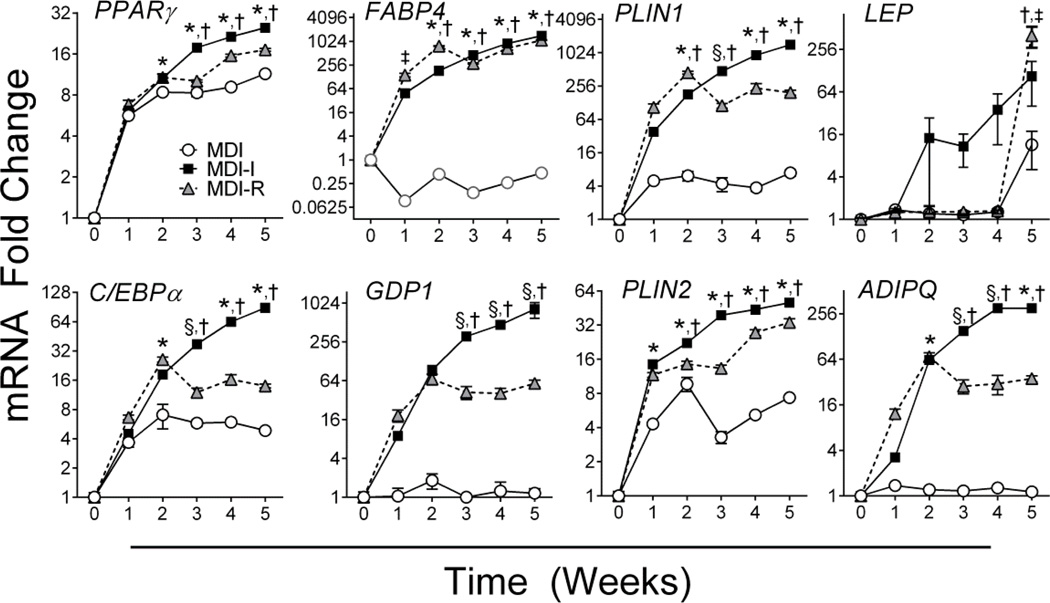
Adipocyte differentiation is characterized by the establishment of a transcriptional program that drives adipocyte-specific gene expression and function. Consistent with adipocyte differentiation, MDI, MDI-I, and MDI-R treatment led to ~6-fold and ~5-fold increase in the expression of adipogenic transcriptional regulators, PPARγ and C/EBPα respectively, by 1 week of treatment (P < 0.001, Figures 2A, B). Additionally, mRNA expression of lipid droplet proteins, PLIN1 and PLIN2, were significantly induced (P < 0.001) by 5-fold and 4-fold with MDI, 38-fold and 14-fold with MDI-I, and 105-fold and 12-fold MDI-R, respectively, following 1 week of differentiation (Figures 2E, F). Repeated measures two-way ANOVA indicated significant differences (P < 0.05) in the gene expression of PPARγ, C/EBPα, FABP4, GPD1, PLIN1, PLIN2, and ADIPOQ between groups at every time point following 2 weeks of differentiation (Figures 2A, B, C, D, E, F, H). For all genes, MDI-I and MDI-R induced significantly greater gene expression than did treatment with MDI alone (P < 0.05). By 5 weeks, gene expression in cells differentiated with MDI-I was significantly greater than in cells differentiated with MDI or MDI-R for all genes (P < 0.001), with the exception of LEP where MDI-R induced the greatest expression (Figure 2G). Interestingly, UCMSCs differentiated in MDI-R had significantly greater mRNA expression of FABP4 (P < 0.001) and PLIN1 (P < 0.001) compared to MDI and MDI-I at 2 weeks of differentiation. Thereafter, a decrease or leveling off in gene expression for many of the adipogenic genes occurred in the MDI-R group.
Figure 2. Time course analysis of adipogenic gene expression following differentiation.
mRNA expression in UCMSCs differentiated with MDI (open circle), MDI-I (black square), and MDI-R (gray triangle) for 0–5 weeks was analyzed via quantitative RT-PCR (n = 6 per treatment group). A: PPARγ, B: C/EBPα, C: FABP4, D: GPD1, E: PLIN1, F: PLIN2, G: LEP, H: ADIPOQ. Values are expressed as mean fold change from undifferentiated UCMSCs (0 week) ± SE. The Y-axis is log-scale. Repeated measures two-way ANOVA followed by Tukey’s multiple comparison test was performed to compare treatment groups. For each time point, * represents a significant difference from MDI, † represents a significant difference between MDI-I and MDI-R, ‡ represents a significant difference between MDI and MDI-R, and § represents a significant difference between MDI and MDI-I (P < 0.05).
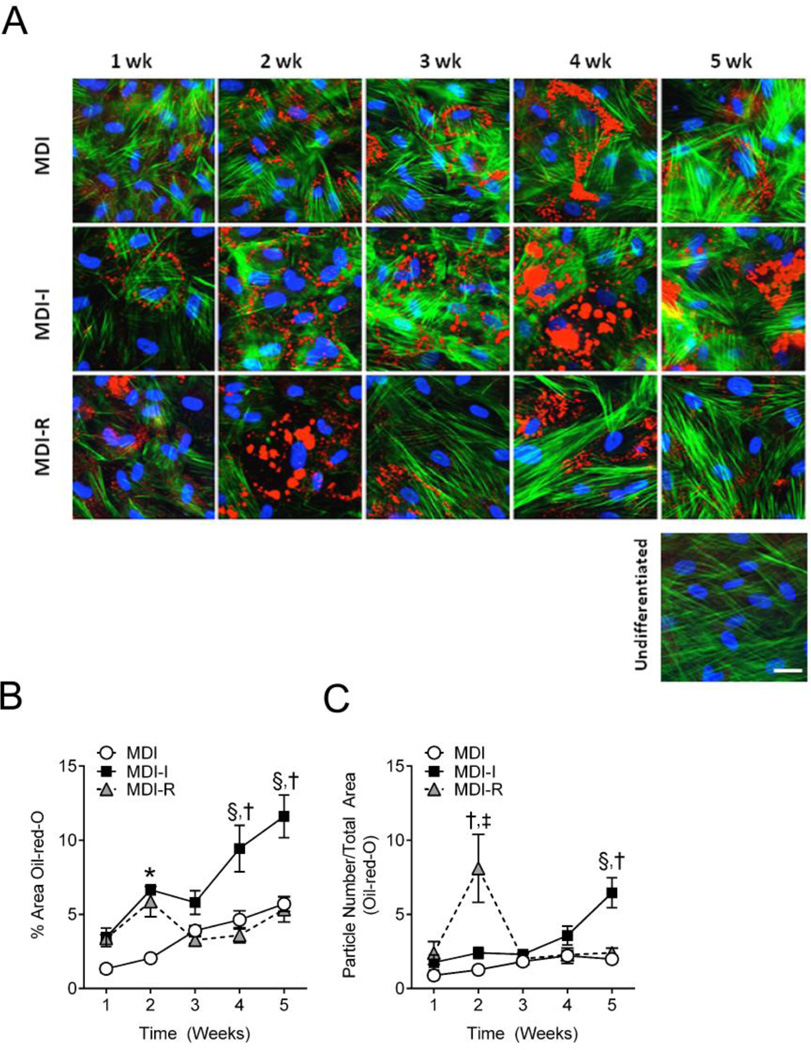
TG accumulation in lipid droplets is indicative of a mature adipocyte and increases throughout the differentiation process. Therefore, the amount of TG present is a common way of measuring the degree of adipocyte differentiation. To measure TG accumulation, we performed semi-quantitative analysis of Oil-red-O stained UCMSCs differentiated in MDI, MDI-I, or MDI-R. Consistent with gene expression changes, lipid staining was apparent in nearly all cells treated with MDI, MDI-I, and MDI-R by 1 week (Figure 3A) and progressively increased over the 5 weeks in both MDI and MDI-I treated cells (Figure 3B). Similar to what was observed with gene expression, cells treated with MDI-R had a drop in lipid staining between 2 weeks and 3 weeks, after which TG storage increased to similar levels as those observed at 1 week post treatment, suggesting a potential loss of differentiation or differentiated cells. Repeated measures two-way ANOVA showed that after 3 weeks of treatment, cells differentiated in MDI-I had significantly more TG compared to either MDI or MDI-R. As differentiation progresses, adipocytes not only accumulate greater amounts of TG, but they also display larger lipid droplets. To semi-quantitatively measure lipid droplet size, we divided the total area of Oil-red-O staining by the number of particles measured. Interestingly at 2 weeks, cells differentiated in MDI-R had significantly larger lipid droplets compared to MDI and MDI-I (Figure 3C), which dramatically decreased by 3 weeks and was no longer different than MDI or MDI-I. By 5 weeks of differentiation, MDI-I had both more TG accumulation (Figure 3B) and significantly larger lipid droplets (Figure 3C) than both MDI and MDI-R, suggesting that MDI-I had the most potent effect on adipocyte differentiation.
Figure 3. Semi-quantitative analysis of triglyceride accumulation and lipid droplet size in differentiated UCMSCs.
UCMSCs differentiated for 1–5 weeks in MDI (open circle), MDI-I (black square), and MDI-R (gray triangle) were fixed and stained for neutral lipids with Oil-red-O (Red), actin filaments with phalloidin (Green), and DNA with DAPI (Blue). The white calibration bar indicates 10 µm. A: Representative images of differentiated UCMSCs were taken at 40x using fluorescent microscopy. B and C: Masking software (Axiovision) was used to quantify the areas stained red from 4–6 images per group, per time point. B: Triglyceride accumulation was determined as the % Area stained red normalized to the number of nuclei per image. C: Lipid droplet size was determined by dividing the total masked area (red) by the number of particles counted. Values are expressed as mean ± SE. Repeated measures two-way ANOVA with Tukey’s posthoc test was performed to compare treatment groups. For each time point, * represents a significant difference between MDI and both MDI-I and MDI-R, † represents a significant difference between MDI-I and MDI-R, ‡ represents a significant difference between MDI and MDI-R, and § represents a significant difference between MDI and MDI-I (P < 0.05).
MDI-I drives adipogenesis by increasing C/EBPα
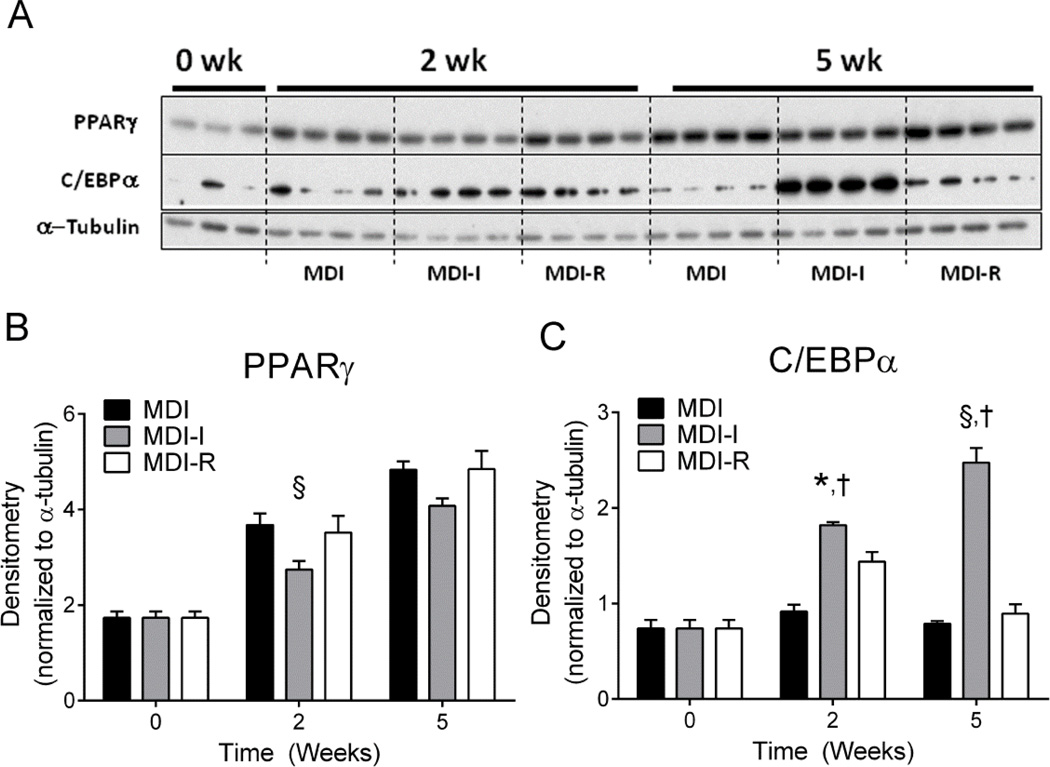
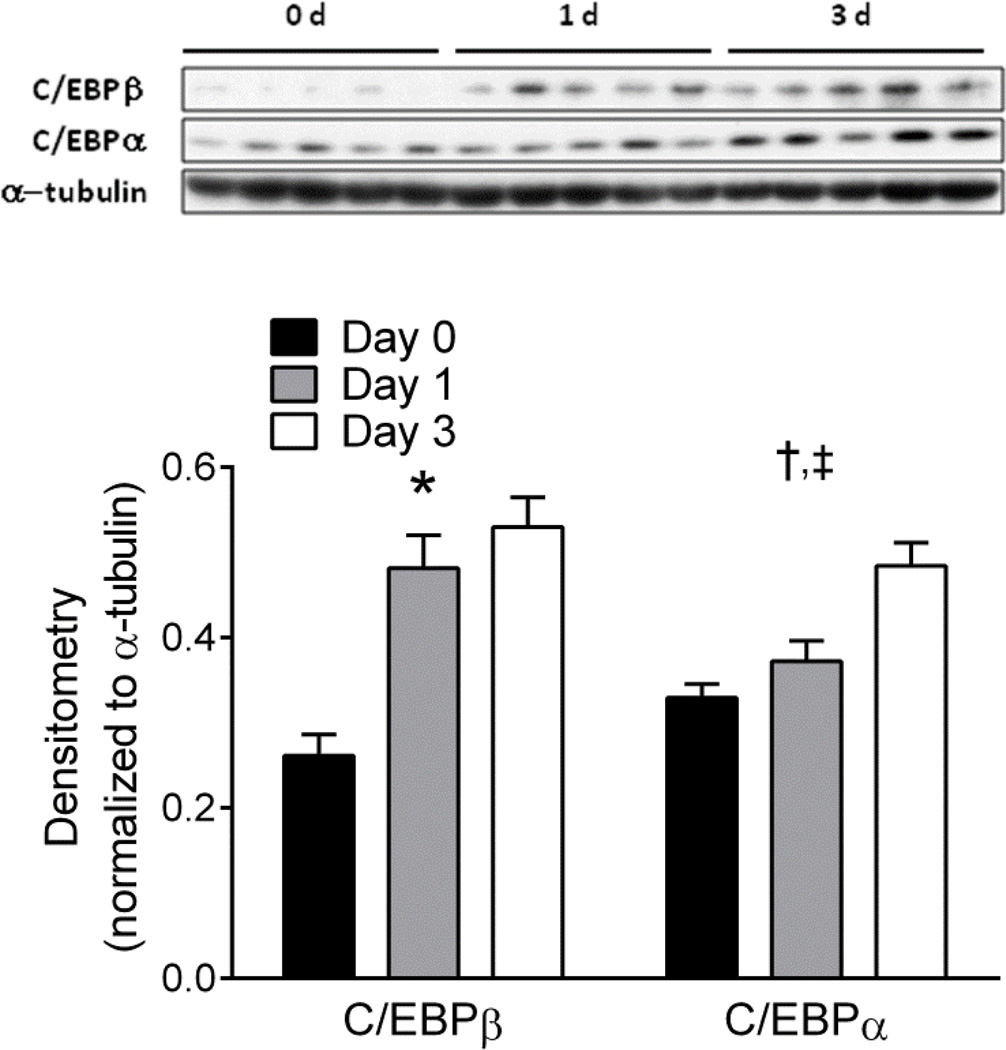
Next we measured protein levels of PPARγ and C/EBPα to ascertain if protein level changes paralleled the observed gene expression changes (Figure 4A). Following 2 and 5 weeks of differentiation, PPARγ protein levels were significantly increased by at least 1.5-fold and 2.5-fold respectively (Figure 4B), in all groups when compared to undifferentiated UCMSCs. However, in contrast to gene expression changes, there were no significant differences in PPARγ protein levels between MDI, MDI-I, or MDI-R groups after 5 weeks of differentiation (Figure 4B). On the other hand, C/EBPα protein levels were more consistent with the observed gene expression changes (Figure 4A, C). Accordingly, MDI-I treatment induced the greatest increase in C/EBPα protein expression by 2 and 5 weeks (3-fold and 6-fold, respectively), whereas there was no significant effect of MDI or MDI-R on C/EBPα levels by 5 weeks (Figure 4C). These data suggest that indomethacin may drive the expression of C/EBPα independently of other adipogenic stimuli. C/EBPα plays a significant role in regulating the genetic program that synchronizes adipogenesis and may therefore be the underlying mechanism for which MDI-I has such potent effects on adipocyte differentiation. To test if indomethacin could directly induce C/EBPα levels, we cultured UCMSCs for 0, 1, and 3 days in DMEM supplemented with 10% FBS and 60 µM indomethacin and measured protein levels of C/EBPα and its upstream regulator C/EBPβ (Figure 5). Following 1 d of indomethacin treatment, we observed a 1.8-fold increase in the amount of C/EBPβ protein but no increase in C/EBPα. By 3 d of treatment there was a significant increase in both C/EBPβ (2-fold) and C/EBPα (1.5-fold) compared to untreated UCMSCs indicating that indomethacin can induce C/EBPα protein in absence of MDI, most likely through stimulation of C/EBPβ expression.
Figure 4. Analysis of PPARγ and C/EBPα protein levels following 2 and 5 weeks of adipocyte differentiation.
Protein levels of PPARγ and C/EBPα in UCMSCs differentiated in MDI, MDI-I, and MDI-R for 2 and 5 weeks were analyzed via immunoblotting. A: Immunoblots probed for PPARγ and C/EBPα. B and C: Densitometric analysis of PPARγ and C/EBPα levels normalized to α-tubulin. Values are expressed as mean ± SE. Significance between groups was determined using a repeated measures two-way ANOVA, followed by Tukey’s multiple comparisons test. For each time point, * represents a significant difference between MDI and both MDI-I and MDI-R, † represents a significant difference between MDI-I and MDI-R, ‡ represents a significant difference between MDI and MDI-R, and § represents a significant difference between MDI and MDI-I (P < 0.05).
Figure 5. Analysis of C/EBPα and C/EBPβ protein levels following 0, 1, and 3 days exposure to indomethacin.
Protein levels of C/EBPα and C/EBPβ in UCMSCs treated with 60 µM indomethacin for 0 (black bar), 1 (gray bar), and 3 (white bar) days were analyzed via immunoblot analysis. The graph depicts densitometric quantitation of immunoblots probed for C/EBPα and C/EBPβ. Values are expressed as mean ± SE. Significance between groups was determined using a repeated measures two-way ANOVA, followed by Tukey’s test for multiple comparisons. For each time point, * represents a significant difference between MDI and both MDI-I and MDI-R, † represents a significant difference between MDI-I and MDI-R, ‡ represents a significant difference between MDI and MDI-R, and § represents a significant difference between MDI and MDI-I (P < 0.05).
Commitment with BMP4 stimulation
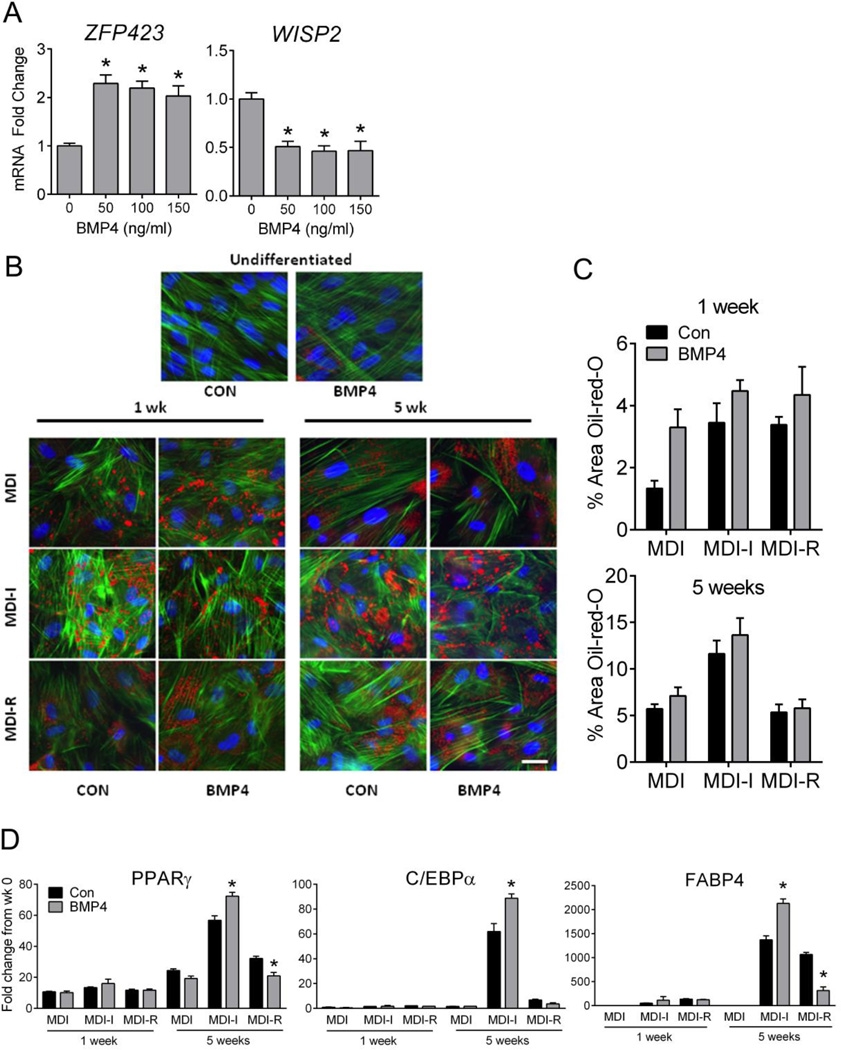
Because UCMSCs treated with MDI showed the least amount of differentiation, we next tested the hypothesis that akin to the mouse clonal MSC cell line C3H10T1/2, commitment to an adipocyte lineage via BMP4 stimulation (a known regulator of adipocyte commitment(35)) would increase the response of UCMSCs to MDI-induced differentiation (Figure 6). Treatment with 50, 100, or 150 ng/ml of BMP4 for 4 days significantly increased mRNA expression of ZFP423 and decreased expression of WISP2 in UCMSCs, however, there was no dose-dependent response to BMP4 treatment (Figure 6A). Pre-treatment with 100 ng/ml of BMP4 showed a trend for increased TG storage in UCMSCs differentiated in MDI by 1 week, however there was no effect on UCMSCs differentiated in MDI-I or MDI-R (Figure 6B, C). By 5 weeks of differentiation, there was no effect of BMP4 treatment on TG storage for any of the groups (Figure 6C).
Figure 6. Analysis of adipocyte commitment in UCMSCS via BMP4 stimulation.
UCMSCs were treated with or without BMP4 for 4 days prior to differentiation with MDI, MDI-I, MDI +R. A: Quantitative RT-PCR analysis of WISP2 and ZFP423 mRNA expressions in UCMSCs following increasing doses (50, 100, 150 ng/ml) of BMP4. Values are expressed as the mean fold change from untreated cells (0) ± SE. Significance between 0 and 50, 100, or 150 was determined using one-way ANOVA, followed by Tukey’s posthoc test (* represents a significant difference from untreated cells, P < 0.05). B: Representative, 40x images of UCMSCs treated with (CON) or without BMP4 (100 ng/ml) following adipocyte differentiation for 1 and 5 weeks. Triglyceride = red (Oil-red-O stain), Actin = green (phalloidin stain), Nuclei = blue (DAPI stain). The white calibration bar indicates 10 µm. C: Semi-quantitative analysis of triglyceride accumulation using masking software (Axiovision) as described in Methods. Values are expressed as mean % Area ± SE following normalization to number of nuclei. A repeated measures two-way ANOVA was used to compare CON (black bar) and BMP4 (gray bar) groups for cells differentiated with MDI, MDI-I, or MDI-R (P < 0.05). D: Quantitative RT-PCR analysis of PPARγ, C/EBPα, and FABP4 mRNA expressions in UCMSCs treated with (CON) or without BMP4 (100 ng/ml) prior to differentiation. Values are expressed as the mean fold change from untreated cells (0 week) ± SE. Significant differences were determined by repeated measures two-way ANOVA followed by Sidak’s multiple comparisons test. * represents a significant difference (P < 0.05) between CON (black bars) and BMP4 (gray bars) at each time point under the varying differentiation conditions (MDI, MDI-I, or MDI-R) (P < 0.05).
In cells pre-treated with BMP4 and differentiated for 1 week in MDI, there was no induction of PPARγ, C/EBPα, or FABP4 mRNA expression (Figure 6D). BMP4 pre-treated cells differentiated in MDI-I for 1 week also had no induction of PPARγ, C/EBPα, or FABP4 mRNA expression compared to control (Figure 6D). By 5 weeks of differentiation, BMP4 treated cells differentiated in MDI-R showed significantly reduced expressions of PPARγ and FABP4 compared to CON. BMP4 treatment led to significantly greater PPARγ C/EBPα, and FABP4 gene expression by 5 weeks of differentiation in MDI-I compared to CON (Figure 6D).
Differentiated UCMSCs are insulin responsive at 2 weeks and resistant by 5 weeks
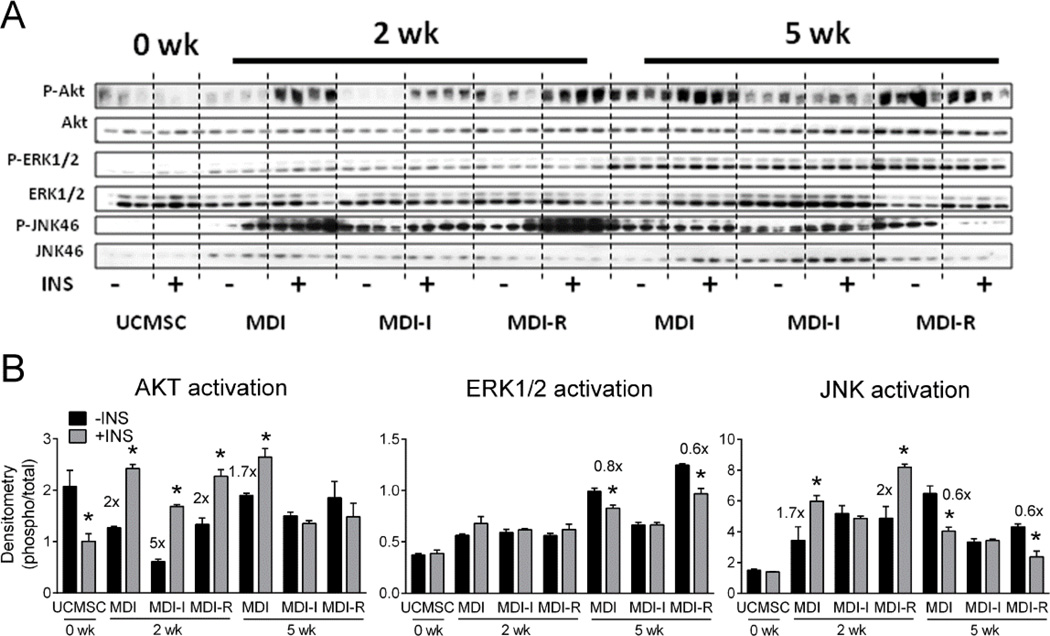
To test if differentiated UCMSCs were insulin responsive, we stimulated cells with 100 nM insulin or vehicle at 2 or 5 weeks post differentiation and measured total and phosphorylated protein levels of AKT, ERK1/2, and JNK (Figure 7A). Following 2 weeks of differentiation, insulin stimulated a significant increase in ratio of phosphorylated AKT (P-AKT) to total AKT in cells treated with MDI, MDI-I, and MDI-R (Figure 7B), suggesting that these cells were insulin sensitive. Cells differentiated in MDI-I had a 5-fold induction of P-AKT to AKT, compared to a 2-fold or a 1.7-fold increase in MDI or MDI-R treated cells, respectively, in response to the insulin challenge, suggesting that UCMSCs differentiated in MDI-I are the most insulin responsive. By 5 weeks of differentiation, only the cells treated with MDI had a significantly increased P-AKT/AKT ratio (Figure 7B) in response to insulin, suggesting that long-term exposure to MDI-I and MDI-R may induce insulin resistance in UCMSCs.
Figure 7. Analysis of AKT, ERK1/2 and JNK activation in differentiated UCMSCs following insulin challenge.
UCMSCs differentiated with MDI, MDI-I, or MDI-R for 2 and 5 weeks were serum starved overnight and stimulated with (+INS) or without (-INS) 100 nM insulin for 10 min. Protein levels of phosphorylated (P-) and total AKT, ERK1/2, and JNK were analyzed via immunoblotting. A: Images of immunoblots probed for P-AKT, AKT, P-ERK1/2, ERK1/2, P-JNK, JNK, and α-tubulin. B-D: Densitometric analysis of (B) AKT, (C) ERK1/2, and (D) JNK levels normalized to α-tubulin. Values are expressed as the mean ratio of phosphorylated to total protein ± SE. Significant differences were determined by repeated measures two-way ANOVA followed by Sidak’s multiple comparisons test. * represents a significant difference (P < 0.05) between -INS (black bar) and +INS (gray bar) at each time point under the varying differentiation conditions (MDI, MDI-I, or MDI-R).
Mitogen activated protein kinases (MAPK) ERK1/2 and JNK are also insulin responsive and become phosphorylated following insulin stimulation (38;39). Interestingly, the ratio of P-ERK1/2 to ERK1/2 was not significantly increased in response to insulin for any of the time points or groups, suggesting that ERK1/2 signaling is not insulin responsive in UCMSCs (Figure 7C). Insulin significantly increased the ratio of P-JNK to JNK in cells differentiated for 2 weeks in MDI and MDI-R by ~1.7-fold (Figure 7D). However, by 5 weeks of differentiation insulin stimulation lead to a significant decrease in JNK activation in the MDI and MDI-R groups. Interestingly, there was no effect of insulin on JNK activation in cells differentiated with MDI-I at either 2 or 5 weeks (Figure 7D).
DISCUSSION
A number of cell culture models have been used to study the mechanisms regulating adipocyte differentiation. The UC is a recently identified source of MSCs that have adipogenic potential (7;26–28;40;41). As a novel source of fetal cells, UCMSCs can be used to determine if the in utero environment affects the differentiation potential of fetal stem cells. However, discrepancies exist regarding the techniques used to study in vitro adipogenic differentiation of these cells (25;42). Furthermore, their functional characterization as adipocytes and cellular commitment to the adipocyte lineage has not been described. In this study, we tested the potential of three adipogenic cocktails used to stimulate adipocyte differentiation in UCMSCs and found that all three induced adipocyte differentiation, albeit to different levels. MDI-I was the most effective at inducing adipocyte differentiation and our data suggest that indomethacin-dependent regulation of C/EBPβ and C/EBPα may be an underlying mechanism for the potent effects of MDI-I on UCMSC adipogenesis. Additionally, the present findings suggest that UCMSCs show minimal adipocyte differentiation in the absence of a PPARγ ligand (MDI-treated), which cannot be rescued by BMP4-induced commitment. Finally, to our knowledge, we are the first to show that differentiated UCMSCs are insulin responsive early in differentiation but have decreased insulin sensitivity by 5 weeks of differentiation.
Cellular commitment to an adipocyte lineage is the first step in adipogenesis, followed by terminal differentiation (43). In cells that have bipotential to differentiate into osteoblasts or adipocytes and in committed pre-adipocytes such as 3T3-L1 cells, MDI is a potent inducer of adipogenesis (44). However, MDI alone is not sufficient to induce adipocyte differentiation in multipotent MSCs (45), suggesting that some level of adipocyte commitment is necessary for MDI-induced adipogenesis. BMP4 is a member of the TGFβ superfamily that promotes commitment of MSCs to an adipocyte lineage (45–47) through regulating Wnt (wingless-type MMTV integration site family) signaling. Specifically, BMP4 induces the dissociation of Zfp423 from WISP2 (a Wnt1 signaling pathway protein) (46) leading to nuclear translocation of Zfp423 and increased PPARγ expression (48). Zfp423 is elevated in preadipocyte cell lines and regulates basal levels of PPARγ, suggesting that it may be a marker for adipocyte commitment (48). We observed sub-maximal differentiation of UCMSCs in the presence of MDI, suggesting that these cells may need some prior adipocyte commitment stimulation. However, BMP4 treatment had minimal effects on increasing the responsiveness of UCMSCs to MDI. While BMP4 stimulation increased Zfp423 and WISP2 gene expression, there was very little effect of BMP4 on lipid accumulation or gene expression following differentiation. Untreated UCMSCs had a relatively high basal level of PPARγ protein and small amounts of lipid accumulation, suggesting that these cells may already be primed for adipocyte commitment. It is possible that a component of the UCMSC growth media induced a preadipocyte competency, making these cells more receptive to adipocyte differentiation.
In C3H10T1/2 pluripotent stem cells, MDI-I induces PPARγ expression (31), therefore it was not surprising that in our UCMSCs, MDI-I had the greatest effects on adipocyte differentiation. However, differences between the effects of MDI-I and MDI-R on UCMSC adipogenesis were not anticipated because both indomethacin and rosiglitazone are PPARγ agonists. PPARγ does not work alone in regulating the adipogenic program, C/EBPα also plays a significant role (49;50) and C/EBPα and PPARγ work in concert to regulate terminal differentiation of adipocytes by increasing the expression of each other and of key genes regulating adipocyte functions (24;51). Because PPARγ protein levels did not differ between groups of UCMSCs, indomethacin- or rosiglitazone-stimulated PPARγ expression cannot explain the phenotypic differences observed in differentiated UCMSCs. Thus, MDI-I-induced expression of C/EBPα may be driving increased differentiation of UCMSCs treated with MDI-I.
Obesity is often accompanied with hyperinsulinemia and insulin resistance of adipose, liver, and muscle tissues. Chronic insulin treatment in differentiated 3T3-L1 cells induces an insulin resistant state that is characterized by impaired PI3-kinase and MAP-kinase signaling (52). To our knowledge, this is the first report testing the insulin responsiveness of UCMSCs differentiated into adipocytes. Although the UCMSCs showed significant activation of AKT following insulin stimulation by 2 weeks of differentiation, by 5 weeks only the cells differentiated with MDI retained insulin sensitivity. These results are surprising, especially since thiazolidinediones, such as rosiglitazone, are insulin sensitizers. Additionally, C/EBPα, which is most elevated in the MDI-I group, is essential for maintaining insulin responsiveness (53). Increased oxidative stress, ER stress, and mitochondrial dysfunction have all been implicated as potential mechanisms underlying the insulin resistance developed from chronic insulin exposure in 3T3-L1 cells (54). Accordingly, indomethacin has been shown to induce oxidative stress and mitochondrial dysfunction in intestinal cells (55), suggesting a potential mechanism for reduced insulin sensitivity in MDI-I differentiated UCMSCs. Alternatively, a more recent report indicates that a certain level of reactive oxygen species (ROS) production is necessary for adipogenesis to take place (56) and perhaps part of the potency of indomethacin as a inducer of adipogenesis is through stimulating ROS production.
Besides its role in regulating glucose uptake, insulin is also responsible for regulating a number of other metabolic and mitogenic pathways. ERK1/2 is a key mediator of the mitogenic actions of insulin (57) and is important for the insulin effects on adipocyte differentiation (58), while JNK activation can induce insulin resistance (59–61) by inhibiting insulin signaling. Interestingly, although ERK1/2 activation increased over time with adipocyte differentiation, UCMSCs did not responds to insulin by activating ERK1/2, suggesting that this mitogenic pathway was inactive. We observed the activation of JNK in response to insulin stimulation earlier during differentiation, however this was reversed by 5 weeks.
In conclusion, the present studies uncover fundamental aspects of UCMSC differentiation capacity towards the adipocytic lineage. Our findings clearly demonstrate that while all three conditions promote adipocyte differentiation, MDI-I was the most effective at inducing adipocyte differentiation. These findings are consistent with indomethacin-dependent regulation of C/EBPβ and C/EBPα as an underlying mechanism for the potent effects of MDI-I on UCMSC adipogenesis. Importantly, the present findings also suggest that UCMSCs demonstrate minimal adipocyte differentiation in the absence of a PPARγ ligand (MDI-alone), which cannot be rescued by BMP4-induced commitment. Finally, these studies reveal that ex vivo differentiated UCMSCs are insulin responsive early in differentiation but have decreased sensitivity by 5 weeks of differentiation. Overall, these data provide primary evidence relating to the functional capacity of ex vivo differentiated UCMSC-derived adipocytes and their adipogenic potential.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the ACNC-Human Studies Core for their assistance. These studies were supported in part by the U.S. Department of Agriculture-ARS-CRIS 6251-51000-005-00D and National Institutes for Health Grant R01-DK084225 (K.S.). Nursing support was provided in part by the UAMS Translational Research Institute funded by the NIH-CTSA program, Grants UL1-TR-000039 and KL2-TR-000063.
Footnotes
AUTHOR CONTRIBUTIONS
JS, AA, TMB and KS conceived and designed the study; JS, FEL and YZ performed laboratory experiments; JS, AA, KS, and TMB analyzed data and interpreted the results; JS created the figures and drafted the manuscript; JS, KS, AA, TMB edited and revised manuscript; All authors approved the final version of the manuscript.
Reference List
- 1.Tsagias N, Koliakos I, Karagiannis V, Eleftheriadou M, Koliakos GG. Isolation of mesenchymal stem cells using the total length of umbilical cord for transplantation purposes. Transfus Med. 2011;21:253–261. doi: 10.1111/j.1365-3148.2011.01076.x. [DOI] [PubMed] [Google Scholar]
- 2.Weiss ML, Medicetty S, Bledsoe AR, Rachakatla RS, Choi M, Merchav S, Luo Y, Rao MS, Velagaleti G, Troyer D. Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson's disease. Stem Cells. 2006;24:781–792. doi: 10.1634/stemcells.2005-0330. [DOI] [PubMed] [Google Scholar]
- 3.Yan M, Sun M, Zhou Y, Wang W, He Z, Tang D, Lu S, Wang X, Li S, Li H. Conversion of human umbilical cord mesenchymal stem cells in Wharton's jelly to dopamine neurons mediated by the Lmx1a and neurturin in vitro: potential therapeutic application for Parkinson's disease in a rhesus monkey model. PLoS One. 2013;8:e64000. doi: 10.1371/journal.pone.0064000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nekanti U, Mohanty L, Venugopal P, Balasubramanian S, Totey S, Ta M. Optimization and scale-up of Wharton's jelly-derived mesenchymal stem cells for clinical applications. Stem Cell Res. 2010;5:244–254. doi: 10.1016/j.scr.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Nekanti U, Rao VB, Bahirvani AG, Jan M, Totey S, Ta M. Long-term expansion and pluripotent marker array analysis of Wharton's jelly-derived mesenchymal stem cells. Stem Cells Dev. 2010;19:117–130. doi: 10.1089/scd.2009.0177. [DOI] [PubMed] [Google Scholar]
- 6.Han Y, Chai J, Sun T, Li D, Tao R. Differentiation of human umbilical cord mesenchymal stem cells into dermal fibroblasts in vitro. Biochem Biophys Res Commun. 2011;413:561–565. doi: 10.1016/j.bbrc.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Karahuseyinoglu S, Cinar O, Kilic E, Kara F, Akay GG, Demiralp DO, Tukun A, Uckan D, Can A. Biology of stem cells in human umbilical cord stroma: in situ and in vitro surveys. Stem Cells. 2007;25:319–331. doi: 10.1634/stemcells.2006-0286. [DOI] [PubMed] [Google Scholar]
- 8.Wang HS, Hung SC, Peng ST, Huang CC, Wei HM, Guo YJ, Fu YS, Lai MC, Chen CC. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 2004;22:1330–1337. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- 9.Esposito M, Lucariello A, Costanzo C, Fiumarella A, Giannini A, Riccardi G, Riccio I. Differentiation of human umbilical cord-derived mesenchymal stem cells, WJ-MSCs, into chondrogenic cells in the presence of pulsed electromagnetic fields. In Vivo. 2013;27:495–500. [PubMed] [Google Scholar]
- 10.Conconi MT, Burra P, Di Liddo R, Calore C, Turetta M, Bellini S, Bo P, Nussdorfer GG, Parnigotto PP. CD105(+) cells from Wharton's jelly show in vitro and in vivo myogenic differentiative potential. Int J Mol Med. 2006;18:1089–1096. [PubMed] [Google Scholar]
- 11.Zeddou M, Briquet A, Relic B, Josse C, Malaise MG, Gothot A, Lechanteur C, Beguin Y. The umbilical cord matrix is a better source of mesenchymal stem cells (MSC) than the umbilical cord blood. Cell Biol Int. 2010;34:693–701. doi: 10.1042/CBI20090414. [DOI] [PubMed] [Google Scholar]
- 12.Anzalone R, Lo Iacono M, Corrao S, Magno F, Loria T, Cappello F, Zummo G, Farina F, La Rocca G. New emerging potentials for human Wharton's jelly mesenchymal stem cells: immunological features and hepatocyte-like differentiative capacity. Stem Cells Dev. 2010;19:423–438. doi: 10.1089/scd.2009.0299. [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Feng XY, Cui BL, Law F, Jiang XW, Yang LY, Xie QD, Huang TH. Human umbilical cord Wharton's Jelly-derived mesenchymal stem cells differentiation into nerve-like cells. Chin Med J (Engl) 2005;118:1987–1993. [PubMed] [Google Scholar]
- 14.Mitchell KE, Weiss ML, Mitchell BM, Martin P, Davis D, Morales L, Helwig B, Beerenstrauch M, Abou-Easa K, Hildreth T, Troyer D, Medicetty S. Matrix cells from Wharton's jelly form neurons and glia. Stem Cells. 2003;21:50–60. doi: 10.1634/stemcells.21-1-50. [DOI] [PubMed] [Google Scholar]
- 15.Jo CH, Kim OS, Park EY, Kim BJ, Lee JH, Kang SB, Han HS, Rhee SH, Yoon KS. Fetal mesenchymal stem cells derived from human umbilical cord sustain primitive characteristics during extensive expansion. Cell Tissue Res. 2008;334:423–433. doi: 10.1007/s00441-008-0696-3. [DOI] [PubMed] [Google Scholar]
- 16.Dorronsoro A, Robbins PD. Regenerating the injured kidney with human umbilical cord mesenchymal stem cell-derived exosomes. Stem Cell Res Ther. 2013;4:39. doi: 10.1186/scrt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Yan Y, Wang B, Qian H, Zhang X, Shen L, Wang M, Zhou Y, Zhu W, Li W, Xu W. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev. 2013;22:845–854. doi: 10.1089/scd.2012.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bays HE, Gonzalez-Campoy JM, Bray GA, Kitabchi AE, Bergman DA, Schorr AB, Rodbard HW, Henry RR. Pathogenic potential of adipose tissue and metabolic consequences of adipocyte hypertrophy and increased visceral adiposity. Expert Rev Cardiovasc Ther. 2008;6:343–368. doi: 10.1586/14779072.6.3.343. [DOI] [PubMed] [Google Scholar]
- 19.Hoffstedt J, Arner E, Wahrenberg H, Andersson DP, Qvisth V, Lofgren P, Ryden M, Thorne A, Wiren M, Palmer M, Thorell A, Toft E, Arner P. Regional impact of adipose tissue morphology on the metabolic profile in morbid obesity. Diabetologia. 2010;53:2496–2503. doi: 10.1007/s00125-010-1889-3. [DOI] [PubMed] [Google Scholar]
- 20.Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 21.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelius P, MacDougald OA, Lane MD. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- 23.MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 24.Mandrup S, Lane MD. Regulating adipogenesis. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 25.Scott MA, Nguyen VT, Levi B, James AW. Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2011;20:1793–1804. doi: 10.1089/scd.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng KW, Liou YM. Differential role of actin-binding proteins in controlling the adipogenic differentiation of human CD105-positive Wharton's Jelly cells. Biochim Biophys Acta. 2012;1820:469–481. doi: 10.1016/j.bbagen.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Ragni E, Vigano M, Parazzi V, Montemurro T, Montelatici E, Lavazza C, Budelli S, Vecchini A, Rebulla P, Giordano R, Lazzari L. Adipogenic potential in human mesenchymal stem cells strictly depends on adult or foetal tissue harvest. Int J Biochem Cell Biol. 2013;45:2456–2466. doi: 10.1016/j.biocel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Kim DW, Staples M, Shinozuka K, Pantcheva P, Kang SD, Borlongan CV. Wharton's Jelly-Derived Mesenchymal Stem Cells: Phenotypic Characterization and Optimizing Their Therapeutic Potential for Clinical Applications. Int J Mol Sci. 2013;14:11692–11712. doi: 10.3390/ijms140611692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciavarella S, Dammacco F, De Matteo M, Loverro G, Silvestris F. Umbilical cord mesenchymal stem cells: role of regulatory genes in their differentiation to osteoblasts. Stem Cells Dev. 2009;18:1211–1220. doi: 10.1089/scd.2008.0340. [DOI] [PubMed] [Google Scholar]
- 30.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 31.Styner M, Sen B, Xie Z, Case N, Rubin J. Indomethacin promotes adipogenesis of mesenchymal stem cells through a cyclooxygenase independent mechanism. J Cell Biochem. 2010;111:1042–1050. doi: 10.1002/jcb.22793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Haile A, Jones LC. Rosiglitazone-induced adipogenesis in a bone marrow mesenchymal stem cell line - biomed 2011. Biomed Sci Instrum. 2011;47:213–221. [PubMed] [Google Scholar]
- 33.Benvenuti S, Cellai I, Luciani P, Deledda C, Baglioni S, Giuliani C, Saccardi R, Mazzanti B, Dal Pozzo S, Mannucci E, Peri A, Serio M. Rosiglitazone stimulates adipogenesis and decreases osteoblastogenesis in human mesenchymal stem cells. J Endocrinol Invest. 2007;30:RC26–RC30. doi: 10.1007/BF03350807. [DOI] [PubMed] [Google Scholar]
- 34.Welters HJ, El Ouaamari A, Kawamori D, Meyer J, Hu J, Smith DM, Kulkarni RN. Rosiglitazone promotes PPARgamma-dependent and -independent alterations in gene expression in mouse islets. Endocrinology. 2012;153:4593–4599. doi: 10.1210/en.2012-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Song TJ, Li X, Hu L, He Q, Liu M, Lane MD, Tang QQ. BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2009;106:12670–12675. doi: 10.1073/pnas.0906266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seshareddy K, Troyer D, Weiss ML. Method to isolate mesenchymal-like cells from Wharton's Jelly of umbilical cord. Methods Cell Biol. 2008;86:101–119. doi: 10.1016/S0091-679X(08)00006-X. [DOI] [PubMed] [Google Scholar]
- 37.Saben J, Zhong Y, Gomez-Acevedo H, Thakali KM, Borengasser SJ, Andres A, Shankar K. Early growth response protein-1 mediates lipotoxicity-associated placental inflammation: role in maternal obesity. Am J Physiol Endocrinol Metab. 2013;305:E1–E14. doi: 10.1152/ajpendo.00076.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazar DF, Wiese RJ, Brady MJ, Mastick CC, Waters SB, Yamauchi K, Pessin JE, Cuatrecasas P, Saltiel AR. Mitogen-activated protein kinase kinase inhibition does not block the stimulation of glucose utilization by insulin. J Biol Chem. 1995;270:20801–20807. doi: 10.1074/jbc.270.35.20801. [DOI] [PubMed] [Google Scholar]
- 39.Lee YH, Giraud J, Davis RJ, White MF. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J Biol Chem. 2003;278:2896–2902. doi: 10.1074/jbc.M208359200. [DOI] [PubMed] [Google Scholar]
- 40.Lu LL, Liu YJ, Yang SG, Zhao QJ, Wang X, Gong W, Han ZB, Xu ZS, Lu YX, Liu D, Chen ZZ, Han ZC. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Haematologica. 2006;91:1017–1026. [PubMed] [Google Scholar]
- 41.Ciavarella S, Dammacco F, De Matteo M, Loverro G, Silvestris F. Umbilical cord mesenchymal stem cells: role of regulatory genes in their differentiation to osteoblasts. Stem Cells Dev. 2009;18:1211–1220. doi: 10.1089/scd.2008.0340. [DOI] [PubMed] [Google Scholar]
- 42.Bosch J, Houben AP, Radke TF, Stapelkamp D, Bunemann E, Balan P, Buchheiser A, Liedtke S, Kogler G. Distinct differentiation potential of "MSC" derived from cord blood and umbilical cord: are cord-derived cells true mesenchymal stromal cells? Stem Cells Dev. 2012;21:1977–1988. doi: 10.1089/scd.2011.0414. [DOI] [PubMed] [Google Scholar]
- 43.Bowers RR, Lane MD. Wnt signaling and adipocyte lineage commitment. Cell Cycle. 2008;7:1191–1196. doi: 10.4161/cc.7.9.5815. [DOI] [PubMed] [Google Scholar]
- 44.Ding J, Nagai K, Woo JT. Insulin-dependent adipogenesis in stromal ST2 cells derived from murine bone marrow. Biosci Biotechnol Biochem. 2003;67:314–321. doi: 10.1271/bbb.67.314. [DOI] [PubMed] [Google Scholar]
- 45.Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101:9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammarstedt A, Hedjazifar S, Jenndahl L, Gogg S, Grunberg J, Gustafson B, Klimcakova E, Stich V, Langin D, Laakso M, Smith U. WISP2 regulates preadipocyte commitment and PPARgamma activation by BMP4. Proc Natl Acad Sci U S A. 2013;110:2563–2568. doi: 10.1073/pnas.1211255110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowers RR, Kim JW, Otto TC, Lane MD. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc Natl Acad Sci U S A. 2006;103:13022–13027. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freytag SO, Paielli DL, Gilbert JD. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 50.Lin FT, Lane MD. CCAAT/enhancer binding protein alpha is sufficient to initiate the 3T3-L1 adipocyte differentiation program. Proc Natl Acad Sci U S A. 1994 Sep 13;:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 52.Ricort JM, Tanti JF, Van Obberghen E, Marchand-Brustel Y. Alterations in insulin signalling pathway induced by prolonged insulin treatment of 3T3-L1 adipocytes. Diabetologia. 1995;38:1148–1156. doi: 10.1007/BF00422363. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 54.Ge X, Yu Q, Qi W, Shi X, Zhai Q. Chronic insulin treatment causes insulin resistance in 3T3-L1 adipocytes through oxidative stress. Free Radic Res. 2008;42:582–591. doi: 10.1080/10715760802158448. [DOI] [PubMed] [Google Scholar]
- 55.Basivireddy J, Vasudevan A, Jacob M, Balasubramanian KA. Indomethacin-induced mitochondrial dysfunction and oxidative stress in villus enterocytes. Biochem Pharmacol. 2002;64:339–349. doi: 10.1016/s0006-2952(02)01067-5. [DOI] [PubMed] [Google Scholar]
- 56.Higuchi M, Dusting GJ, Peshavariya H, Jiang F, Hsiao ST, Chan EC, Liu GS. Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and Forkhead box O1 mediated upregulation of antioxidant enzymes. Stem Cells Dev. 2013;22:878–888. doi: 10.1089/scd.2012.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 58.Porras A, Santos E. The insulin/Ras pathway of adipocytic differentiation of 3T3 L1 cells: dissociation between Raf-1 kinase and the MAPK/RSK cascade. Int J Obes Relat Metab Disord. 1996;20:S43–S51. [PubMed] [Google Scholar]
- 59.Aguirre V, Uchida T, Yenush L, Davis R, White MF. The c-Jun NH(2)-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307) J Biol Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- 60.Nguyen MT, Satoh H, Favelyukis S, Babendure JL, Imamura T, Sbodio JI, Zalevsky J, Dahiyat BI, Chi NW, Olefsky JM. JNK and tumor necrosis factor-alpha mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J Biol Chem. 2005;280:35361–35371. doi: 10.1074/jbc.M504611200. [DOI] [PubMed] [Google Scholar]
- 61.Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes. 2003;52:1–8. doi: 10.2337/diabetes.52.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.