Abstract
The three-nucleotide mRNA reading frame is tightly regulated during translation to ensure accurate protein expression. Translation errors that lead to aberrant protein production can result from the uncoupled movement of the tRNA in either the 5′ or 3′ direction on mRNA. Here, we report the biochemical and structural characterization of +1 frameshift suppressor tRNASufJ, a tRNA known to decode four, instead of three, nucleotides. Frameshift suppressor tRNASufJ contains an insertion 5′ to its anticodon, expanding the anticodon loop from seven to eight nucleotides. Our results indicate that the expansion of the anticodon loop of either ASLSufJ or tRNASufJ does not affect its affinity for the A site of the ribosome. Structural analyses of both ASLSufJ and ASLThr bound to the Thermus thermophilus 70S ribosome demonstrate both ASLs decode in the zero frame. Although the anticodon loop residues 34–37 are superimposable with canonical seven-nucleotide ASLs, the single C31.5 insertion between nucleotides 31 and 32 in ASLSufJ imposes a conformational change of the anticodon stem, that repositions and tilts the ASL toward the back of the A site. Further modeling analyses reveal that this tilting would cause a distortion in full-length A-site tRNASufJ during tRNA selection and possibly impede gripping of the anticodon stem by 16S rRNA nucleotides in the P site. Together, these data implicate tRNA distortion as a major driver of noncanonical translation events such as frameshifting.
Keywords: ribosome, mRNA, reading frame, anticodon stem–loop, decoding, X-ray crystal structure
INTRODUCTION
The accurate translation of the genetic code into properly folded and active proteins is crucial to cellular survival. Errors in protein synthesis can result in misfolded or truncated proteins that trigger protein degradation pathways and even cellular apoptosis (Manley et al. 1978; Nangle et al. 2002; Lee et al. 2006). Translation of a triplet nucleic acid sequence on mRNA into 20 different amino acids is carried out by the ribosome with high fidelity (10−4–10−3) where missense errors, or tRNA misincorporation, account for the majority of this error rate (1 in 3000 residues) (Edelmann et al. 1977; Bouadloun et al. 1983; Kramer et al. 2007). While processivity errors, such as changes in the mRNA reading frame, occur much less frequently (1 in ∼30,000 amino acids incorporated), they are considered to be more detrimental than missense errors because they often prevent the production of a full length, functional protein (Kurland 1992). In organisms with a nearly equal genomic GC and AT nucleotide frequency, like Escherichia coli, out-of-frame stop codons occur roughly every 20 codons resulting in the rapid termination of translation following a frameshift error (Jorgensen and Kurland 1990; Kurland 1992). While much is known about the prevention of missense errors by ribosomal proofreading mechanisms (Zaher et al. 2009; Rodnina 2012), how the ribosome maintains the mRNA reading frame is still poorly understood.
Genetic suppressor experiments in Saccharomyces cerevisiae and Salmonella typhimurium first demonstrated the three-nucleotide genetic code could be altered (Riyasaty and Atkins 1968; Riddle and Roth 1970; Yourno 1970, 1972; Sherman et al. 1974). Extragenic mutations in tRNAs were found to compensate for insertions or deletions in the genetic code by noncanonical decoding of a non-three-nucleotide codon (for review, see Roth 1974). The first sequenced external suppressor of a genetically encoded frameshift (sufD) was a cytosine insertion immediately 5′ of the tRNAGly anticodon (5′-CCC-3′; all codons and anticodons are shown 5′–3′) between position 33 and 34 (Riddle and Carbon 1973). Because this particular frameshift suppressor expanded the glycine codon to GGG-G (first three nucleotides denote a glycine codon with the additional nucleotide preceded by a hyphen and underlined), the Watson–Crick complementarity between the cytosine insertion in the anticodon loop and the extra guanine in the codon suggested a four-base interaction between the tRNA–mRNA pair could form (Bossi and Roth 1981). Many frameshift suppressor tRNAs were subsequently found to contain similar complementarity between insertions in anticodon stem–loops (ASLs) and suppressible four-nucleotide codons, providing strong evidence for a quadruplet or four-base decoding model (Fig. 1A; Riyasaty and Atkins 1968; Riddle and Roth 1972; Yourno 1972; Prather et al. 1981; Cummins et al. 1982). In addition, optimal frameshift suppression resulting in higher frameshift efficiencies occurred from a four-nucleotide Watson–Crick base-pair interaction (Yarus 1982; Gaber and Culbertson 1984; Curran and Yarus 1987; Moore et al. 2000; Anderson et al. 2002).
FIGURE 1.
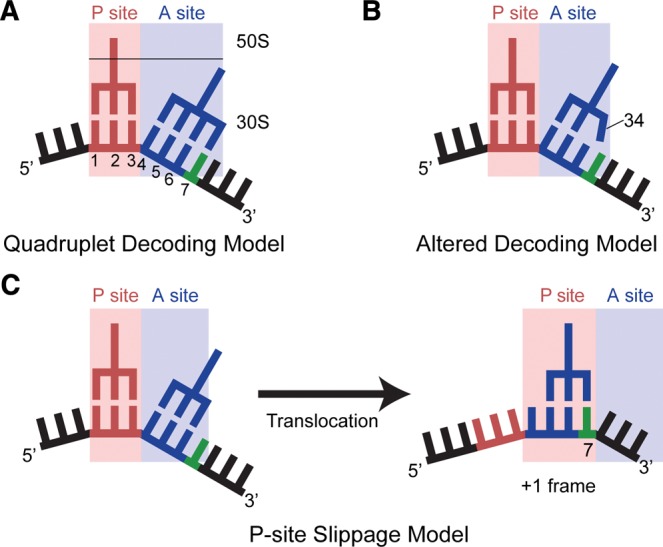
Possible models for +1 frameshifting resulting from an eight-nucleotide anticodon stem–loop. (A) The quadruplet decoding model posits that insertions in the anticodon stem–loop of a frameshift suppressor tRNA leads to a four-nucleotide anticodon capable of decoding and translocating a four-nucleotide mRNA codon (with the extra nucleotide shown in green). The numbering of the mRNA begins with the first position in the P site. (B) An alternative model is that the nucleotide insertion in the anticodon stem–loop causes a widening of the loop, allowing the anticodon nucleotide 34 to interact with the fourth nucleotide of the A-site codon (green; numbered as 7 in A). (C) In the P-site slippage model, normal decoding in the zero frame occurs in the A site; however, the transition into the +1 frame occurs after translocation to the P site due to a weakened interaction between the anticodon and codon.
The first exception to this quadruplet decoding model came with the identification of frameshift suppressor tRNASufJ, a modified tRNA3Thr (GGU) containing a cytosine insertion in the anticodon loop between nucleotides C31 and U32, rather than adjacent to the anticodon (Fig. 2; Bossi and Smith 1984). The C31.5 insertion allows tRNASufJ to decode four nucleotide codons (ACC-A, ACC-C, and ACC-U) as a single threonine, to restore the correct reading frame in a number of histidine biosynthesis gene derivatives such as hisA, hisC, hisF, and hisG (Bossi and Roth 1981). While the variability in the additional nucleotide of the codon that frameshift suppressor tRNASufJ decodes strongly suggests that a direct interaction between the additional nucleotide of the tRNA is not necessary for +1 suppression, it is unclear how an insertion within the anticodon loop, but distant from the anticodon, could expand the size of the codon. Because of the unique location of the insertion and apparent lack of Watson–Crick complementarity, it was proposed that tRNASufJ mediated +1 frameshifting through a distinct mechanism (Bossi and Smith 1984).
FIGURE 2.
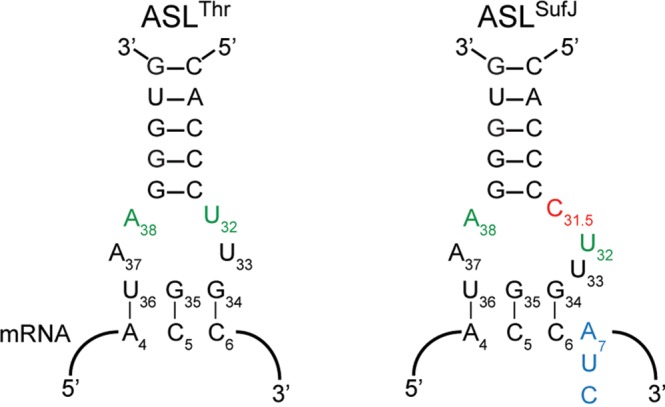
Frameshift suppressor tRNASufJ is a derivative of tRNAThr. Secondary structure representation of the anticodon stem–loops of tRNAThr and frameshift suppressor tRNASufJ interacting with their respective codons. The C31.5 insertion (red) in ASLSufJ is 5′ to the anticodon nucleotides 34, 35, and 36 causing the codon to increase from three to four nucleotides (blue). The conserved U32·A38 interaction (green) in tRNAThr may be altered in tRNASufJ due to the insertion.
More recent work has resulted in two additional models for +1 frameshifts. The first model was derived from X-ray crystal structures of 30S bound to ASLs containing eight-nucleotide loops. These ASLs were not identified by suppressor studies but rather, were optimized for unnatural amino acid incorporation exploiting +1 frameshift decoding (Hohsaka et al. 2001). These structures revealed a noncanonical ASL conformation where widening of the anticodon loop allows for three nucleotides of the ASL to extend over four nucleotides of the codon in the A site (Fig. 1B; Dunham et al. 2007). An additional model was proposed after the identification of tRNASufJ and other subsequent suppressors such as tRNASufA6 and suf16 tRNA mutations, where the shift into the new mRNA frame was not dependent upon forming Watson–Crick interactions in the A site (Fig. 1C; Gaber and Culbertson 1984; Qian et al. 1998). Instead, these studies indicated that frameshift efficiencies were heavily influenced by the next incoming tRNA, implying the frameshift in these particular cases, occurs in the P site (Farabaugh 2000; Atkins and Bjork 2009; Nasvall et al. 2009). Given these distinct models, it has remained unclear whether a single, unified model could explain all types of +1 frameshifting facilitated by suppressor tRNAs.
Here, we present biochemical and structural studies of frameshift suppressor tRNASufJ to elucidate how this suppressor decodes a four-nucleotide codon in the A site. We performed affinity experiments to determine the effect of the C31.5 insertion on the ability of either ASLSufJ or tRNASufJ to recognize an A-site codon. We next solved five X-ray crystal structures of the ASLs of tRNASufJ and tRNAThr bound to the A site of the Thermus thermophilus (Tth) 70S ribosome. Our molecular insights reveal that the conformational distortions present in frameshift suppressor tRNAs likely promote +1 frameshifting.
RESULTS
The nucleotide insertion 5′ of the anticodon minimally alters affinity for the A site
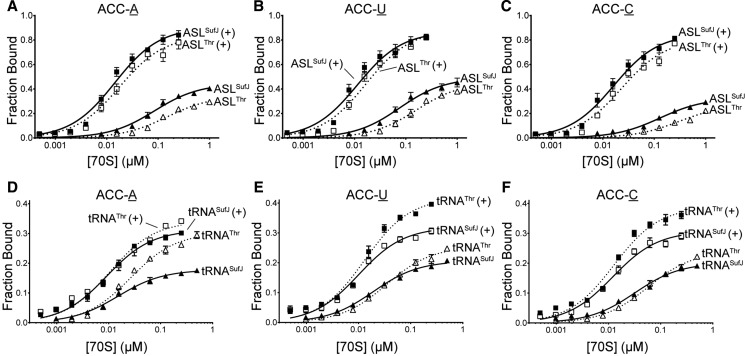
Expanded ASLs containing insertions 5′ of the anticodon show lower affinities than wild-type tRNAs for the ribosomal A site (Walker and Fredrick 2006). Therefore, as a first step in understanding the behavior of tRNASufJ on the ribosome, we asked whether the expanded ASL of tRNASufJ affects its ability to form stable, high affinity codon–anticodon interactions in the A site, typical of a canonical tRNA interacting with a cognate codon. We performed affinity binding experiments with ASLSufJ and ASLThr using E. coli 70S ribosomes programmed with a P-site tRNAfMet containing either an ACC-A, ACC-U or ACC-C codon in the A site (Fig. 3). [32P]-ASLSufJ was incubated with increasing amounts of 70S ribosomes and applied to nitrocellulose filters to determine the amount of ASLSufJ bound. While ASLSufJ bound to the A site of the ribosome with an apparent dissociation constant (KD) of 85 and 76 nM for the ACC-A and ACC-U codons, respectively, ASLSufJ had a slight decrease in affinity for the ACC-C codon (130 nM) (Fig. 3A–C; Table 1). These KD values are all within the previously reported range for tRNA or ASL binding to the A site (KDs between 33 and 500 nM) (von Ahsen et al. 1997; Phelps et al. 2002; Walker and Fredrick 2006). The antibiotic paromomycin preferentially enhances the affinity of cognate ASLs (∼15 fold) while only modestly enhancing the affinity of near-cognate ASLs (approximately twofold) (Ogle et al. 2002). Our results indicate that paromomycin increases the KD approximately six- to eightfold for all three +1 suppressible codons, consistent with a cognate interaction between the anticodon of ASLSufJ and the +1 codons. If the interaction in the A site was in the +1 mRNA frame, then the addition of paromomycin should not affect the KD for the ACC-A codon given that the interaction would be noncognate (U36-C5 and G34-A7 mismatches) (Fig. 2). These data provide initial support for a model where the interaction between ASLSufJ and all three +1 suppressible codons is cognate suggesting ASLSufJ is bound in the zero or normal frame.
FIGURE 3.
A-site binding of ASLSufJ, ASLThr, tRNASufJ, and tRNAThr. Increasing concentrations of 70S ribosomes were programmed with A-site codons (A) ACC-A, (B) ACC-U, or (C) ACC-C and mixed with 2 nM [32P] ASLThr (□/▵ dashed lines) or ASLSufJ (▪/▴; solid lines). The presence of the antibiotic paromomycin is indicated by “+”. Similar binding experiments were also performed with tRNAThr (□/▵ dashed lines) or tRNASufJ (▪/▴; solid lines) with A-site codons (D) ACC-A, (E) ACC-U, or (F) ACC-C in the presence or absence of paromomycin (indicated by “+”).
TABLE 1.
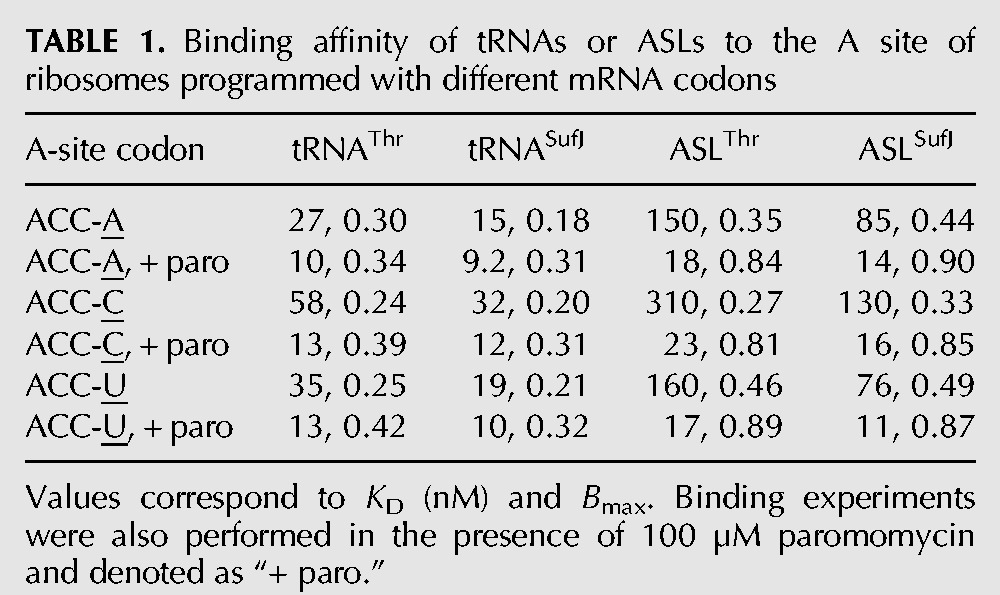
To assess whether the 31.5 insertion in ASLSufJ affects A-site binding, we next measured the binding of ASLThr to all three +1 suppressible codons in the presence or absence of paromomycin (Table 1). Similar KD trends were observed, with ASLThr having a lower affinity for ACC-C (310 nM) compared with the ACC-A and ACC-U codons (150 and 160 nM, respectively) (Fig. 3D–F). These data indicate the C31.5 insertion in ASLSufJ does not lower the affinity as might have been expected of the ASL for the three codons, but instead, slightly increases the affinity.
The binding of tRNA to the A site of the ribosome occurs with slightly higher affinities than ASLs due to other tRNA features that interact with the 50S subunit such as the TΨC loop, the D loop and the CCA tail (Jorgensen et al. 1985; Khade and Joseph 2010). To test whether tRNASufJ exhibits the same trends in A-site binding as ASLSufJ, namely an affinity indicative of a cognate codon–anticodon interaction slightly higher than tRNAThr, we again performed binding assays. Consistent with the observed ASL binding trends, tRNASufJ binds to each of the ACC-A, ACC-C, and ACC-U codons with approximately twofold tighter KD (15–32 nM) than tRNAThr (27–58 nM) (Table 1). Also consistent with our ASL binding studies, both tRNAs had similar dissociation constants for ACC-A and ACC-U codons (tRNASufJ 15 and 19 nM, respectively; tRNAThr 27 and 35 nM, respectively), and a slightly weaker affinity for the ACC-C codon (tRNASufJ 32 nM; tRNAThr 58 nM). These results indicate that both suppressor ASLSufJ and tRNASufJ follow general trends previously seen for other canonical tRNAs and provide further evidence that the expansion of the anticodon loop to eight nucleotides by the C31.5 insertion does not impede binding to cognate codons in the ribosomal A site.
Structural determination of ASLSufJ bound to +1 suppressible codons in the 70S A site
To determine how tRNASufJ decodes a four-nucleotide codon and understand how an expanded ASL is accommodated in the A site, we solved three X-ray crystal structures of Tth 70S programmed with a P-site tRNAfMet and ASLSufJ bound to each of the following +1 suppressible codons: ACC-A, ACC-C, and ACC-U (Fig. 4A; Supplemental Fig. S1; Table 2). The resolutions of the three structures ranged from 3.5 to 3.6 Å and, in all three structures, unbiased Fo–Fc difference electron density maps showed a clear signal for mRNA, P-site tRNAfMet, and A-site ASLSufJ nucleotides 29–42 (Supplemental Fig. S2).
FIGURE 4.
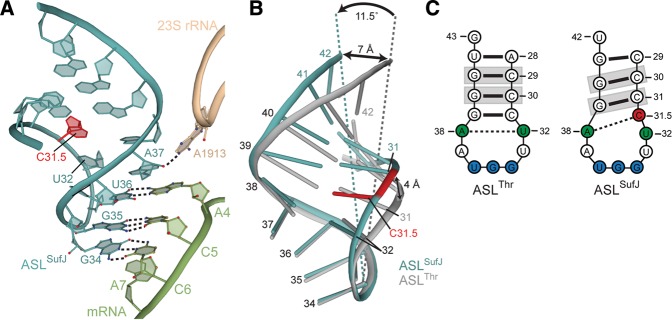
Conformation of ASLSufJ in the 70S A site. (A) ASLSufJ (blue) forms three Watson–Crick base pairs with three nucleotides of the ACC-A codon (green) in the zero frame (no paromomycin). Although the C31.5 insertion (red) increases the anticodon loop to eight nucleotides, the ASL is recognized as cognate with 23S rRNA nucleotide A1913 (tan) maintaining a hydrogen bond interaction with the 2′-OH of A37. (B) Overlay of ASLSufJ and ASLThr (gray) bound in the 70S A site shows the C31.5 insertion displaces the 5′ and 3′ phosphate backbone by 4 Å and 7 Å, respectively. This narrowing of the major and minor grooves of ASLSufJ also results in the entire stem tilting 11.5° toward the back of the A-site decoding center. (C) Secondary structure representation of ASLThr and ASLSufJ shows that the 31.5 insertion changes the conserved interaction between the U32·A38 (green) to C31.5 (red)·A38. The nucleotides that are gripped by 16S rRNA residues G1338 and A1339 upon translocation to the P site are highlighted in gray.
TABLE 2.
Data collection and refinement statistics
In the 70S structure of ASLSufJ decoding an ACC-A codon in the absence of paromomycin, the three-nucleotide anticodon of ASLSufJ (34, 35, and 36) forms Watson–Crick base pairs with the first three nucleotides of the ACC codon in the zero frame (Fig. 4A). The structures of ASLSufJ bound to the ACC-C and ACC-U codons in the presence of paromomycin reveal the same three Watson–Crick base-pair interactions between the codon and anticodon, indicating the antibiotic does not alter the A-site conformations (paromomycin was used to enhance the diffraction of these crystals) (Supplemental Fig. S1). In all three structures, there are no interactions with the fourth nucleotide of the A-site codon (position 7 of the mRNA) (Figs. 1, 4A; Supplemental Fig. S1). This lack of interaction is despite the potential to form a Watson–Crick base pair between the codon nucleotide A7 in the ACC-A codon and ASL nucleotide U33 (Fig. 2). Additionally, the presence of either an A, C, or U 3′ to the codon (fourth position of the A-site codon or denoted as A7, C7, or U7) does not alter the mRNA path (Supplemental Fig. S3). ASLSufJ models in all three structures superimpose well, with a root mean square deviation of 0.25 Å, providing additional evidence that the ASL does not adopt different conformations depending upon the identity of the fourth nucleotide of the A-site codon. In summary, these structures confirm that the +1 frameshift mediated by tRNASufJ does not occur during decoding in the A site and therefore, excludes the quadruplet decoding model (Fig. 1A).
During decoding, 16S rRNA nucleotides A1492, A1493, and G530 monitor the minor groove of the codon–anticodon helix to probe for Watson–Crick base-pair geometry (Ogle et al. 2001; Demeshkina et al. 2013). These 16S rRNA residues form hydrogen bonds with the first and second nucleotide pairs of the codon–anticodon helix during cognate tRNA decoding (Ogle et al. 2001). In addition to canonical interactions with 16S rRNA, 23S rRNA residue A1913 also forms a hydrogen bond with the 2′-OH of ASLSufJ nucleotide 37. All of these interactions are seen upon ASLSufJ decoding the ACC-A codon (in the absence of paromomycin), indicating the ribosome recognizes this interaction as cognate (Fig. 4A). The 70S structures of ASLSufJ bound to the ACC-C and ACC-U codons additionally contain paromomycin, which was added to enhance the diffraction properties of these complexes (Supplemental Fig. S1). As expected, the structures containing paromomycin also display the same characteristic cognate interactions as the 70S-ACC-A structure lacking paromomycin. Therefore, since all three structures contain the same A-site interactions between the ACC codon and the anticodon, this provides strong evidence that all three +1 suppressible codons are recognized as cognate by the ribosome.
To accurately compare ASLSufJ to other ASLs that contain seven nucleotide anticodon loops, we solved two additional 70S complexes containing ASLThr bound to either ACC-A or ACC-C codons in the A site of the 70S Tth ribosome to 3.6 Å resolution (Supplemental Fig. S4; Table 2). Both structures also contained paromomycin. These two codons were selected because of the approximately twofold higher affinity of both ASLSufJ and ASLThr for the ACC-A codon as compared with the ACC-C codon (Fig. 3; Table 1). Unbiased Fo–Fc difference electron density maps showed a clear signal for mRNA, P-site tRNAfMet, and A-site ASLThr nucleotides 28–43. Structural analyses revealed that ASLThr recognizes both ACC-A and ACC-C codons in the zero frame as cognate, with A1492, A1492, and G530 interacting with the first two base pairs of codon–anticodon helix.
The C31.5 insertion in ASLSufJ results in conformational rearrangements in the stem of the ASL
The C31.5 insertion in tRNASufJ was previously predicted to form a new stacking interaction with U32 allowing a widening of the unpaired loop of the ASL, while preserving the anticodon structure (Bossi and Smith 1984). Our results indicate that the anticodon loop nucleotides 33–37 are superimposable with other ASLs containing seven nucleotides, including ASLThr (Fig. 4B). The C31.5 insertion neither forces a bulge in the stem to allow for stacking with U32 nor causes a widening of the loop; in fact, the loop of ASLSufJ is narrower than normal ASLs (Figs. 4B, 5A). In contrast, large, concerted movements of both the 5′ and 3′ phosphate backbone of the anticodon stem facilitate accommodation of the C31.5 insertion (Fig. 4B). The 5′ stem, in which C31.5 is located, displaces the phosphate backbone by ∼4 Å toward the 3′ ASL stem (Fig. 4B). Likewise, the 3′ stem undergoes an even larger displacement of ∼7 Å in the same direction away from the 5′ stem. These combined conformational rearrangements result in the tilting of the anticodon stem ∼11.5° away from the P-site tRNAfMet, toward the back of the 30S A site (Fig. 4B).
FIGURE 5.
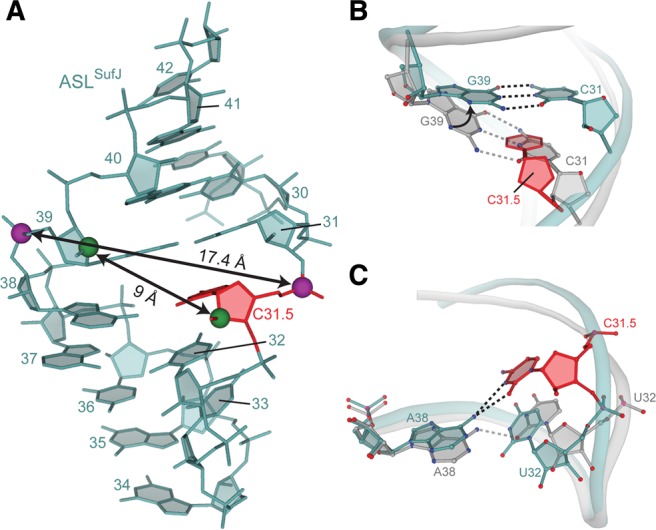
Narrowing of ASLSufJ alters anticodon stem base-pair interactions. (A) Both the major (as measured by C31.5·G39 phosphate–phosphate distances; purple spheres) and minor grooves (as measured by C31.5·G39 C2′–C2′ distances; green spheres) are narrowed in ASLSufJ as compared with canonical major and minor groove distances of 18.4 Å and 14.2 Å, respectively (e.g., ASLThr). The C31.5 insertion is shown as red. (B) The C31.5 insertion (red) causes C31 in ASLSufJ (blue) to shift in the 5′ direction; however, the interaction with G39 (blue) is maintained via a 25° rotation of its base around the glycosidic bond. The canonical C31–G39 interaction of a canonical seven-nucleotide ASLThr is shown for comparison (gray). (C) The 5′ phosphate backbone movement shifts U32 (blue) closer to the opposite RNA stem while A38 rotates 8° to form a bifurcated hydrogen bond with the inserted C31.5 (red). This new interaction prevents the formation of the conserved U32·A38 base pair of ASLThr (gray).
The concerted movement of the anticodon stems of ASLSufJ narrows the major groove from 18.4 to 17.4 Å (phosphate–phosphate distances) and the minor groove from 14.2 to 9 Å (C2′–C2′ distances) (Fig. 5A). Collectively, these changes cause a reduction in the average base-pair incline (or the angle between the base pairs) and the helical axis from 14° in ASLThr to 11.5° in ASLSufJ with a concomitant increase in the overall helical twist (29° in ASLThr versus 33° in ASLSufJ). An additional consequence of the ASL stem rotation toward the back of the A site, is that this movement prevents a conserved interaction between Lys121 of ribosomal protein S13 and the 2′-OH of G40. While the electron density of the Lys121 side chain is not interpretable, the conformational rearrangement of nucleotide G40 of ASLSufJ would prevent any possibility of forming a hydrogen bonding interaction with this residue given the larger distance.
Anticodon stem register is maintained in ASLSufJ despite alteration of the conserved 32–38 base pair
The C31.5 nucleotide occupies the physical position of C31 in the 70S-ASLThr structures, thereby shifting the 5′ side of the ASLSufJ stem by one nucleotide (Fig. 5A). However, the base-pair register is maintained with the opposite side of the stem via a rotation of the G39 base (25°) about its N-glycosidic bond (Fig. 5B). While this change in torsion angle preserves the C31–G39 interaction of the stem, it disrupts the conserved U32·A38 base pair (Fig. 5C). Although the position of U32 is largely consistent between the 70S-ASLThr and 70S-ASLSufJ structures, A38 rotates 8° to form a new bifurcated hydrogen bond between its N6 position and both the N3 and O2 atoms of the inserted C31.5 (Fig. 5C). The C31.5 insertion in ASLSufJ structurally replaces the U32 in the U32·A38 interaction as seen in ASLThr, therefore forming a new C31.5·A38 pair (Fig. 5C).
Projection of tRNASufJ in the A and P sites indicates potential rearrangements due to steric clashes
Previously determined 70S and 30S structures containing A-site tRNAs or ASLs, all contain anticodon stem–loops that adopt a conformation that closely approximates an accommodated or pre-peptidyl transferase state (Voorhees et al. 2009). However, in special cases such as an insertion in the anticodon or modifications at nucleotide 34, the A-site ASL can adopt an alternate conformation but only at the anticodon or the unpaired anticodon loop (Murphy et al. 2004; Phelps et al. 2006; Cantara et al. 2013). The conformational changes of the anticodon stem that we observe in ASLSufJ have never been seen previously in any 70S structure. To understand how the 11.5° tilt would affect the position of ASLSufJ in the context of a full-length tRNA, we aligned the anticodons of ASLSufJ and tRNAPhe (PDB code 2WDH) and then projected a full-length tRNASufJ using fully accommodated Phe-tRNAPhe as a guide (Fig. 6A; Voorhees et al. 2009). This alignment revealed the tilted stem domain of ASLSufJ repositions the acceptor arm >20 Å distant from the position of a canonical accommodated state tRNA. Moreover, tRNASufJ would clash with components of the 50S (Fig. 6A). Clearly tRNASufJ does not bind in this manner to the 70S and must instead undergo structural remodeling to be positioned properly for tRNA accommodation and peptide bond formation. In summary, the 11.5° tilt of the anticodon stem of ASLSufJ suggests that the C31.5 insertion changes the energetic landscape that governs the transition of tRNA between its various functional conformations.
FIGURE 6.
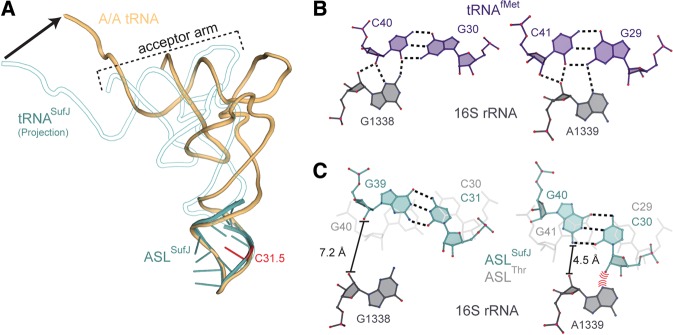
Modeling tRNASufJ interactions in the A and P sites. (A) Modeling studies extending our structure of ASLSufJ (blue) to a full-length tRNASufJ (white outlined with blue) reveal that the tilting position of the stem region would result in the CCA end of tRNASufJ >20 Å distant from the position of an accommodated A-site tRNA (gold) (arrow at the CCA end indicates the difference). We predict that conformational rearrangements of tRNASufJ are required to prevent this interaction with the ribosome. (B) The P-site tRNAfMet (purple) is gripped by 16S rRNA G1338 and A1339 (gray) by the formation of A-minor interactions with both C40-G30 and C41-G29 (PDB code 2J02). (C) Superposition of ASLSufJ on the anticodon of tRNAfMet in the P site suggests that the deformation of the stem domain would prevent the formation of canonical G1338 and A1339 interactions. A semitransparent overlay of the superpositioning of ASLThr is shown for comparison (light gray).
In the 30S A site, the ribosome exclusively interacts with the anticodon of the tRNA to ensure high fidelity tRNA selection. Upon translocation to the P site by EF-G, the anticodon is no longer closely monitored. Instead, the ribosome interacts with the anticodon stem and other regions of the tRNA body to optimally orient the acceptor arm for peptide bond formation. It has been proposed that the P site has evolved to tightly grip the tRNA in order to maintain the proper mRNA reading frame (Selmer et al. 2006; Atkins and Bjork 2009; Nasvall et al. 2009; Jager et al. 2013). For example, initiator tRNAfMet is “gripped” by 16S rRNA residues G1338 and A1339 through A-minor motif interactions with anticodon stem base pairs (Fig. 6B; Lancaster and Noller 2005; Selmer et al. 2006). Superpositioning of A-site ASLSufJ into the P site by alignment of the anticodon to tRNAfMet reveals the 5′ stem adjacent to the insertion site and the C31.5 phosphate would sterically clash with 16S rRNA residue A1339. However, the twist in the ASLSufJ stem prevents the formation of A-minor interactions with 16S rRNA nucleotide G1338 (Fig. 6C). This predicted lack of interaction indicates that either the gripping of the anticodon stem of tRNASufJ is different from wild-type tRNAs or a conformational rearrangement of the stem occurs. The remodeling of the anticodon stem could possibly occur during translocation or after translocation into the P site, which in turn, facilitates the movement of mRNA by one nucleotide into the +1 frame.
DISCUSSION
Extragenic +1 frameshift suppressors were predominately identified as nucleotide insertions in the anticodon loops of tRNAs (for review, see Atkins and Bjork 2009). Although one interpretation of this phenomenon was that efficiency of +1 frameshifting was dependent on the Watson–Crick complementarity between the insertions located in both the tRNA and codon, the identification of tRNASufJ necessitated alternative hypotheses (Bossi and Smith 1984; Farabaugh 2000). While tRNASufJ does contain an insertion in its anticodon loop, the extra nucleotide is located adjacent to the anticodon stem, distal from the anticodon and therefore it was unclear whether this insertion actually expanded the anticodon to more than three nucleotides. The structures here reveal that ASLSufJ binds to three +1 suppressible codons in the zero frame allowing only a three-nucleotide codon–anticodon cognate interaction in the A site. The C31.5 insertion does not appear to alter the structural integrity of the ASL in the same manner as other frameshift suppressors where disordering of the conserved U turn, the entire 5′ stem or nucleotide 32 occurs (Phelps et al. 2006; Dunham et al. 2007; Maehigashi et al. 2014). Instead, ASLSufJ accommodates the insertion through concerted movements of both the 5′ and 3′ phosphate backbones of the anticodon stem, causing a reorganization of key nucleotide interactions in the anticodon loop. These results indicate that insertions either 5′ or 3′ of the anticodon that result in +1 frameshifting may adopt different structural changes with an underlying theme being that tRNA plasticity, rather than the codon–anticodon interaction, drives +1 frameshifting.
Our 70S-A-site ASLSufJ structures reveal that the C31.5 insertion does not widen the anticodon loop but, rather, causes a narrowing of both the major and minor grooves of the ASL. Narrowing of the anticodon loop was also observed in a recent 70S structure containing an A-site +1 frameshift suppressor tRNASufA6 (Maehigashi et al. 2014). In this case, tRNASufA6 contains an insertion 3′ to the anticodon between nucleotides 37 and 38, on the opposite side of the loop to C31.5 in tRNASufJ. In contrast, a previously solved 30S structure bound to a +1 frameshift suppressor ASL shows an insertion at 33.5 causes a widening of the loop (Dunham et al. 2007). It remains unclear whether a narrowing or a widening of frameshift suppressor tRNA anticodon loops indicates a different mechanism by which a +1 reading of the mRNA occurs. However, a common emerging theme is that insertions in the anticodon stem–loops of tRNAs are accommodated in different ways thus reinforcing the idea of structural plasticity as a driver of +1 frameshifting.
Although the reorganization of the anticodon loop interactions allows for maintenance of the base-pair registry in the anticodon stem, the major consequence of this preservation and the C31.5 insertion is that the identity of the 32–38 interaction is altered from U32·A38 to C31.5·A38. Anticodon stem nucleotides 32 and 38 are important for tRNA affinity and selection on the ribosome with the identity of this pair directly correlated to both the Watson–Crick base-pair strength of the codon–anticodon interaction and the aminoacyl group attached to the CCA 3′ end (Olejniczak and Uhlenbeck 2006; Ledoux and Uhlenbeck 2008). A strong C32·A38 interaction is predicted to counteract a weak codon–anticodon interaction such as in the case of tRNALys (UUU), whereas a weak U32·A38 interaction is normally paired with a strong codon–anticodon interaction such as in the case of tRNAPro (GGG) (Olejniczak and Uhlenbeck 2006). This correlation of the 32–38 identity and the codon–anticodon strength allows for the fine tuning of tRNA selection by the ribosome. Surprisingly, compensatory mutations of the rare A32·U38 pair in tRNAAla to the more common U32·A38 displayed no effects on tRNA binding or incorporation when decoding cognate codons (Ledoux et al. 2009). However, tRNAAla with the common U32·A38 pair is more rapidly accommodated when a near-cognate codon is present in the A site, indicating a loss of fidelity. Additional evidence for the importance of the 32–38 pairing comes from tRNA suppressor experiments using an amber stop codon. Here, stop codon readthrough efficiencies were increased when 32–38 was mutated to a strong C32·A38 pair, again strongly suggesting a loss of fidelity (Yarus et al. 1986a,b; Kleina et al. 1990; McClain et al. 1998; Tsai et al. 1998). We predict that, in general, the insertion of a nucleotide within an anticodon loop reduces its binding affinity to the ribosomal A site. However, in the case of ASLSufJ where the insertion results in the formation of a new, strong C31.5·A38 interaction, our interpretation is that the predicted lower affinity resulting from expanding the ASL to eight nucleotides is counterbalanced by the increased strength of the 31.5·38 interaction. Indeed, our affinity measurements with ASL/tRNASufJ indicate that the C31.5 insertion slightly increases A-site affinity as compared with wild-type ASL/tRNAThr. Likewise we would predict that if C31.5 were mutated to an uridine resulting in a U31.5·A39 pairing, the binding affinity for this weaker pairing in the ribosomal A site would be reduced.
Our results examining ASLSufJ in light of our recent studies of ASLSufA6 (Maehigashi et al. 2014) lead us to propose that +1 frameshifting by these suppressors does not occur by quadruplet decoding in the A site. Although the structures of ASLSufJ and ASLSufA6 indicate the insertions alter the anticodon loops in distinct manners, both structures involve either the rearrangement or disruption of the 32–38 pair. This reorganization of the 32–38 pairing occurs despite ASLSufJ forming a cognate interaction while ASLSufA6 forms a near-cognate interaction with their +1 suppressible codons in the A site. What remains unclear is if the frameshift occurs during translocation or after translocation of the mRNA–tRNA pair to the P site. Unlike ASLSufA6, the 5′ ASLSufJ stem nucleotide 32 is ordered. Disordering may be an important stimulus for the +1 frameshift event because EF-G directly interacts with the 5′ stem in the A site before translocation (Brilot et al. 2013). Interestingly, the narrowing of the stem and 11.5° tilting of ASLSufJ suggests that adjustment of P-site interactions with conserved 16S rRNA nucleotides G1338 and A1339 could occur upon translocation. Another possibility is that ASLSufJ would be required to undergo a conformational rearrangement of its anticodon loop to maintain this important gripping interaction, which could then facilitate the shifting into the new +1 mRNA reading frame. Taken together, these results help to begin to unravel mechanistic details of +1 frameshifting.
MATERIALS AND METHODS
E. coli ribosome purification
70S ribosomes were purified as previously described with a few modifications (Powers and Noller 1991). Briefly, E. coli MRE600 cells were grown in LB to an OD600 of 0.6–0.8 and then cooled on ice for 20 min to increase the concentration of run-off 70S ribosomes. Cultures were then pelleted and resuspended in buffer A (20 mM HEPES pH 7.5, 100 mM NH4Cl, 10.5 mM MgOAc, 0.5 mM EDTA, 0.1 mM benzamidine, 0.1 mM phenylmethanesulfonylfluoride (PMSF) and 6 mM β-mercaptoethanol (β-Me)) and lysed using a EmulsiFlex cell disruptor. The lysate was clarified by centrifugation (30 min at 70,000g) and pelleted over a sucrose cushion (1.1 M sucrose, 20 mM HEPES pH 7.5, 500 mM NH4Cl, 10.5 mM MgOAc, 0.5 mM EDTA) for 17 h at 158,000g at 4°C. The ribosome pellet was resuspended in buffer C (20 mM Tris–Cl pH 7.5, 400 mM KCl, 10 mM MgOAc, 1.5 M (NH4)2SO4, 0.1 mM benzamidine, 0.1 mM PMSF, and 6 mM β-Me) and purified over a Butyl-650S HIC column (Toyopearl) using a reverse (NH4)2SO4 gradient. 70S ribosomes were further separated over a 10%–40% sucrose gradient in buffer E (10 mM HEPES pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10.25 mM MgOAc, 0.25 mM EDTA, 0.1 mM benzamidine, 0.1 mM PMSF and 6 mM β-Me). Fractions were pooled and concentrated by pelleting over a sucrose cushion, resuspended and dialyzed overnight in buffer G (5 mM HEPES pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM MgOAc, and 6 mM β-Me). Lastly, purified 70S ribosomes were concentrated using an Amicon 100K molecular weight cut-off concentrator (Millipore) and stored at −80°C.
In vitro transcription
Salmonella typhimurium tRNASufJ, along with a 5′ T7 promoter site, were subcloned into a pUC19 vector using overlapping DNA oligos (IDT). Plasmid overexpression, purification and in vitro transcription reactions were performed as previously described using BstNI (NEB) to linearize the plasmid (Linpinsel and Conn 2012). The 5′ triphosphate was removed by Calf Intestinal Phosphatase (NEB) and purified by phenol–chloroform and chloroform extractions followed by ethanol precipitation and stored in Tris–EDTA buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA) at −20°C. For the Salmonella typhimurium tRNAThr, we encountered difficulties purifying tRNAThr away from a contaminating RNA band of a similar size after in vitro transcription. Therefore we PCR amplified the tRNAThr gene and a 5′ T7 promoter site from overlapping DNA oligos (IDT) and purified the in vitro transcribed RNA product as previously described (Linpinsel and Conn 2012).
RNA sequences
The mRNA sequence used in both the filter binding experiments and crystallization trials was 5′-GGCAAGGAGGUAAAAAUGACC HAAA-3′, where H represents an A, U, or C nucleotide (threonine codon is in bold and follows the italicized AUG start site). ASLThr sequence was 5′-CACCCUUGGUAAGGGUG-3′ and for ASLSufJ was 5′-CACCCCUUGGUAAGGGUG-3′ where the anticodon is underlined and nucleotide insertion in ASLSufJ is indicated in bold (IDT).
5′-Labeling of ASLs and tRNAs
tRNAs or ASLs (1 µM) were incubated with [γ-32P]-ATP (1 µCi/µL, 3000 Ci/mmol, PerkinElmer) and T4 polynucleotide kinase (NEB) for 1 h at 37°C. Free nucleotide was removed by purification over a Sephadex G-25 spin column (GE Healthcare) and the labeling efficiency was determined by scintillation counting (Beckman LS-5000TD). A typical filter binding reaction contained 200,000–700,000 cpm/µL of tRNA or ASL. Labeled tRNA or ASL were stored at a concentration of 10 µM at −20°C.
70S binding assays
The affinity of tRNAs or ASLs for the 70S A site was measured using filter binding assays as previously described (Olejniczak et al. 2005). Briefly, purified E. coli 70S ribosomes (500 nM) were incubated with mRNA (1 µM) in buffer G at 37°C for 5 min, followed by the addition of P-site tRNAfMet (1 µM) at 37°C for 30 min. For some experiments as indicated, paromomycin (100 µM) was incubated for an additional 25 min at room temperature. Twofold dilutions were made, resulting in a range of ribosome concentrations from 0.98 to 1 μM which was experimentally determined to provide full coverage of the equilibrium dissociation curve; overall the ribosome concentration range were measured from 5 nM to 1 µM. A-site tRNA (2 nM) or ASL (2 nM) was added to each solution and the reaction was allowed to come to equilibrium at room temperature. A 3-h incubation was required for tRNA binding to reach equilibrium, however with the addition of paromomycin only a 2 h incubation was necessary. The ribosome reaction (30 µL) was then filtered through a 0.45 µm nitrocellulose membrane and washed with buffer G (1 mL). The nitrocellulose filters were dissolved in Filtron-X (National Diagnostics) and counted using a Beckman LS-5000TD scintillation counter. Dissociation constants (KD) and Bmax values were obtained by fitting the data to a one site specific binding nonlinear regression using GraphPad Prism as has been done for previous equilibrium binding experiments (Ledoux and Uhlenbeck 2008).
70S complex formation and crystallization
Thermus thermophilus ribosomes were purified and crystallization trials were performed as previously described with a few minor modifications (Selmer et al. 2006). Briefly, 70S ribosomes (4.4 µM) were incubated with CC-puromycin (Dharmacon; 22 µM), an aminoacyl mimic consisting of the RNA dinucleotide CC covalently attached to puromycin-5′-monophosphate, for 30 min at 55°C. This was followed by incubation with a two molar excess of mRNA (IDT; 8.8 µM) for 5 min. Four molar excess of P-site tRNAfMet (Chemical Block; 17.6 µM) was incubated for 30 min at 55°C and finally, five molar excess of the appropriate ASL (IDT; 22 µM) was incubated for 30 min at 55°C. The complexes were cooled to room temperature and then incubated with antibiotic paromomycin (0.1 mM) for an additional 20 min at room temperature. Deoxy BigCHAP (Hampton Research; 2.8 µM) was added just prior to crystallization. Crystals were grown by sitting-drop vapor diffusion in 4%–5% polyethylene glycol (PEG) 20K, 4%–5% PEG 550 MME, 0.1 M Tris–Acetate pH 7.0, 0.2 M KSCN and 10 mM MgCl2, and cryoprotected by increasing PEG 550 MME in a stepwise manner to a final concentration of 30%. Crystals were flash frozen in liquid nitrogen for data collection.
X-ray data collection and structure determination
X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline and the Northeastern Collaborative Access Team (NE-CAT) 24-IDC beamline at the Advanced Photon Source, Argonne National Laboratory. Each data set was integrated and scaled using the XDS software package (Kabsch 2010). A search model composed of the Tth 70S ribosome (PDB codes 2WDG, 2WDH, 2WDI, and 2WDJ) with all mRNA and tRNA ligands removed was used for crystallographic refinement with the PHENIX software suite (Adams et al. 2010). Additional rounds of coordinate refinement were performed with rigid groups defined by the head, body, platform, and 3′-minor domain of the 30S subunit, along with mobile elements of the 50S subunit: 5S rRNA, L1 arm, protein L9, A-site finger, and the central protuberance. Modeling of mRNA, tRNA, and conformational changes in rRNA and ribosomal proteins along with the placement Mg2+ ions were performed using Coot (Emsley et al. 2010). Iterative rounds of model building were followed by positional and group atomic displacement parameter (ADP) refinement in PHENIX, yielding a final model with the statistics reported in Table 2. Figures were generated using PyMOL (www.pymol.org).
DATA DEPOSITION
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.pdb.org) under the PDB codes 4TUA, 4TUB, 4TUC, 4TUD, and 4TUE.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program and National Institutes of Health (NIH) Training Grant T32 GM8367 (to C.E.F.), and National Institute of General Medical Sciences of the NIH Award R01GM093278 (to C.M.D.). C.M.D. is a Pew Scholar in the Biomedical Sciences (Pew Charitable Trusts). We thank G.L. Conn and members of the Dunham lab for helpful discussions throughout the project and critical reading of the manuscript, and staff members at both SER-CAT and NE-CAT beamlines for assistance during data collection. This work is based on research conducted at the Advanced Photon Source on the NE-CAT beamlines (supported by National Center for Research Resources [NIH] Award RR-15301) and SER-CAT beamline. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by US DOE Contract DE-AC02-06CH11357.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.046953.114.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66(Pt 2): 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Magliery TJ, Schultz PG 2002. Exploring the limits of codon and anticodon size. Chem Biol 9: 237–244. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Bjork GR 2009. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev 73: 178–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Roth JR 1981. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell 25: 489–496. [DOI] [PubMed] [Google Scholar]
- Bossi L, Smith DM 1984. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci 81: 6105–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouadloun F, Donner D, Kurland CG 1983. Codon-specific missense errors in vivo. EMBO J 2: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N 2013. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci 110: 20994–20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantara WA, Murphy FV IV, Demirci H, Agris PF 2013. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci 110: 10964–10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins CM, Donahue TF, Culbertson MR 1982. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci 79: 3565–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M 1987. Reading frame selection and transfer RNA anticodon loop stacking. Science 238: 1545–1550. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G 2013. New structural insights into the decoding mechanism: translation infidelity via a G·U pair with Watson–Crick geometry. FEBS Lett 587: 1848–1857. [DOI] [PubMed] [Google Scholar]
- Dunham CM, Selmer M, Phelps SS, Kelley AC, Suzuki T, Joseph S, Ramakrishnan V 2007. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA 13: 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann P, Gallant J 1977. Mistranslation in E. coli. Cell 10: 131–137. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ 2000. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol 64: 131–170. [DOI] [PubMed] [Google Scholar]
- Gaber RF, Culbertson MR 1984. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol Cell Biol 4: 2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohsaka T, Ashizuka Y, Taira H, Murakami H, Sisido M 2001. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry 40: 11060–11064. [DOI] [PubMed] [Google Scholar]
- Jager G, Nilsson K, Bjork GR 2013. The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PLoS One 8: e60246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F, Kurland CG 1990. Processivity errors of gene expression in Escherichia coli. J Mol Biol 215: 511–521. [DOI] [PubMed] [Google Scholar]
- Jorgensen T, Siboska GE, Wikman FP, Clark BF 1985. Different conformations of tRNA in the ribosomal P-site and A-site. Eur J Biochem 153: 203–209. [DOI] [PubMed] [Google Scholar]
- Kabsch W 2010. XDS. Acta Crystallogr D Biol Crystallogr 66(Pt 2): 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khade P, Joseph S 2010. Functional interactions by transfer RNAs in the ribosome. FEBS Lett 584: 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina LG, Masson JM, Normanly J, Abelson J, Miller JH 1990. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol 213: 705–717. [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG 1992. Translational accuracy and the fitness of bacteria. Annu Rev Genet 26: 29–50. [DOI] [PubMed] [Google Scholar]
- Lancaster L, Noller HF 2005. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell 20: 623–632. [DOI] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC 2008. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell 31: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC 2009. A sequence element that tunes Escherichia coli tRNAAlaGGC to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL 2006. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443: 50–55. [DOI] [PubMed] [Google Scholar]
- Linpinsel JL, Conn GL 2012. General protocols for preparation of plasmid DNA template, RNA in vitro transcription, and RNA purification by denaturing PAGE. Methods Mol Biol 941: 43–58. [DOI] [PubMed] [Google Scholar]
- Maehigashi T, Dunkle JA, Miles SJ, Dunham CM 2014. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Natl Acad Sci 111: 12740–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Gesteland RF 1978. Suppression of amber mutants in vitro induced by low temperature. J Mol Biol 125: 433–447. [DOI] [PubMed] [Google Scholar]
- McClain WH, Schneider J, Bhattacharya S, Gabriel K 1998. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc Natl Acad Sci 95: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Persson BC, Nelson CC, Gesteland RF, Atkins JF 2000. Quadruplet codons: implications for code expansion and the specification of translation step size. J Mol Biol 298: 195–209. [DOI] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V, Malkiewicz A, Agris PF 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191. [DOI] [PubMed] [Google Scholar]
- Nangle LA, De Crecy Lagard V, Doring V, Schimmel P 2002. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J Biol Chem 277: 45729–45733. [DOI] [PubMed] [Google Scholar]
- Nasvall SJ, Nilsson K, Bjork GR 2009. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. J Mol Biol 385: 350–367. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Uhlenbeck OC 2006. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88: 943–950. [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC 2005. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat Struct Mol Biol 12: 788–793. [DOI] [PubMed] [Google Scholar]
- Phelps SS, Jerinic O, Joseph S 2002. Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol Cell 10: 799–807. [DOI] [PubMed] [Google Scholar]
- Phelps SS, Gaudin C, Yoshizawa S, Benitez C, Fourmy D, Joseph S 2006. Translocation of a tRNA with an extended anticodon through the ribosome. J Mol Biol 360: 610–622. [DOI] [PubMed] [Google Scholar]
- Powers T, Noller HF 1991. A functional pseudoknot in 16S ribosomal RNA. EMBO J 10: 2203–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather NE, Murgola EJ, Mims BH 1981. Nucleotide insertion in the anticodon loop of a glycine transfer RNA causes missense suppression. Proc Natl Acad Sci 78: 7408–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Li JN, Zhao H, Hagervall TG, Farabaugh PJ, Bjork GR 1998. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell 1: 471–482. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Carbon J 1973. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol 242: 230–234. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Roth JR 1970. Suppressors of frameshift mutations in Salmonella typhimurium. J Mol Biol 54: 131–144. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Roth JR 1972. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. J Mol Biol 66: 495–506. [DOI] [PubMed] [Google Scholar]
- Riyasaty S, Atkins JF 1968. External suppression of a frameshift mutant in salmonella. J Mol Biol 34: 541–557. [DOI] [PubMed] [Google Scholar]
- Rodnina MV 2012. Quality control of mRNA decoding on the bacterial ribosome. Adv Protein Chem Struct Biol 86: 95–128. [DOI] [PubMed] [Google Scholar]
- Roth JR 1974. Frameshift mutations. Annu Rev Genet 8: 319–346. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942. [DOI] [PubMed] [Google Scholar]
- Sherman F, Stewart JW, Jackson M, Gilmore RA, Parker JH 1974. Mutants of yeast defective in iso-1-cytochrome c. Genetics 77: 255–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F, Curran JF 1998. tRNA2Gln mutants that translate the CGA arginine codon as glutamine in Escherichia coli. RNA 4: 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen U, Green R, Schroeder R, Noller HF 1997. Identification of 2′-hydroxyl groups required for interaction of a tRNA anticodon stem-loop region with the ribosome. RNA 3: 49–56. [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Fredrick K 2006. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol 360: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M 1982. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science 218: 646–652. [DOI] [PubMed] [Google Scholar]
- Yarus M, Cline S, Raftery L, Wier P, Bradley D 1986a. The translational efficiency of tRNA is a property of the anticodon arm. J Biol Chem 261: 10496–10505. [PubMed] [Google Scholar]
- Yarus M, Cline SW, Wier P, Breeden L, Thompson RC 1986b. Actions of the anticodon arm in translation on the phenotypes of RNA mutants. J Mol Biol 192: 235–255. [DOI] [PubMed] [Google Scholar]
- Yourno J 1970. Nature of the compensating frameshift in the double frameshift mutant hisD3018 R5 of Salmonella typhimurium. J Mol Biol 48: 437–442. [DOI] [PubMed] [Google Scholar]
- Yourno J 1972. Externally suppressible +1 “glycine” frameshift: possible quadruplet isomers for glycine and proline. Nat New Biol 239: 219–221. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.