Abstract
The TATA box represents one of the most prevalent core promoters where the pre-initiation complexes (PICs) for gene transcription are assembled. This assembly is crucial for transcription initiation and well regulated. Here we show that some cellular microRNAs (miRNAs) are associated with RNA polymerase II (Pol II) and TATA box-binding protein (TBP) in human peripheral blood mononuclear cells (PBMCs). Among them, let-7i sequence specifically binds to the TATA-box motif of interleukin-2 (IL-2) gene and elevates IL-2 mRNA and protein production in CD4+ T-lymphocytes in vitro and in vivo. Through direct interaction with the TATA-box motif, let-7i facilitates the PIC assembly and transcription initiation of IL-2 promoter. Several other cellular miRNAs, such as mir-138, mir-92a or mir-181d, also enhance the promoter activities via binding to the TATA-box motifs of insulin, calcitonin or c-myc, respectively. In agreement with the finding that an HIV-1–encoded miRNA could enhance viral replication through targeting the viral promoter TATA-box motif, our data demonstrate that the interaction with core transcription machinery is a novel mechanism for miRNAs to regulate gene expression.
Keywords: microRNAs, TATA-box motif, core transcription machinery, transcription activation
INTRODUCTION
miRNAs represent an important class of regulators during many important biological processes in animals, plants, fungus and viruses (Bartel 2004; Bushati and Cohen 2007). In the nucleus, miRNA precursors (pri-miRNAs) are transcribed and processed by Drosha and its cofactor DGCR8 to generate pre-miRNAs (Lee et al. 2003). The pre-miRNAs are then exported to the cytoplasm (Yi et al. 2003) wherein they are further sliced by Dicer to generate ∼22 nt mature miRNAs (Hutvagner et al. 2001) and bound by Argonaute (AGO) proteins and loaded into the RNA-induced silence complexes (RISCs) (Gregory et al. 2005). It is widely accepted that most miRNAs function through guiding the cytoplasmic RISCs to target 3′ UTR of mRNAs to suppress translation or degrade mRNAs (Hutvagner and Zamore 2002; Martinez et al. 2002; Bushati and Cohen 2007; Bartel 2009). The regulatory mode of miRNAs has been extended by the observation that 5′ UTR (Lytle et al. 2007) or coding region (Tay et al. 2008) of mRNA could also be the target for miRNA-mediated translation repression. Moreover, miRNAs could switch from suppression to activation of translation by targeting 5′ UTR (Orom et al. 2008) or 3′ UTR (Vasudevan et al. 2007) of mRNAs. It is noteworthy that the role of miRNAs as post-transcriptional activators is reported only in a few studies and is not broadly applicable now. These findings reveal that the mechanism of miRNA-mediated gene regulations is quite complicated in metazoans.
It has been demonstrated that a fraction of cellular miRNAs is enriched in the nucleus (Hwang et al. 2007; Liao et al. 2010; Taft et al. 2010) although their functions are largely unknown. Several studies have suggested that miRNAs could modulate gene expression at transcriptional level. MiR-373 can readily induce the expression of E-cadherin and cold-shock domain-containing protein C2 (CSDC2) through targeting a site in their promoters (Place et al. 2008). Another miRNA, miR-423-5p, induces transcriptional silencing by targeting a highly conserved region in the promoter of progesterone receptor (PR) gene (Younger and Corey 2011). MiR-744 and miR-1186 induce Ccnb1 expression and manipulate mouse cell proliferation with putative binding sites in the gene promoter (Huang et al. 2012). The target sites of these previously reported miRNAs are located in a wide range (∼1000 bp) of promoter without any unique feature and the underlying molecular mechanism was considered to be through altering epigenetic modifications of the promoter, including acetylation and/or methylation at histones (Li et al. 2006; Portnoy et al. 2011; Huang et al. 2012). Promoter-associated RNA (paRNA) transcripts overlapped with the promoter are possible mediators of these modifications, but they are reported only in a few cases (Schwartz et al. 2008; Younger and Corey 2011). These findings suggest that the mechanism underlying miRNA-mediated transcriptional regulation is also diverse.
The key RNA interfering (RNAi) components such as AGO proteins have been found in the nucleus (Tan et al. 2009; Weinmann et al. 2009; Nishi et al. 2013) and associated with the Pol II core transcription machinery (Guang et al. 2008, 2010; Cernilogar et al. 2011). In Schizosaccharomyces pombe, centromeric repeats region–derived siRNAs associate with Argonaute 1 and form the RNA-induced transcriptional silencing (RITS) complex to induce the heterochromatin assembly (Volpe et al. 2002; Motamedi et al. 2004; Verdel et al. 2004). In Caenorhabditis elegans, siRNAs and piRNAs can enter the nucleus and trigger co-transcriptional silencing of genes (Guang et al. 2008, 2010; Le Thomas et al. 2013). SiRNA-directed recruitment of the nuclear RNAi components, the NRDE factors, could inhibit RNA Pol II during the elongation phase of transcription (Guang et al. 2010). A recent report suggests that the key protein components of the RNAi pathway, i.e., DCR2 and AGO2, could directly interact with the transcription machinery and control the processivity of RNA Pol II in Drosophila (Cernilogar et al. 2011). These findings raise the possibility that small noncoding RNAs could directly interact with the Pol II core transcription machinery and affect gene transcription in metazoans.
Our recent study has revealed that a novel HIV-1–encoded miRNA, miR-H3, could target the TATA-box motif in HIV-1 5′ LTR and enhance viral replication (Zhang et al. 2014). We wondered whether this novel pattern of gene regulation could be extended to the cellular miRNAs. In the present study, we find that many cellular miRNAs interact with the pol II core transcription machinery. In silico prediction indicates that many of these miRNAs are complementary to the TATA-box core promoters. Subsequent experiments reveal that let-7i can induce IL-2 expression in primary CD4+ T cells and in mice. Unlike previous reports, this regulation is through the sequence-specific interaction between miRNA and TATA-box core promoter to facilitate the assembly of PIC and accelerate transcription initiation. In addition, many other cellular miRNAs that target the TATA-box motif also up-regulate the promoter activities of many important genes. Our findings reveal a novel and broad regulatory mechanism of cellular miRNAs.
RESULTS AND DISCUSSION
Some cellular miRNAs and Argonaute proteins are associated with RNA Pol II core transcription machinery
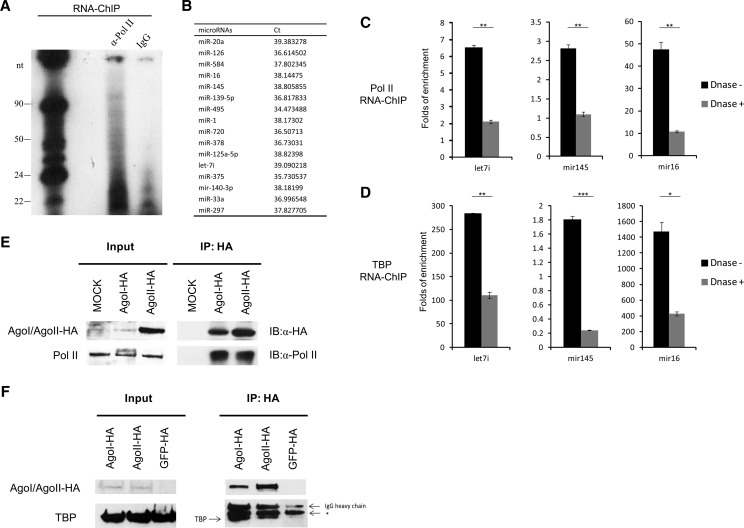
TATA-box motif represents one of the most prevalent core promoters, wherein the pre-initiation complexes (PICs) are assembled. The PICs comprise several general transcription factors, including RNA polymerase II core unit, TFIID (or TATA-box–binding protein, TBP), TFIIA, TFIIB, TFIIE, and TFIIF (Kornberg 2007). During transcription initiation, TBP firstly binds to the TATA box of a gene and nucleates the assembly of PICs (Smale and Kadonaga 2003). We initiated our work by investigating cellular miRNAs that directly interacted with the general transcription factors. Through an RNA-ChIP method, we found that a fraction of small RNAs of ∼22 nt length was significantly enriched by an anti-Pol II antibody (Fig. 1A) in human PBMCs, suggesting an association between small RNAs and Pol II. The Pol II-associated small RNAs were further analyzed by an miRNA-array assay and a number of miRNAs were detected (Fig. 1B). The RNA-ChIP followed by quantitative real-time RT–PCR (qRT-PCR) was used to confirm the association of Pol II/TBP with some miRNAs, including let-7i, mir-145 and mir-16 (Fig. 1C,D). Moreover, when treated with DNase (Supplemental Fig. S1A), the association of miRNAs with both Pol II and TBP were significantly reduced, suggesting that this association is DNA dependent (Fig. 1C,D). We also investigated whether the passenger strand of the miRNA was associated with the general transcription factors, but they were not enriched by these proteins (Supplemental Fig. S1B). By normalizing to the input, we found that a fraction of 1%–40% of the nucleus localized miRNAs was associated with the general transcription factors (Supplemental Fig. S1C). Given that TBP binds to TATA box which is close to the transcription start site (TSS), these data also imply that these miRNAs interact with the DNA sequences closed to the TSS. We further investigated whether the miRNA-binding proteins (the key RNAi components) are associated with the core transcription machinery in human cells. Through co-immunoprecipitation assay, we found that both Ago1 and Ago2 bound to Pol II (Fig. 1E) and TBP (Fig. 1F) in HEK293T cells. These results suggest cellular miRNAs and their binding proteins are associated with the RNA Pol II core transcription machinery in human cells.
FIGURE 1.
Cellular miRNAs and AGO proteins are associated with RNA Pol II core transcription machinery. (A) Autoradiogram of 32P-labeled RNA associated with human RNA Pol II purified by RNA chromatin immunoprecipitation (RNA-ChIP). The PBMCs were isolated from healthy individuals and activated with anti-CD3 (1 μg/mL) and anti-CD28 (5 μg/mL) antibodies for 48 h. RNA-ChIP assay was then performed. The chromatin-associated RNA was extracted and radiolabeled. These data represent three independent experiments. (B) Identification of RNA Pol II-associated miRNAs with miRNA expression microarray. The table only shows the detected miRNAs. qRT-PCR quantification of the miRNAs associated with RNA Pol II (C) and TBP (D) with or without DNase treatment. The enriched miRNAs were determined by real-time qRT-PCR and normalized to that of IgG. P-values were calculated using the two tailed unpaired Student's t-test with equal variances, n = 3 biological replicates. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. Co-immunoprecipitation (co-IP) assay to detect the association of HA-tagged Ago proteins with RNA Pol II (E) or TBP (F). HA-tagged Ago1 and Ago2 constructs were transfected into HEK293T cells. After 48 h, cell lysates were immunoprecipitated with anti-HA antibody, and subsequently detected with anti-RNA Pol II or anti-TBP antibody. (*) Nonspecific band. These data represent at least three independent experiments.
Let-7i up-regulates IL-2 promoter activity in human and mouse CD4+ T-cells
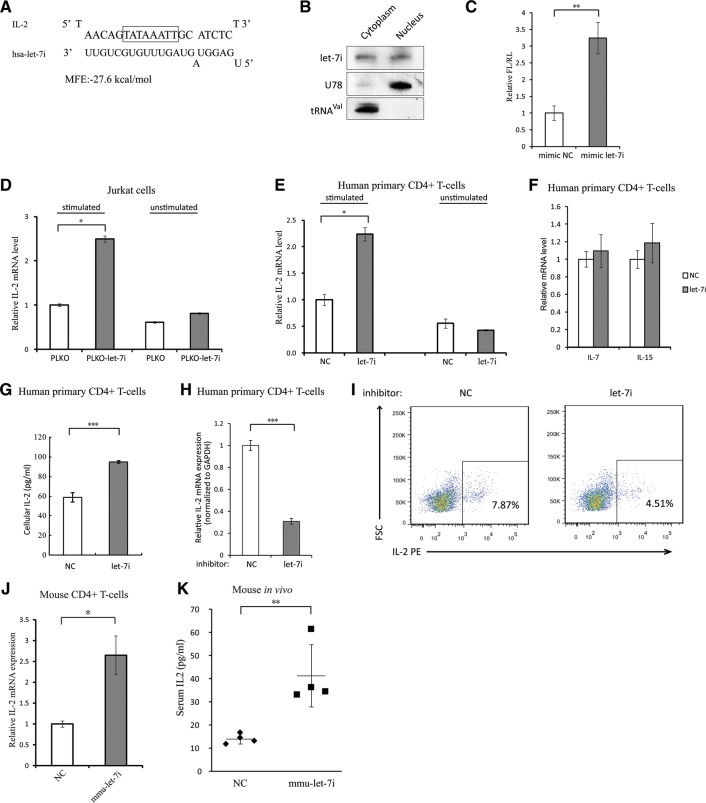
Computer prediction of miRNA-binding sites on gene core promoter region was then performed with the RNA-hybrid web server (Kruger and Rehmsmeier 2006). Interestingly, a nearly perfect binding between interleukin-2 (IL-2) core promoter region and let-7i was predicted with a remarkable MFE value (−27.6 kcal/mol) (Fig. 2A), suggesting that let-7i might interact with the core transcription machinery and be involved in IL-2 transcription regulation. Northern blot assay indicated that let-7i miRNA had nearly equal distributions in the cytoplasm and the nucleus of a lymphocyte cell line Sup-T1 cells (Fig. 2B), which was also confirmed by qRT-PCR assay (Supplemental Fig. S2A). The nuclear accumulations of let-7i miRNA enable its regulatory function(s) at transcriptional level. IL-2 is a key cytokine for T-lymphocyte activation and plays important roles in many immune functions. Overexpression of let-7i significantly enhanced the IL-2 promoter activity in HEK293T cells (Fig. 2C). To determine whether let-7i regulates the endogenous IL-2 expression, Jurkat cells were stably transduced with lentiviral vectors expressing GFP (PLKO-GFP) or let-7i precursor-GFP (PLKO-let-7i) (Supplemental Fig. S2B). The expression level of IL-2 mRNA was examined in the successfully infected cell populations. We found that the IL-2 mRNA levels of activated Jurkat cells overexpressing let-7i were significantly higher than those cells transduced with the empty vectors carrying gfp gene only (Fig. 2D). Alternatively, CD4+ T-lymphocytes isolated from PBMCs of healthy donors were treated with synthetic agomir-let-7i or negative control for 48 h (Supplemental Fig. S2C). After activation with anti-CD3/anti-CD28 antibodies for 24 h, let-7i also potently up-regulated IL-2 mRNA expression level (Fig. 2E). As controls, the mRNA levels of other interleukins, i.e., IL-7 and IL-15, were not affected, arguing against a possible nonspecific global response to the overexpression of let-7i (Fig. 2F). ELISA (enzyme-linked immunosorbent assay) analysis revealed that the cells over-expressing let-7i produced more IL-2 compared to the negative control (Fig. 2G). Moreover, when let-7i miRNA was blocked by a specific inhibitor (Supplemental Fig. S2D), the IL-2 mRNA level in CD4+ T-lymphocytes was significantly reduced (Fig. 2H), and so was the cellular IL-2 protein level (Fig. 2I).
FIGURE 2.
Let-7i enhances IL-2 promoter activity in human and mouse primary CD4+ T cells. (A) Predicted binding between let-7i and IL-2 core promoter region. TATA-box motif was highlighted. (B) Northern blot demonstrating the relative nuclear and cytoplasmic abundance of endogenous let-7i in Sup-T1 cells. U78 small nucleolar RNA and valine-tRNA (tRNAval) served as controls for the nuclear and cytoplasmic fractionation. (C) The let-7i mimic or negative control was co-transfected with the IL-2 promoter-driven firefly luciferase (FL) construct into HEK293T cells, a CMV-renilla luciferase (RL) plasmid served as a transfection control. Dual-luciferase assay was performed at 24 h after transfection. (D) The effect of overexpressing let-7i on IL-2 mRNA expression. The PLKO-let-7i cells which expressed let-7i miRNA plus GFP and PLKO cells which just expressed GFP were generated by infecting Jurkat cells with lentiviral vector containing let-7i precursor or the PLKO empty vector and sorted with FACS. The gfp-positive cells were subsequently treated with (left) and without (right) phorbolmyristate acetate (PMA, 50 ng/mL) and ionomycin (1 μM) for 12 h. IL-2 mRNA was then evaluated by qRT-PCR and normalized to GAPDH mRNA. (E) Levels of IL-2 mRNA in primary human CD4+ T cells after treatment with agomir-let-7i. The CD4+ T-cells were treated with agomir-let-7i or control for 48 h, and then cells were activated with anti-CD3 and anti-CD28 antibodies. The levels of IL-2 mRNA were evaluated by qRT-PCR and normalized to β-actin mRNA. (F) Levels of IL-7 and IL-15 mRNA in primary human CD4+ T cells after the treatment of agomir-let-7i as described above. (G) Cellular IL-2 protein was determined by ELISA. The samples were collected from the cell lysates of primary human CD4+ T cells transfected with let-7i mimic or negative control and activated as described above. (H) IL-2 mRNA levels in primary human CD4+ T cells transfected with let-7i miRNA inhibitor or negative control. The CD4+ T cells were transfected with let-7i inhibitor or negative control at a final concentration of 200 nM and then activated as described above. The levels of IL-2 mRNA were evaluated by qRT-PCR and normalized to GAPDH mRNA. (I) Intracellular staining of IL-2 in the CD+ cells transfected with let-7i miRNA inhibitor or negative control. The cells were transfected with let-7i miRNA inhibitor or negative control at 200 nM and activated with anti-CD3/anti-CD28 for 48 h before IL-2 intracellular staining and flow cytometry assay. These data represent three independent experiments. (J) IL-2 mRNA levels in mouse CD4+ T cells transfected with mmu-let-7i mimic or negative control. (K) Quantification of serum IL-2 levels of mice treated with agomir-mmu-let-7i or negative control (25 nmol per animal) for 48 h by ELISA. P-values were calculated using the two-tailed unpaired Student's t-test with equal variances, n = 3 biological replicates unless specified. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
As both the let-7 family and IL-2 gene are highly conserved across the species (Kaiser et al. 2004; Roush and Slack 2008), we investigated whether this regulatory pathway is conserved in other species. Computer prediction indicated the presence of a possible binding between let-7i and IL-2 promoter in other mammals, including mice, pigs and cows (Supplemental Fig. S2E). The endogenous transcription of mouse IL-2 could also be enhanced by the murinelet-7i mimic (Fig. 2J). An in vivo assay suggested that mice serum IL-2 level could also be significantly up-regulated by agomir-mmu-let-7i, a cholesterol-modified miRNA mimic, with single injection (Fig. 2K).
Histone epigenetic modification or promoter-associated RNA is dispensable for let-7i–induced transcriptional activation
Several studies have suggested that the transcriptional activation by small noncoding RNAs involves histone modifications (Li et al. 2006; Janowski et al. 2007). To determine the potential role of histone modifications in the activation of IL-2 promoter activity, we added trichostatin A (TSA), a histone deacetylase inhibitor, to the cells transfected with either activating let-7i mimic or a negative control (Yoshida et al. 1990, 1995). The treatment of TSA could increase the mRNA expression of COX-2 gene in HEK293T cells (Supplemental Fig. S3A), indicating that an effective concentration of TSA was used. However, TSA had no effect on the miRNA-mediated activation of IL-2 gene by let-7i (Supplemental Fig. S3B), suggesting that the deacetylation of histone proteins is unlikely to be required for the activation process. We also treated HEK293T cells with 5′-deoxy-5′-(methylthio)adenosine (dMTA), a protein methyltransferase inhibitor known to alter the methylation status of histones and other nuclear proteins (Williams-Ashman et al. 1982; Song and Ghosh 2004; Janowski et al. 2007). Treatment with dMTA had no effect on the basal and activated levels of IL-2 promoter activity (Supplemental Fig. S3C), suggesting that histone-methylation changes are not essential for the IL-2 activation either.
To further investigate whether the histone modifications that have been reported to be involved in RNAa mediated by small RNAs, such as reduced H3K9 tri-methylation (Li et al. 2006), increased H3K4 tri-methylation (Turunen et al. 2009) or loss of acetylation at H3K9 (Janowski et al. 2007), were effected in this study, we performed ChIP assay to determine the level of these modifications on IL-2 promoter region upon the overexpression of let-7i. The data showed that these epigenetic modifications on IL-2 promoter were not significantly altered by let-7i (Supplemental Fig. S3D).
As noncoding RNAs derived from the promoter region have been found to be the targets for various small noncoding RNAs, it is possible that these TATA-box motif–associated miRNAs could bind to these sense-stranded noncoding RNAs and therefore induce epigenetic modifications (Schwartz et al. 2008; Napoli et al. 2009; Chu et al. 2010). To examine this possibility, we tried to detect IL-2 promoter–associated RNAs (paRNAs) in both activated and resting human CD4+ T-lymphocytes, but we did not get any positive signal for the existence of the paRNA(s) (Supplemental Fig. S3E). Additionally, overexpression of a sense-stranded noncoding RNA covering -200 to the TSS of IL-2 promoter did not affect the enhancement effect of let-7i on IL-2 promoter activity (Supplemental Fig. S3F). Taken together, the noncoding transcripts from IL-2 promoter region, which have not actually been detected, are not responsible for the up-regulation of IL-2 promoter activity induced by let-7i.
Sequence-specific interaction between miRNA and TATA-box motif facilitates IL-2 PIC assembly and transcription initiation
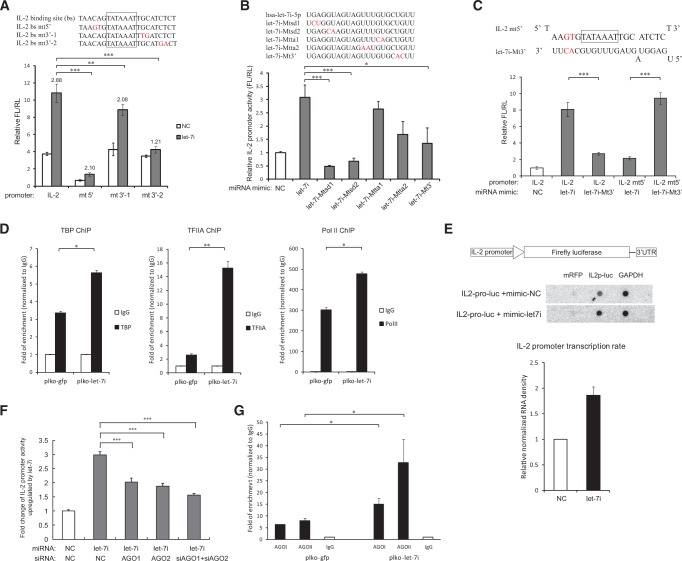
To investigate whether the identified miRNA-binding site is sequence-specifically required for the up-regulation of promoter activities, we introduced mutations into the IL-2 core promoter (Fig. 3A, top). We found that the mutations at the 3′ end of the promoter-binding site impaired the enhancement effect most significantly (Fig. 3A, bottom). Alternatively, a series of mutations were introduced to the synthetic let-7i miRNA mimic. The mutations did not significantly change the nuclear localization of these miRNAs (Supplemental Fig. S4A). Figure 3B indicates that mutations in the 5′ “seed” region mostly impaired the enhancement effect of let-7i, which is consistent with the result of promoter mutations. Interestingly, a let-7i mutant that restored the complementary base-pairing with one mutant of IL-2 core promoter was able to restore the enhancement effect of miRNA on promoter activity (Fig. 3C). These results demonstrate that the targeting of let-7i to the TATA-box region, rather than any other region(s), is required and sufficient for its regulation of IL-2 promoter activity. Most importantly, the data in Figure 3C also show strong evidence that let-7i directly binds to the TATA-box motif within the cells, given that the promoter-associated RNA was not detected. We then investigated whether other members of the let-7 family could also activate IL-2 promoter. Among the family members, the sequence of let-7b and let-7g is most similar to let-7i and their activating effects were most significant (Supplemental Fig. S4B), suggesting that the efficiency of the promoter activation is closely related to their sequence.
FIGURE 3.
Sequence-specific binding between let-7i miRNA and the TATA-box motif facilitates IL-2 PIC assembly and transcription initiation. (A, top) Mutations were introduced into the predicted let-7i–binding site in IL-2 core promoter region (in red). (Bottom) Wild-type and mutated IL-2 promoter–driven reporter constructs were transfected into HEK293T cells with let-7i mimic or negative control. Cells were harvested at 36 h post transfection and the promoter activities were determined by dual-luciferase assay. Up-regulation folds of the promoter activities relative to the control by let-7i were shown. n = 5 biological replicates. (B) A series of mutations were generated in the 5′ seed region, the sequence complementary to IL-2 TATA box, or 3′ region of let-7i miRNA, respectively (mutated nucleotide in red). The wild-type let-7i mimic or these mutated mimics were transfected to HEK293T cells, respectively, with the IL-2 promoter–driven luciferase construct. The IL-2 promoter activities were examined as described above. n = 5 biological replicates. (C) A mutated let-7i mimic was co-transfected with an IL-2 promoter–driven reporter which harbored mutations base-pairing complementary to the mutated let-7i mimic, and promoter activities were measured as described above. n = 5 biological replicates. (D) ChIP assay with antibody against TBP, TFIIA, or Pol II in Jurkat cells expressing GFP only (plko-gfp) or let-7i precursor (plko-let-7i) to examine the IL-2 promoter DNA associated with the general transcription factors. The enrichment of the promoter DNA by the transcription factors was normalized to IgG. These data represent three independent experiments. (E) Nuclear run-on assay of IL-2 promoter activity. The IL-2 promoter–driven luciferase construct was co-transfected into HEK293T cells with let-7i precursor or the empty vector. GAPDH was selected as an input control and mRFP as negative control. This figure represents three independent experiments. (F) Effects of inhibiting Ago1, Ago2, or both genes on the activation of IL-2 promoter by let-7i miRNA mimics. Ago proteins were knockdown with specific siRNAs in HEK293T cells, and the promoter activities of IL-2 in the presence of let-7i or control mimic were measured as described above. n = 3 biological replicates. (G) ChIP experiments were performed with antibody against AGO1 or AGO2 in Jurkat cells expressing GFP (plko-gfp) or let-7i precursor (plko-let-7i) to examine their association with the IL-2 promoter. The plko-gfp cells and plko-let-7i cells were generated as described above. The primers covering -146 to -8 relative to the TSS of IL-2 promoter were used to detect the associated promoter DNA. The enrichments of the promoter DNA by Ago proteins were normalized to IgG. This figure represents three independent experiments. P-values were calculated using the two-tailed unpaired Student's t-test with equal variances. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Our data suggest that cellular miRNAs could directly interact with TATA-box motifs, and are associated with the core transcription factors such as Pol II and TBP. Since the TBP–TATA-box binding is the first step of PIC assembly, it is important to determine whether let-7i affects the assembly of IL-2 PICs. To this end, ChIP assay was conducted to examine the binding of several general transcription factors to the IL-2 core promoter. The results indicated that, in addition to Pol II, let-7i significantly enhanced the binding of TBP and TFIIA to the IL-2 promoter (Fig. 3D). Consistently, the HIV-1–derived miRNA (miR-H3), which targets the HIV-1 TATA-box motif, also enhances the binding of the general transcription factors TBP and Pol II to the viral promoter (Zhang et al. 2014). Then we investigated whether the binding of let-7i affected the transcription initiation of IL-2 promoter. A nuclear run-on assay indicated that the overexpression of let-7i increased the transcription initiation of IL-2 promoter compared to the negative control (Fig. 3E). To explore the effect of let-7i on the transcription elongation of IL-2, initiated versus elongated IL-2 transcripts were measured by qRT-PCR with probe sets targeting 5′ UTR, exon 2–4, or 3′ UTR of IL-2 mRNA, respectively. We found that let-7i induced similar up-regulation of both initiated and elongated transcripts (Supplemental Fig. S4C), consistent with the elevated transcription initiation.
Since miRNA-binding proteins AGO1/2 have been found in the nucleus and reported to participate in transcriptional regulation mediated by small noncoding RNAs, it is intriguing to examine whether these activating miRNAs are recruiting AGO proteins to the promoters for their regulations. When Ago1 and Ago2 proteins were knocked down by siRNAs (Supplemental Fig. S4D), the activating effect of IL-2 promoter by let-7i was significantly impaired (Fig. 3F), indicating that both Ago1 and Ago2 may be recruited to the IL-2 promoter by let-7i for its regulation. To further test this hypothesis, we performed ChIP assay to determine the association of Ago1 and Ago2 to the IL-2 promoter in the Jurkat cell lines stably expressing GFP (PLKO-GFP) or let-7i precursor-GFP (PLKO-let-7i). Significant enrichment of Ago1 and Ago2 were detected in the region of -146 to -8 of the IL-2 promoter, which covers the identified let-7i target site (Fig. 3G), rather than an intergenic region downstream from IL-2 gene (Supplemental Fig. S4E). Moreover, the overexpression of let-7i (PLKO-let-7i) potently increased the enrichment of these proteins in the IL-2 promoter region (Fig. 3G).
Cellular miRNAs targeting TATA-box motif up-regulate the promoter activities of many important genes
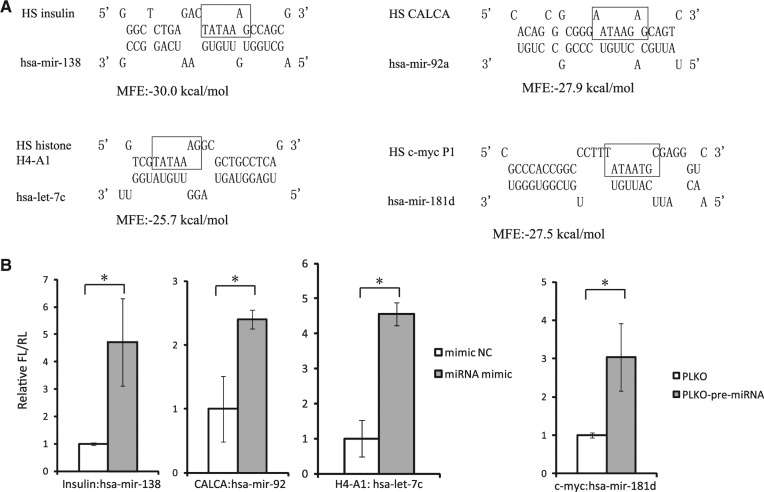
To determine whether the binding of miRNA to TATA-box core promoter is an universal phenomenon, we further performed a comprehensive computational screen of the putative bindings between 354 TATA-box containing human gene promoters and 1223 hsa-miRNAs from miRBase (ver. 16). When the cutoff of MFE was set to −27 kcal/mol, 13,323 putative bindings were found in the ±20-bp region centered with the TATA-box motif. From 16 gene promoters experimentally examined, four miRNAs, including mir-138, mir-92a, let-7c, or mir-181d, were identified to enhance the promoter activity of insulin, calcitonin, histone H4-A1, or c-myc, respectively, in HEK293T cells (Fig. 4A,B). Among these miRNAs, let-7c and miR-138 had a nearly equal distribution in the nucleus and the cytoplasm, while mir-92a and mir-181d were relatively enriched in the cytoplasm (Supplemental Fig. S5). In the Liao et al. study, the nuclear/cytoplasmic expression ratio of all these miRNAs was >1 (Liao et al. 2010), suggesting that they are a little enriched in the nucleus. The nuclear/cytoplasmic ratio of miRNAs is not very consistent among different reports, which may be due to the different cell lines or tissues used for the examinations. For instance, Jeffries et al. (2011) used neural stem cells, while Liao et al. (2010) used human nasopharyngeal carcinoma cells to investigate the nucleus-enriched miRNAs, but their top nucleus-localized miRNAs lists have little overlap. These observations also reflect the cell- or tissue-specific preferential expression of miRNAs. For the miRNAs in Figure 4, we detected their subcellular distribution only in lymphocyte cell lines, which may not be their preferentially expressed cells. Their subcellular distributions and regulatory functions in the nucleus need to be further explored in the context of physiological conditions.
FIGURE 4.
TATA-box motif-specific miRNAs enhance the promoter activity of many important cellular genes. (A) Predicted interactions between the core promoter of four human genes and four human miRNAs. TATA-box motifs were highlighted. (B) The promoter activities of indicated genes were measured with dual-luciferase assay upon the overexpression of their corresponding miRNAs. The promoter-driven luciferase reporters were transfected into HEK293T cells with synthesized miRNA mimics or miRNA precursors containing construct. P-values were calculated using the two-tailed unpaired Student's t-test with equal variances, n = 3 biological replicates. (*) P < 0.05.
In this report, we show that several endogenous miRNAs could target the TATA-box core promoters to up-regulate gene transcription. Unlike previous studies of the small RNAs (miRNAs or siRNAs) induced gene expression activation (a phenomenon named RNAa) (Li et al. 2006; Janowski et al. 2007; Place et al. 2008), the miRNA target sites of the present work and the HIV-1–encoded miRNA miR-H3 (Zhang et al. 2014) are quite unique and within 50 bp upstream of the TSS. Moreover, these miRNAs and their binding proteins AGO1 and AGO2 are associated with the general transcription factors such as Pol II and TBP, suggesting an interaction between the “nuclear RISC” and the RNA Pol II core transcription machinery. Our results also demonstrate that the gene activation is through direct interaction between miRNA and the TATA-box core promoter, which could facilitate the PIC assembly and transcription initiation. Sequence mutation experiments in both IL-2 promoter and let-7i miRNA indicate that their interactions are sequence-specific (Fig. 3C). Although some reports suggest that noncoding RNAs derived from the promoter region are the mediator of small RNA–induced chromatin epigenetic modifications (Schwartz et al. 2008; Napoli et al. 2009; Chu et al. 2010), the attempt to find IL-2 promoter-associated noncoding transcript did not get any positive signal of them and overexpression of the speculated paRNA did not affect IL-2 promoter activity either in our study. Thus, our findings reveal a novel transcription activation approach which is through the direct interaction between miRNAs and the core transcription machinery (Zhang et al. 2014). A recent report suggested that the key protein components of RNAi pathway, i.e., DICER2 and AGO2, are associated with RNA Pol II core transcription machinery (Cernilogar et al. 2011), while our study further reveals that cellular miRNAs could directly interact with the core promoters. These findings together suggest that the miRNA–Argonaute complexes in the nucleus (nuclear RISCs) could interact with the core transcription machine and participate in different steps of the transcription process.
The RNA polymerase II transcription initiation is a quite complicated process. Assembly of the preinitiation complexes and its post-assembly control are critical early steps in the transcription of eukaryotic genes. TATA box represents the most conserved and widespread core promoter which is enriched in the genes of tissue-specific expression or viral origin (Sandelin et al. 2007; Juven-Gershon et al. 2008). The TBP turnover on TATA-containing promoters is significantly higher than that on non-TATA promoters in yeast (van Werven et al. 2009), indicating that it is a highly regulated process. The TATA-box–TBP binding is negatively regulated by cellular factors such as nucleosome, NC2 and BTAF1 (Mot1p) (Thomas and Chiang 2006). On the contrary, the TATA-box–TBP binding could be enhanced by TFIIA, TFIIB, and SWI/SNF (Thomas and Chiang 2006). All these regulatory factors are proteins. Our results reveal that miRNAs could directly interact with the TATA-box motifs and associate with TBP (Figs. 1 and 3) to enhance transcription initiation, implying that the miRNAs are involved in the TBP turnover on TATA-containing promoter and affect the assembly of PICs. The formation of TFIID–TFIIA–promoter DNA complex (D-A complex) is important for transcription activation (Wang et al. 1992). It is also reported that the isomerized D-A complex is sufficient for gene activation (Chi and Carey 1996). Certain gene activators and coactivators could facilitate the formation of D-A complex, but the mechanism has not been fully clarified (White et al. 1992; Chi and Carey 1993; Ge and Roeder 1994; Shykind et al. 1995). In this study, we indicate that miRNAs enhance the binding of TBP and TFIIA to gene core promoter, and facilitate the assembly of the PICs containing these general transcription factors (Fig. 3D). These data suggest that certain cellular miRNAs might have facilitated the D-A complex formation to activate transcription via binding to the core promoters.
Although several studies suggest that small RNAs induced transcriptional gene silence or activation is through epigenetic programming in the promoters, the details on how these small RNAs sequence-specifically guide these modifications are still elusive (Kawasaki and Taira 2004; Morris et al. 2004; Place et al. 2008; Younger and Corey 2011; Huang et al. 2012, 2013; Rajasethupathy et al. 2012). In any model, the RNA:DNA interaction seems to be an unavoidable issue to ensure the specificity of the epigenetic modifications on the target promoter. However, by what form(s) they interact needs to be further discussed. Given that the fully assembled PIC is over 2 million Daltons, it is likely that a small RNA could be included within such a large complex. During transcription initiation, the TATA-box binding protein (TBP) binds to the TATA-box motif and unwinds the DNA double helix (Burley and Roeder 1996), which could enable the pairing between a complementary small RNA with its single-strand DNA target in the cells. The interaction of small noncoding RNA with the single-stranded DNA may stabilize the PICs under certain physiological conditions. It is also possible that miRNA function as a “wedge” to maintain the “open” status of the transcription initiation site and to improve the processivity of pol II by facilitating the loading of new pol II units to the vicinity of TSS. Moreover, small RNAs might interact with the double-strand promoter DNAs as suggested before (Huang et al. 2013). The binding of small RNA–AGO complexes to the TATA-box motif could recruit TBP and other general transcription factors to the core promoter (as suggested by Figs. 3 and 1), and facilitate the assembly of PICs. After TBP has bound to the TATA box, the small RNAs may be released from this special site. However, these hypotheses need to be clarified by further studies. A crystal structure analysis will be quite useful to verify whether small RNAs are incorporated into the PICs assembled on the TATA-box core promoter.
Since the binding between miRNA and TATA-box motif is sequence specific, we believe that the regulation of transcription by miRNAs is much more specific and accurate than that by protein transcription factors. Accumulating evidence has demonstrated that the biogenesis and function of miRNAs are also regulated by many transduction signals (Cullen 2004; O'Donnell et al. 2005; Krol et al. 2010). Therefore, our findings indicate a novel signal pathway to specifically regulate the gene expression at transcriptional level. This is in accordance with the observation that a number of miRNAs are found in the nucleus. Moreover, since the binding between let-7i miRNA and IL-2 promoter TATA box was also predicted in other mammals and validated in mice, it is likely that the enhancement of gene transcription by cellular miRNAs via targeting the TATA-box motif is conserved during evolution, at least in mammals and in viruses (Zhang et al. 2014).
MATERIALS AND METHODS
Plasmids, miRNA mimics, and siRNAs
The reporter vector containing IL-2 promoter–driven firefly luciferase was constructed by replacing the CMV promoter of pMIR-REPORT vector (Applied Biosystems) with the sequence of −400 to +1 bp relative to the TSS of the human IL-2 gene promoter. A series of mutations on the IL-2 core promoter region were generated by polymerase chain reaction (PCR) and inserted into the pMIR-REPORT vector. The vectors containing the insulin, CALCA, c-myc, histone H4-A1 gene promoter, respectively, were generated in the same way. The construct for overexpressing IL-2 promoter–associated RNA (paRNA) was generated by replacing the EGFP gene in the pEGFP-C1 vector (Clontech) with a 200-bp fragment upstream of the TSS of IL-2 in sense orientation. The PLKO-let-7i construct was generated by inserting the hsa-let-7i precursor sequence into multiple cloning sites of the PLKO.1 vector (Sigma), and the puromycin gene therein was replaced by a gfp gene. Wild-type hsa-let-7i, mmu-let-7i, and mutated hsa-let-7i miRNA mimics were purchased from Genepharma or Ribobio. Agomir-hsa-let-7i and negative control were purchased from Ribobio. The siRNAs against human Ago 1-2 genes were purchased from Dharmacon.
Antibodies and reagents
Anti-HA (MMS-101P) and anti-Pol II (8WG16) monoclonal antibodies were purchased from Covance Inc. Anti-TBP monoclonal antibody (ChIP Ab+) was purchased from Upstate (Millipore) or prepared by ourselves. Anti-AGOI and AGOII antibodies were purchased from Abcam. Anti-TFIIA-γ (D6) monoclonal antibody was purchased from Santa Cruz Biotechnology. Anti-actin antibody (D6A8) was purchased from Cell Signaling Technology. Anti-human CD3 and anti-human CD28 antibodies were purchased from BD Biosciences. Anti-mouse CD3 and anti-mouse CD4 antibodies were purchased from eBioscience. Phorbolmyristate acetate (PMA) and ionomycin were purchased from Sigma. EZ-Magna ChIP A/G kit (10086) for RNA-ChIP assay was purchased from Millipore.
Cell culture
Jurkat and HEK293T cells were obtained from ATCC (American Type Culture Collection) and cultured according to the ATCC recommendations. Human PBMCs were isolated from the whole blood of healthy donors with Ficoll-Hypaque Solution (HAO YANG). Resting primary CD4+ T lymphocytes were then isolated from PBMCs with CD4+ T Cell Isolation Kit II (BD Biosciences). Human primary PBMCs and CD4+ T cells were grown in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 50 units/mL penicillin and 50 μg/mL streptomycin. Mouse splenic CD4+ T cells were sorted from single-cell suspensions by using Mouse CD4+ T Cell Isolation Kit II (BD Biosciences). The sorted cells were grown in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 50 units/mL penicillin, and 50 μg/mL streptomycin.
Transfection and infection
miRNA mimics and miRNA inhibitors were transfected into human or mouse CD4+ T cells with Lipofectamine RNAiMAX (Invitrogen) at final concentrations of 50–200 nM. After 48–72 h of transfection, the CD4+ T lymphocytes were activated by stimulation with anti-CD3 (1 μg/mL) and anti-CD28 (5 μg/mL) for 12–24 h. The treatment of CD4+ T cells by agomir was performed by adding agomir-hsa-let-7i or negative control to cell culture medium directly at a final concentration of 50 nM, and then stimulated as described above. Transfection of HEK293T cells was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For AGO proteins knockdown assay, the siRNAs against AGO1 or AGO2 were transfected into HEK293T cells, after 24 h 5- to 10-ng IL-2 promoter–driven firefly luciferase (FL) reporter and 2-ng renilla luciferase (RL) constructs were cotransfected with let-7i miRNA mimic or negative control mimic at final concentrations of 25 nM into HEK293T cells. After another 48 h, FL and RL activities were measured.
Recombinant lentiviruses were generated by cotransfecting noninfectious lentiviral vectors including pCMV-ΔR8.2, VSV-G, and PLKO.1 constructs into 60% confluent HEK293T cells (100-mm cell cultural dish) using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Viral supernatant was collected at 48 h post transfection and used to infect Jurkat cells in the presence of 4 μg/mL polybrene. The successfully infected cells (GFP+ cells) were then sorted with a high-speed cell sorter device (BD Biosciences), and subsequently activated with 50 ng/mL PMA and 1-μM ionomycin.
MicroRNA array and data analysis
To study the small RNA fraction of ∼22 nt associated with Pol II, miRNA expression profiling was performed using the Serum/Plasma Focus miRNA PCR Panel, 384 well (V1.R) (Exiqon). Each panel comprises 168 LNA miRNA primer sets focusing on serum/plasma-relevant human miRNAs which are highly expressed in T-lymphocytes and seven reference miRNAs. The experimental and data analysis procedures were conducted following manufacturer's instructions. Several miRNAs were selected for further confirmation of their expressions by stem–loop qRT-PCR method, as previously described (Chen et al. 2005).
Quantitative real-time RT-PCR analysis
Total RNAs from Jurkat or CD4+ T cells was isolated with TRIzol reagent (Invitrogen) and then subjected to cDNA synthesis using PrimeScript RT reagent Kit (Takara). All primers were annealed at 37°C and reverse transcription proceeded at 42°C. Quantitative PCR was performed with SYBR Premix ExTaq II Kit (Takara) following the manufacturer's instructions. Quantification was normalized to the housekeeping gene GAPDH or β-actin. The sequences of the primers used in this study are listed in the Supplemental Material.
Computational prediction of miRNA-targeting site in the core promoter region
The core promoter sequences (−49 to +1) of Pol II–transcribed genes of human were downloaded from the Eukaryotic Promoter Database (EPD, http://epd.vital-it.ch/). TATA-box containing core promoters were identified by the presence of “ATAA” conserved motif (Smale and Kadonaga 2003). The mature miRNA sequences of human were downloaded from miRBase (http://www.mirbase.org/). The miRNA-binding sites on gene core promoters were predicted with RNA-hybrid web server (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). The MFE cut-off for candidates was less than −25 kcal/mol.
Dual-luciferase reporter assay
HEK293T cells were seeded in 48-well plates (Corning) at a density of 20,000 cells per well one day before transfection. Five to ten nanograms of gene promoter–driven firefly luciferase (FL) reporter and 2 ng renilla luciferase (RL) constructs were cotransfected with miRNA precursor construct/the empty vector, or miRNA mimic/negative mimic control into 293T cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After 24–48 h, FL and RL activities were measured with the Dual-Glo luciferase assay system according to the manufacturer's instructions (Promega).
Co-immunoprecipitation
Co-immunoprecipitation was performed as described previously with minor modifications (Yang et al. 2001). Briefly, HEK293T cells were transfected with pcDNA3.1-Ago1-HA, pcDNA3.1-Ago2-HA, or pcDNA3.1-GFP as control. After 48 h, the cells were disrupted in lysing buffer (150 mM NaCl, 10 mM Tris-HCl, 0.5% NP-40, 1% Triton X-100, 1 mM NaF, 1 mM Na5VO4, 2 mM EDTA, and 1% glycerol supplemented with PMSF and protease inhibitors) for 30 min on ice. Each lysate was immunoprecipitated with 40 μL of anti-HA agarose (Sigma) for 4 h. The beads were washed four times with STN buffer [10 mM Tris–HCl (pH 7.5), 0.25% NP-40 and 50 mM NaCl], and then boiled in SDS-containing buffer at 100°C for 10 min. The supernatants were then used for immunoblotting. Anti-Pol II, anti-TBP, or anti-β-actin antibodies were used to detect human Pol II, TBP, or β-actin, respectively.
Enzyme-linked immunosorbent assay
The human and mouse IL-2 ELISA test was performed with IL-2 ELISA kit (Xibosheng) following the manufacturer's instruction.
RNA- and DNA-ChIP assay
RNA-ChIP (chromatin immunoprecipitation) was performed as described previously in Sun et al. (2006) with some modifications. The fresh PBMCs were stimulated with anti-CD3 and anti-CD28 for 48 h, and were then harvested and washed with cold PBS. Cells were cross-linked with 1% formaldehyde and stopped by 0.125 M glycine. Cells were lysed and then sonicated on ice. The supernatants were added with the following antibodies plus 50 µL fully resuspended protein A/G magnetic beads (Millipore) for immunoprecipitation (∼3 × 107 cells for each immunoprecipitation): anti-RNA Pol II (8WG16, 5 µg), anti-TBP (5 µg), or seronegative mouse IgG (5 µg), respectively. The mixtures were incubated overnight at 4°C. The protein A/G bead-antibody/chromatin/RNA complex was then washed once by 0.5 mL various cold buffers in the order listed below: low-salt washing buffer, high-salt washing buffer, LiCl washing buffer, and TE washing buffer (Millipore). After washing, RNAs were then extracted by phenol:chloroform:isoamyl alcohol and followed by ethanol precipitation. The isolated RNAs were treated with TURBO DNAfree kit (Ambion) to remove DNA and then labeled with 32P-γ-ATP using T4 kinase. The reaction mixtures containing 1 µL of RNA, 1 µL of 32P-γ-ATP (3000 Ci/mmol, 10 mCi/mL, PerkinElmer), 2 µL 10× kinase reaction buffer, 1 µL T4 polynucleotide kinase (Takara), and 15 µL dH2O were incubated at 37°C for 1 h. The labeled RNAs were then loaded into 10% denatured PAGE gel for electrophoresis. After gel-drying, the gel was exposed to a blanked X-ray film for 3 h. DNase treatment was performed by adding 10 units of DNase I (Promega) to the samples during the antibody/magnetic beads incubation step. DNA-ChIP assay was performed with Magna ChIP A/G Kit (Millipore) by following the manufacturer's instruction.
Nuclear run-on assay
HEK 293T cells were transfected with IL-2 promoter–driven firefly luciferase reporter and let-7i mimics or negative control in a final concentration of 50 nM. After 20 h, cells were collected for nuclear run-on assay as previously described in Smale (2009) with minor modifications. Nuclear run-on transcription was performed in the presence of 5 μL of 10 mCi/mL [α-32P] UTP. After treatment of the reaction with RNase-free DNase I and protease K, RNA was extracted with phenol/chloroform/isoamyl alcohol and hybridized to denatured IL-2 promoter–driven firefly luciferase plasmid immobilized on a nylon membrane. Approximately 200-bp probes of GAPDH or RFP gene were selected as input and negative control, respectively. The hybridization was conducted with hybridization buffer (50% formamide, 6× SSC, 10× Denhardt's solution, 0.2% SDS) at 37°C overnight. After washing with 2× SSC and 0.2% SDS for three times, the membranes were wrapped in a plastic wrap and exposed to PhosphorImager screen (GE Healthcare).
Subcellular fractions isolation and Northern blot
The nuclear and cytoplasmic fractions of Sup-T1 cells were isolated with the PARIS Kit (Ambion) by following the manufacturer's instructions. Then the isolated nuclear and cytoplasmic RNAs were subjected to Northern blot assay. The procedures of Northern blot were performed by following the description by Varallyay et al. (2008). The LNA-modified oligo-nucleotide probe for miRNA detection was purchased from Exiqon.
Animals
The female mature BALB/cA mice (8–10 wk) were treated with Agomir-mmu-let-7i or negative control through tail-vein injections at doses of 25 nmol per animal. Measurements of serum IL-2 levels were performed 48 h after the injection. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Introduction of Innovative R&D Team Program of Guangdong Province (No. 2009010058), National Special Research Program for Important Infectious Diseases (No. 2013ZX10001004), National Basic Research Program of China (973 Program) (No. 2010CB912202), National Natural Science Foundation of China (No. 30972620), Natural Science Foundation of Guangdong (No. 9251008901000022), Research Fund for the Doctoral Program of Higher Education of China (No. 20090171110083) to H.Z., National Natural Science Foundation of China (No. 81301431), China Postdoctoral Science Foundation (No. 2012M511866 and No. 2013T60824) to Y.Z.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045633.114.
REFERENCES
- Bartel DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Roeder RG 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65: 769–799. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Carey M 1993. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol 13: 7045–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Carey M 1996. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes Dev 10: 2540–2550. [DOI] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA, Corey DR 2010. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 38: 7736–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2004. Transcription and processing of human microRNA precursors. Mol Cell 16: 861–865. [DOI] [PubMed] [Google Scholar]
- Ge H, Roeder RG 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78: 513–523. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC 2012. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR 2007. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 3: 166–173. [DOI] [PubMed] [Google Scholar]
- Jeffries CD, Fried HM, Perkins DO 2011. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA 17: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Rothwell L, Avery S, Balu S 2004. Evolution of the interleukins. Dev Comp Immunol 28: 375–394. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211–217. [DOI] [PubMed] [Google Scholar]
- Kornberg RD 2007. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci 104: 12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34: W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R 2006. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci 103: 17337–17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH 2010. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5: e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci 104: 9667–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802. [DOI] [PubMed] [Google Scholar]
- Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV 2009. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J 28: 1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Nishi A, Nagasawa T, Ui-Tei K 2013. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 19: 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843. [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci 105: 1608–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy V, Huang V, Place RF, Li LC 2011. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA 2: 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER 2012. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149: 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ 2008. The let-7 family of microRNAs. Trends Cell Biol 18: 505–516. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA 2007. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev 8: 424–436. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA 2008. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15: 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shykind BM, Kim J, Sharp PA 1995. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev 9: 1354–1365. [DOI] [PubMed] [Google Scholar]
- Smale ST 2009. Nuclear run-on assay. Cold Spring Harb Protoc 2009: doi: 10.1101/pdb.prot5329. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT 2003. The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–479. [DOI] [PubMed] [Google Scholar]
- Song MR, Ghosh A 2004. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci 7: 229–235. [DOI] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 21: 617–628. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. 2010. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol 17: 1030–1034. [DOI] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JP IV, Cuellar TL, McManus MT, Kiriakidou M 2009. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res 37: 7533–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178. [DOI] [PubMed] [Google Scholar]
- Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Vaisanen S, Yla-Herttuala S 2009. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res 105: 604–609. [DOI] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Varallyay E, Burgyan J, Havelda Z 2008. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3: 190–196. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wang W, Gralla JD, Carey M 1992. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev 6: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507. [DOI] [PubMed] [Google Scholar]
- White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P 1992. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J 11: 2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Ashman HG, Seidenfeld J, Galletti P 1982. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol 31: 277–288. [DOI] [PubMed] [Google Scholar]
- Yang S, Sun Y, Zhang H 2001. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem 276: 4889–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179. [PubMed] [Google Scholar]
- Yoshida M, Horinouchi S, Beppu T 1995. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17: 423–430. [DOI] [PubMed] [Google Scholar]
- Younger ST, Corey DR 2011. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39: 5682–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan M, Geng G, Liu B, Huang Z, Luo H, Zhou J, Guo X, Cai W, Zhang H 2014. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.