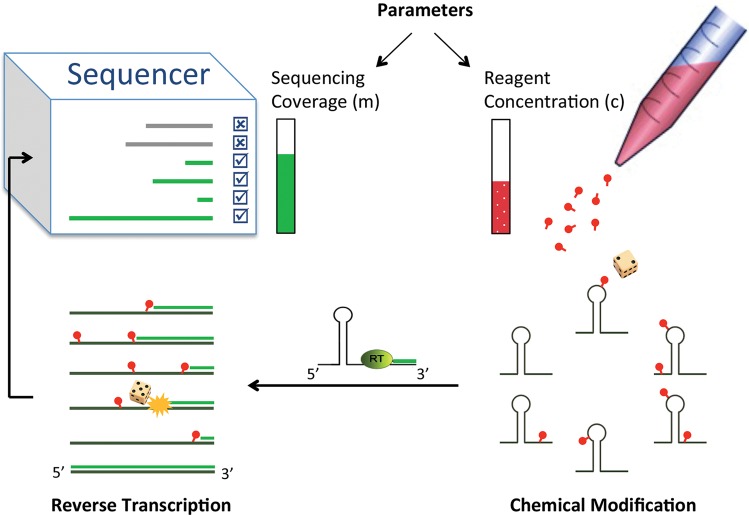
FIGURE 1.
Overview of chemical structure mapping followed by next-generation sequencing. Reagent molecules preferentially react with unconstrained nucleotides to modify them. Reverse transcriptase (RT) traverses the RNA and drops off upon encountering the first modification. RT may occasionally drop off prior to the modification in what is termed natural drop off. Sequencing of the resulting cDNA fragments reveals the sites of modification. When RT starts at a single predetermined primer binding site, one can control two parameters: average degree of modification, which depends on reagent concentration and reaction duration, and number of sequenced fragments, which depends on choice of sequencing coverage. Stochasticity in the composition of sequencing readouts arises from randomness in modification patterns, transcription termination events, and fragment sequencing.