Abstract
Although Piwi proteins and Piwi-interacting RNAs (piRNAs) genetically repress transposable elements (TEs), it is unclear how the highly diverse piRNA populations direct Piwi proteins to silence TE targets without silencing the entire transcriptome. To determine the capacity of piRNA-mediated silencing, we introduced reporter genes into Drosophila OSS cells, which express microRNAs (miRNAs) and piRNAs, and compared the Piwi pathway to the Argonaute pathway in gene regulation. Reporter constructs containing several target sites that were robustly silenced by miRNAs were not silenced to the same degrees by piRNAs. However, another set of reporters we designed to enable a large number of both TE-directed and genic piRNAs to bind were robustly silenced by the PIWI/piRNA complex in OSS cells. These reporters show that a bulk of piRNAs are required to pair to the reporter's transcripts and not the reporter's DNA sequence to engage PIWI-mediated silencing. Following our genome-wide study of PIWI-regulated targets in OSS cells, we assessed candidate gene elements with our reporter platform. These results suggest TE sequences are the most direct of PIWI regulatory targets while coding genes are less directly affected by PIWI targeting. Finally, our study suggests that the PIWI transcriptional silencing mechanism triggers robust chromatin changes on targets with sufficient piRNA binding, and preferentially regulates TE transcripts because protein-coding transcripts lack a threshold of targeting by piRNA populations. This reporter platform will facilitate future dissections of the PIWI-targeting mechanism.
Keywords: Piwi, piRNAs, gene silencing
INTRODUCTION
Piwi proteins and piRNAs are best known to suppress transposable elements (TEs) in animal germ cells, as supported by TE transcript up-regulation in Piwi pathway mutants (Aravin et al. 2004; Kalmykova et al. 2005; Saito et al. 2006; Vagin et al. 2006), a genetic role in hybrid dysgenesis studies (Brennecke et al. 2008; Chambeyron et al. 2008; Khurana et al. 2011; Grentzinger et al. 2012), and perfect complementarity between piRNA sequences and TE coding transcripts (Juliano et al. 2011; Siomi et al. 2011). In Drosophila, the precursors to TE-associated piRNAs can begin as long single-stranded transcripts from loci that trap TE relics in a biased orientation so that transcription generates piRNAs complementary to TEs (Brennecke et al. 2007; Lau et al. 2009a; Malone et al. 2009). One such piRNA cluster locus on the X chromosome, called flamenco (flam), genetically affects TE mobilization in the ovarian follicle cells (Bucheton 1995; Mevel-Ninio et al. 2007). Finally, PIWI can also potentially regulate genes via generating abundant genic piRNAs such as from the 3′ UTR of traffic jam (tj) (Robine et al. 2009; Saito et al. 2009).
Both transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) as well as gene activation mechanisms have previously been proposed for Piwi/piRNA-mediated function (Pal-Bhadra et al. 2004; Grivna et al. 2006; Brower-Toland et al. 2007; Klenov et al. 2007; Yin and Lin 2007; Aravin et al. 2008). However, it is unclear which of these mechanisms dominate in gonadal cells, and how many piRNAs are required to manifest target regulation. Despite the fact that Drosophila PIWI retains the catalytic residues which exhibit “Slicer” endonucleolytic activity (Lau et al. 2006; Saito et al. 2006; De Fazio et al. 2011; Reuter et al. 2011), this activity is not required for TE repression nor loading of piRNAs into PIWI (Saito et al. 2010; Darricarrere et al. 2013). If piRNAs require perfect base-pairing for efficient targeting and silencing, this would partially explain the preference of piRNAs targeting TEs over genes. Nevertheless, additional mechanistic studies are required to address how well PIWI and piRNAs regulate gene and TE expression at both the TGS and PTGS levels.
Argonaute (Ago) proteins, which bind microRNAs (miRNAs) and small interfering RNAs (siRNAs) are homologs of Piwi proteins. AGO1/miRNA complexes can inhibit thousands of transcripts by base-pair interactions between the miRNA “seed” elements and a single or a few target sites in the 3′ untranslated regions (3′ UTRs) of genes (Bartel 2009). However, it is unclear which transcripts interact with Piwi proteins and piRNAs; and whether metazoan piRNAs routinely bind targets with base mismatches. The enormous diversity of piRNAs in gonadal cells dwarf the smaller number of miRNA sequences (greater than millions of unique piRNA sequences [Betel et al. 2007] versus <1000 miRNAs [Chiang et al. 2010]). If PIWI/piRNA-targeting mechanisms can operate like AGO1/miRNA complexes in tolerating base-pairing mismatches, the entire germ cell transcriptome would be predicted to be targeted. How then are piRNA targets determined?
RESULTS
Allowing mismatches while mapping diverse piRNA sequences pinpoints the entire transcriptome
To illustrate the challenge in bioinformatically predicting piRNA targets beyond expected TEs, we conducted a straightforward prediction of Drosophila OSS cell piRNAs that could base-pair to Drosophila RefSeq transcripts with different degrees of mismatches (Supplemental Fig. S1). When one or fewer mismatches are allowed, the majority of TEs are predictably matched to piRNAs, but very few piRNAs (<2%) are accounted for in targeting a modest number of mRNA targets (<4% of transcriptome). Although ∼50% of Drosophila mRNAs can be paired with two base mismatches to a small fraction of piRNAs (<5%), allowing three base mismatches in predictions resulted in nearly every mRNA transcript predicted to pair with a piRNA. In these types of target predictions, we assumed each small RNA sequence has an equal propensity to base-pair with a transcript target, and this assumption can be extended to miRNA target predictions which deal with a much smaller diversity of miRNA sequences. However, the >4.2 million unique piRNA sequences expressed in OSS cells lends to the prediction that the entire germ cell transcriptome could be targeted by piRNAs. Therefore, we require a deeper understanding of what features may dictate the effectiveness of piRNA sequences in target regulation before we can devise more sophisticated target prediction models.
Target silencing capacity is different between miRNAs and piRNAs
Many sequence features for miRNA target prediction have been classically tested with several tandem miRNA-binding sequences inserted into the 3′ UTR of the luciferase reporter gene (Grimson et al. 2007). We extended this platform to examine whether individual piRNAs could regulate analogous reporter constructs (Fig. 1). We chose the four most-abundant piRNAs in OSS cells (nominally named piR-1–4) to compare with two modestly expressed miRNAs (miR-14 and miR-12). These small RNAs showed similar relative abundances in OSS cells from deep-sequencing as well as by quantitative Northern blots (Fig. 1B), although the minor differences between the measuring approaches could be technical or due to different passage numbers of the OSS cell sample. We designed three sets of reporter constructs per small RNA—a perfectly complementary target, a target with three mismatched bases downstream from an intact “seed” sequence, and a mutated target with three more mismatches that completely disrupts the “seed” sequence to serve as a baseline for reporter normalization. These plasmid reporters were transiently transfected into an early isolate of OSS cells described in Lau et al. (2009b) and then luciferase activity was assayed 2 d later.
FIGURE 1.
AGO1/miRNA and PIWI/piRNA complexes act differently on gene silencing reporter constructs. (A) Diagram of the reporter construct design, (RBS) RNA binding site, repeated in four tandem copies (4×). (B) Relative abundance of the small RNAs that were determined from early cultures of OSS cells used in these assays (Lau et al. 2009b). Relative abundance from deep-sequencing was calculated from reads per million, while similar relative abundance levels were confirmed by quantitative Northern blots (below graphs, standards are 10, 50, 100, 500, and 1000 pg). Only two miRNA and piRNA Northern blots are shown as representatives. (C) Reporter gene expression levels normalized to the respective mutated site reporter. Error bars are the standard error of biological triplicates. Asterisk indicates statistical significance relative to the mutated sites control, at P < 0.05, Wilcoxon rank-sum test. (D) The tethered construct reporter system (Pillai et al. 2004; Djuranovic et al. 2010). (E) This reporter design is sufficient to induce AGO1-mediated repression, but in a cell line with an endogenous Piwi pathway, it is unable to trigger silencing by tethered PIWI (which behaves similar to the LacZ negative control). The asterisk marks statistically significant λN-HA-AGO-mediated repression (P < 0.05, Wilcoxon rank-sum test). (F) The upper Western blot shows each of the λN-HA- and HA-fusion transgene constructs were similarly expressed. The lower Western blot shows the λN-HA-PIWI is nuclear localized like endogenous PIWI in OSS cells. Sub-cellular fractions from OSS cells transfected plasmid expressing λN-HA-PIWI, probed with anti-HA antibody.
Although the miRNAs displayed a typical strong gene silencing effect for perfectly complementary as well as bulged sites containing “seed” matching sequence, the piRNA reporters displayed more modest gene silencing with perfectly complementary targets, and little regulation with bulged target sites (Fig. 1C). These results hinted at a difference in target silencing capacity between miRNAs and piRNAs.
To test if the targeting capacity difference is due to the distinctions between small RNA types or instead by differences between Ago and Piwi proteins themselves, we generated a B-box loop reporter construct that tethers Drosophila AGO1 or PIWI bearing an N-terminally fused λ-N protein, thereby bringing AGO1/PIWI to the reporter gene in a small RNA-independent manner (Fig. 1D). Consistent with previous studies (Djuranovic et al. 2011), the λ-N-DmAGO1 could robustly silence the reporters which contain five B-box loops (Fig. 1E). Although all λ-N- and HA-tagged proteins were expressed from the transgenes, and λ-N-HA-PIWI was nuclear localized (Fig. 1F), the λ-N-HA-PIWI was still unable to trigger gene silencing (Fig. 1E) and instead resembled the negative control of λ-N-LacZ in promoting the B-box-luciferase reporter expression by a side effect of the λ-N protein binding the stem–loop and possibly stabilizing the mRNA (Pillai et al. 2004). Thus, while these reporters with five B-box loop binding sites can recapitulate gene silencing by tethering AGO1 complexes independent of the miRNA, tethering of PIWI to transcripts in this manner is insufficient for gene silencing.
Silencing by piRNAs has a bulk base-pairing/targeting requirement
We hypothesized that many more PIWI/piRNA complexes may be required to bind to the reporter transcript in order to trigger gene silencing. Therefore, we constructed an array of Renilla luciferase plasmids that could be targeted by many TE-directed piRNAs derived from the flam locus or genic tj 3′-UTR-derived piRNAs (Fig. 2; Robine et al. 2009; Saito et al. 2009). A 2.4-kb flam and a 1.5-kb tj 3′-UTR element were cloned into the luciferase 3′ UTR in either the same “Sense” orientation or in the opposite “Antisense” orientation as the luciferase transcript. In the “Sense” reporters, the transcript's sequence is in the same orientation as endogenous OSS cell piRNAs, and is predicted not to base-pair to the piRNAs; whereas the “Antisense” reporters can base-pair perfectly to endogenous piRNAs. We also constructed a series of 5′ and 3′ end truncations down to 300 bp of both the flam and tj piRNA-binding elements so that each truncation progressively reduced the number of piRNAs predicted to target the reporter.
FIGURE 2.
Reporter gene silencing requires a bulk of PIWI/piRNA complexes. (A) Design of the piRNA reporter assay in OSS cells. (B) A representative assay result from C of a piRNA reporter showing strong silencing of the “Antisense” reporter compared with the “Sense” reporter. Although it appears there is less expression from the Antisense reporter which could be targeted by piRNAs, this Antisense reporter cannot be meaningfully compared with the different sequence composition of the Sense reporter. The siPIWI/siGFP normalization is critical because it shows only the Antisense reporter is derepressed upon PIWI knockdown. (C) (Left) Reporter genes bearing segments of flamenco elements inserted into the 3′ UTR of luciferase. (Middle) Graph of the amounts of endogenous piRNAs in OSS cells that target the Antisense reporters. Sense reporters have the same sequence as these piRNAs. Relative numbers of piRNAs were determined from early cultures of OSS cells used in these assays, based on reads per million (RPM) (Lau et al. 2009b). (Right) Graph of normalized ratios of luciferase expression between PIWI knockdown with siPIWI and the control siGFP siRNA. (D) Graphics and graphs analogous to C showing the effect on reporters bearing a segment of tj 3′ UTR. Reporter assay values depict ratio of each reporter transgene tested by transfecting OSS cells with PIWI knockdown (siPIWI) and normalized over control OSS cell (siGFP). Error bars represent standard error of mean of biological triplicates. Asterisks indicate statistical significance in the difference between the Antisense reporters compared with the Sense reporters (P < 0.05, Wilcoxon rank-sum test).
To demonstrate that our reporters were specifically silenced by piRNAs and not by other processes, we first nucleofected OSS cells with a PIWI-targeting siRNA (siPIWI) or a control (siGFP), and then 48 h later transfected the reporter plasmids (Fig. 2A). After confirming robust PIWI knockdown (Supplemental Fig. S2A), cells were harvested 2–3 d after reporter transfection and luciferase activity was measured. It was apparent that this reporter system was now responsive to piRNA targeting and PIWI-dependent gene silencing (Fig. 2B). Although the Antisense element reporter was expressed much lower than the Sense element reporter in siGFP-treated cells, this result likely reflected piRNA targeting but by itself was not rigorous enough to establish PIWI-dependent gene silencing because of different base sequence compositions between Antisense and Sense elements. Therefore, we normalized the ratio of luciferase activities for the same individual reporter between siGFP and siPIWI-treated cells, which then meaningfully revealed a PIWI-mediated regulatory effect and controls for each reporter having different mRNA stabilities from unique sequence compositions. These experiments were performed with an earlier isolate of OSS cells similar to what was described in Lau et al. (2009b), where small RNAs were deeply sequenced.
Both the full-length, “Antisense” flam and tj 3′-UTR reporters were robustly de-repressed upon PIWI knockdown when compared with the siGFP control, whereas “Sense” reporters barely fluctuated between the two siRNA treatments (Fig. 2C,D). The strongest, approximately fivefold derepression was observed with all Antisense flam reporters that could bind at least 3000 RPM of piRNAs, whereas reporters targeted by the ∼2000 RPM of piRNAs from the full tj 3′ UTRs were maximally derepressed at ∼2.5-fold. The series of truncations did not indicate whether one end of each piRNA-targeted element was more amenable to silencing than the other, but rather suggested that at least a minimum of ∼200 RPM of piRNAs was required to exhibit a cutoff of 1.5-fold derepression. When plotting the gene silencing capacity of each reporter element against the relative amount (RPMs) of piRNAs capable of targeting each reporter (Supplemental Fig. S2B), we could fit a hyperbolic curve to these data. This analysis implied that the number of targeting piRNAs was the main indicator of gene silencing capacity rather than a specific region or sequence in these two major piRNA generating elements. This also suggested that the most robust PIWI silencing of reporters required at least 2000 RPM of targeting piRNAs, a threshold that was consistent with our observations of endogenous TE transcript PIWI repression in OSS cells (Sytnikova et al. 2014). Since nearly all the Sense reporters were not significantly changed between siPIWI and siGFP conditions, this suggests that the derepression of Antisense reporters can be attributed to endogenous piRNAs pairing to the Antisense reporter transcript rather than to the plasmid DNA.
We examined the steady-state mRNA levels of the piRNA-targeted reporters to see if these mRNAs were being silenced through translation inhibition with minor mRNA destabilization, such as for mRNAs whose 3′ UTR is targeted by certain miRNAs (Doench and Sharp 2004; Eulalio et al. 2008; Djuranovic et al. 2012). Northern blots of the Renilla luciferase mRNA of Antisense reporters bearing either the full-length flam and tj 3′-UTR elements showed strong reduction in control siGFP cells but were derepressed to the same levels of the Sense reporter upon PIWI knockdown (Supplemental Fig. S2C,D). This indicated that reporter silencing was due to suppressed mRNA levels rather than translational regulation.
Assessing the regulation of endogenous PIWI-associated transcripts
Since our reporter assay platform could specifically determine PIWI-mediated regulation by both TE-directed and genic piRNAs, we deployed this system to examine OSS cell transcripts we had identified by PIWI Cross Linking Immuno Precipitation Deep Sequencing (CLIP-seq) and transcriptome profiling in the accompanying study (Sytnikova et al. 2014). For PIWI-associated genic transcripts, we tested elements from Akap200, larp, and Ef2b because these genes displayed the most significant PIWI CLIP-seq scores and displayed changes in cytoplasmic RNA levels after PIWI knockdown (Sytnikova et al. 2014). We cloned into the 3′ UTR of the luciferase reporters 300–400-bp long elements corresponding to the PIWI-associated regions of these genes. In a similar fashion to the flam and tj reporters, we tested the plasmids in siPIWI knockdown and siGFP control OSS cells (Supplemental Fig. S3), however, the reporters bearing these specific PIWI-associated transcript elements were not significantly affected by PIWI knockdown (Supplemental Fig. S3B). Although these transcripts were associated with PIWI as shown by CLIP-seq and further confirmed by RNA Immunoprecipitation (Sytnikova et al. 2014), no piRNAs could be mapped with antisense complementarity to these transcripts. Thus, the absence of gene expression change from these reporters bearing the PIWI-associated transcript elements is consistent with a requirement for a sufficient number of complementary-pairing piRNAs to trigger gene silencing.
We also constructed a set of reporters to test a genomic element that exemplifies a second group of genes which were strongly up-regulated but did not appear to directly interact with PIWI in CLIP-seq experiments (Sytnikova et al. 2014). We chose to test a de novo inserted 297 TE at the Mec2 locus because, like the other genes in OSS cells, the 297 TE was generating a novel nascent RNA being targeted by PIWI. We cloned the Mec2 promoter region that included the 297 TE into the 3′ UTR of luciferase in one set of plasmids; and in another set, downstream from the Poly-A termination signal of luciferase (Supplemental Fig. S3C), thus separating the 297 TE element from the luciferase transcript. When we tested these reporters in siGFP- and siPIWI-treated OSS cells, all of the 297-Mec2 segment reporters were derepressed upon PIWI knockdown in a statistically significant and reproducible manner (Supplemental Fig. S3D). Since both the sense-oriented elements and elements placed downstream from the luciferase Poly-A signal were derepressed, these results are consistent with an RT-PCR analysis showing that the 297-Mec2 elements can express a transcript independently of the luciferase transcript and plasmid promoter, and this transcript can then recruit PIWI silencing (Supplemental Fig. S3E). This contrasts with the flam reporter, which can only be silenced by PIWI when the flam element is oriented to be transcribed antisense to piRNAs. Compared with the flam reporter which is mainly comprised of Idefix sequences, the 297-Mec2 reporters have half as many predicted targeting piRNAs if the entire 297 was transcribed. However, the RT-PCR analysis also revealed a transcript pattern that was more complicated than we had expected, because this transcript only covered a portion of the 297-Mec2 segment (Supplemental Fig. S3E) and this could explain the lower than expected levels of derepression after PIWI knockdown. Additional TE segments will need to be tested in the future to fully understand the range of PIWI-targeting mechanisms.
PIWI-mediated TGS is robust and dependent on base-pairing to the nascent transcript
In OSS and OSC cells, PIWI operates at the nascent RNA level to silence endogenous TEs and genes adjacent to a transcriptionally active TE (Sienski et al. 2012; Sytnikova et al. 2014). To determine whether the PIWI-dependent regulation of our reporter transgenes was also due to TGS or PTGS, we inserted the flam element into the 5′ UTR of the luciferase reporter, either juxtaposed to the transcriptional start site (TSS) or within a small and efficiently spliced intron in the 5′ UTR that enhances the translation of the luciferase (Supplemental Fig. S4A). The piRNA-targeted elements were cloned either in the “Sense” or “Antisense” orientation, similar to the configuration of the 3′-UTR-based reporters. The piRNA-targeting elements juxtaposed to the TSS behaved similarly to the 3′-UTR-based reporters, where only the “Antisense” reporters were strongly derepressed upon PIWI knockdown (Supplemental Fig. S4B). This experiment served as a control for PIWI targeting being still dependent on piRNA binding to the element in the 5′ UTR. However, the significant extension of the 5′ UTR with these TSS-adjacent elements strongly dampened translation efficiency (Supplemental Fig. S4C). Since the transfection efficiency of OSC cells was also much less compared with OSS cells, we continued to test for PIWI TGS on reporters only in OSS cells (Supplemental Fig. S4D).
In addition, we moved the flam and the tj 3′-UTR elements into an intron of luciferase that is efficiently spliced out the piRNA-binding element. This configuration restored luciferase activity levels to the same level as when the elements were cloned in the luciferase 3′ UTR (Fig. 3A and Supplemental Fig. S4D). The efficiency of splicing was also confirmed by RT-PCR of mRNA (Fig. 3B), and we observed the “Antisense” intronic flam element reporter mRNA to be strongly repressed in the siGFP-treated cells and up-regulated in the siPIWI knockdown cells. Both the intronic flam-piRNA and tj 3′-UTR-piRNA-targeted Antisense elements were strongly derepressed upon PIWI knockdown (Fig. 3C). Since the spliced mature mRNAs would lack the piRNA-binding elements, these data support a model that PIWI and both genic and TE piRNAs are targeting the nascent transcript of Renilla luciferase even prior to splicing, and PIWI-mediated TGS can occur so robustly that even transiently transfected plasmid reporters can be efficiently silenced.
FIGURE 3.
PIWI/piRNA complexes robustly mediate TGS on transgenes. (A) Reporter constructs with piRNA-binding sequences placed into the intron within the 5′ UTR of the Renilla luciferase gene. (B) RT-PCR analysis of the reporter mRNAs from transfected OSS cells. Specificity of spliced transcripts was confirmed next to negative amplification controls and the plasmid DNA which contained the intron and piRNA-binding element sequence. (C) Reporter assay values depict ratio of each reporter tested by transfecting OSS cells with PIWI knockdown (siPIWI) and normalized over control OSS cell (siGFP). Error bars represent standard error of mean of biological triplicates. Asterisks indicate statistical significance in the difference between the Antisense reporters compared with the Sense reporters (P < 0.05, Wilcoxon rank-sum test).
The chromatin state of endogenous genes and reporter plasmids silenced by the PIWI/piRNA complex
To examine how the PIWI/piRNA complex mediates TGS, we deployed Chromatin ImmunoPrecipitation (ChIP) analysis of RNA Polymerase II (RNA Pol II) and Histone H3 lysine 9 tri-methylation (H3K9me3) enrichment at endogenous OSS cell genomic loci in addition to the transfected flamenco piRNA reporter (Fig. 4A). We observed expected enrichment of the silenced chromatin mark of H3K9me3 at Blood, an endogenous multicopy TE, and this mark decreased after PIWI knockdown. RNA Pol II was depleted from Blood loci and enriched on the promoter region of Actin5C, a constitutively expressed housekeeping gene locus. RNA Pol II was minimally present at the Mec2 locus because the de novo inserted 297 TE was triggering PIWI-mediated gene silencing (Sytnikova et al. 2014) and the enrichment of H3K9me3. However, RNA Pol II recruitment was increased at Mec2 after PIWI knockdown while the H3K9me3 levels were decreased, which is consistent with the TGS mechanism proposed for PIWI-mediated gene silencing (Wang and Elgin 2011; Sienski et al. 2012; Huang et al. 2013; Le Thomas et al. 2013; Rozhkov et al. 2013). These results confirm that our ChIP protocol is properly interrogating the chromatin marks on OSS cell loci.
FIGURE 4.
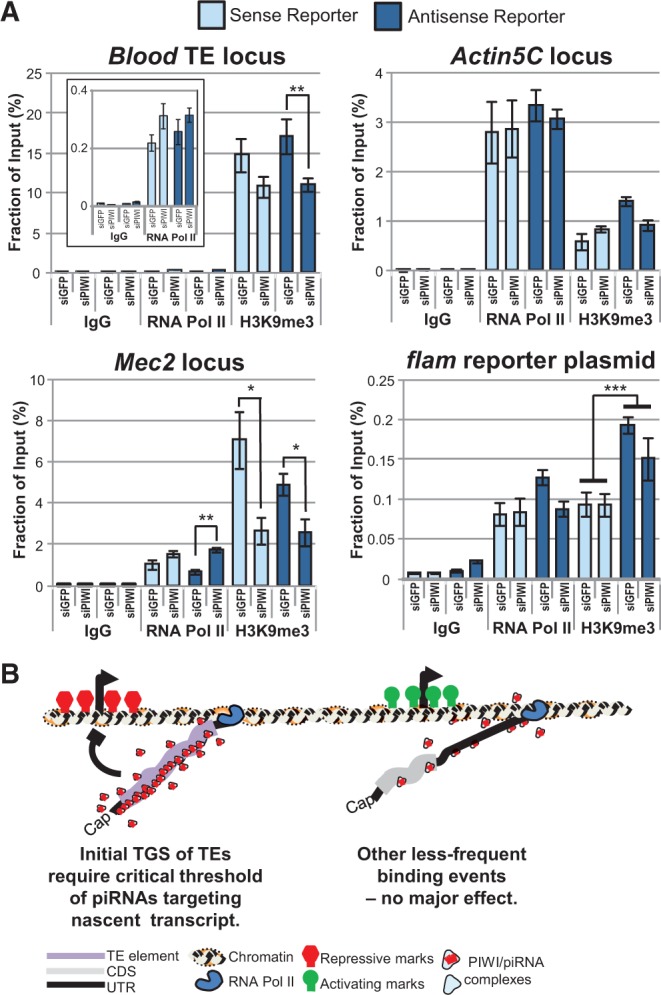
PIWI-mediated TGS operates through chromatin changes. (A) Chromatin immunoprecipitation (ChIP) assays measuring RNA Polymerase II (RNA Pol II) and Histone H3 lysine 9 tri-methylation (H3K9me3) enrichment at OSS cell genomic loci and the flamenco piRNA reporter. Blood is an endogenous TE that is epigenetically silenced, Actin5C is a constitutively expressed housekeeping gene locus, Mec2 is a locus with a de novo 297 TE and under PIWI-mediated silencing in OSS cells (Sytnikova et al. 2014), and the flam element 3′-UTR reporter constructs were transiently transfected plasmids. Error bars represent standard error of mean of biological triplicates. Wilcoxon rank-sum tests for certain comparisons of siPIWI versus siGFP conditions are represented by (***) P < 0.05, (**) P < 0.1, and (*) P < 0.2. The inset for the Blood ChIP is a different scale of the main figure to show the RNA Pol II levels. (B) A model that suggests the dominant mechanism for PIWI directed gene silencing is TGS and this mechanism requires a bulk of piRNAs interacting with the nascent transcript. Most protein-coding transcripts are unaffected by less-frequent PIWI/piRNA interactions.
Since the flam element 3′-UTR reporter constructs were transiently transfected as plasmids, we expected chromatin mark enrichment levels to be dampened by a bulk of plasmids being automatically silenced by other non-PIWI pathway heterochromatin silencing processes (Suzuki et al. 2006; Riu et al. 2007). Indeed, the relative quantitation of chromatin lysate inputs indicated there was 1024- and 32-fold more copies of plasmid DNA and Blood TE loci, respectively, when compared with Actin5C and Mec2 loci which were equivalent to each other (data not shown). Nevertheless, our ChIP analysis managed to detect RNA Pol II and H3K9me3 on both the Sense and Antisense flam reporters (Fig. 4A, lower right panel). Since only the flam Antisense reporters were sensitive to PIWI/piRNA targeting, we expected to detect greater H3K9me3 levels on the Antisense reporter compared with the Sense reporter, and H3K9me3 levels only decreased on the Antisense reporter upon PIWI knockdown. However, we also observed an Antisense-reporter specific decrease in RNA Pol II levels upon PIWI knockdown, which contrasts with the increased levels of reporter transcripts and luciferase expression of the reporter as well as the RNA Poll II increase at Mec2 following PIWI knockdown. We can conclude that some proportions of plasmid reporters are properly incorporated with facultative chromatin to allow enough nascent RNA transcription of the flam piRNA element for the recruitment PIWI/piRNA complex to initiate TGS.
DISCUSSION
The extreme diversity of piRNAs, i.e., in Drosophila follicle and OSS cells, creates a target prediction challenge because most piRNAs are sequenced with much lower frequency compared with miRNAs (Girard et al. 2006; Lau et al. 2006, 2009a,b; Brennecke et al. 2007). Therefore, it is not straightforward to choose individual “abundant” piRNAs to carry out target predictions. When we used a mainstay approach to validate miRNA target regulation—reporters with individual target sites—for a subset of the most-abundant single piRNAs, we did not observe a robust gene silencing effect. The difference in gene silencing mechanisms between AGO1/miRNA complexes and PIWI/piRNA complexes was further emphasized by a tethered reporter assay that bypasses the requirement for small RNA-mediated target binding. This is consistent with the literature indicating that, in contrast to Ago proteins, Piwi proteins do not interact with the translation regulating GW182 proteins (Miyoshi et al. 2009).
Genetically, PIWI affects the levels of chromatin marks like H3K9Me3 and Heterochromatin Protein 1 (HP1) deposition (Pal-Bhadra et al. 2004; Brower-Toland et al. 2007; Klenov et al. 2007; Yin and Lin 2007; Wang and Elgin 2011). Several recent genome-wide studies examining mutants and genetic perturbations of the Piwi pathway in flies have confirmed the transcriptional silencing mechanism of TEs by PIWI via a transcription-repressive chromatin state (Sienski et al. 2012; Huang et al. 2013; Le Thomas et al. 2013; Ohtani et al. 2013; Rozhkov et al. 2013). Here, we present a reporter assay platform that shows a dependence on PIWI and piRNA complementarity for triggering robust gene silencing, and the silencing capacity appeared to be present in both TE-directed piRNAs as well as genic piRNAs. Whereas, TEs are obviously the targets of TE-directed piRNAs, the targets of genic piRNAs are still mysterious because we have not been able to determine particular up-regulated genes that can pair to a large enough bulk of genic piRNAs. Our data in OSS cells indicates that the robust TE-repression by PIWI and piRNAs requires base-pairing to the nascent transcript rather than the DNA, although it is possible that upon extended time periods during fly development the PIWI/piRNA complex may also directly interact with DNA.
From our data, we propose a model (Fig. 4B) that can explain how the tremendous diversity of different piRNAs primarily direct PIWI complexes to silence TE transcripts without affecting most other protein-coding transcripts. Since the largest proportions of Drosophila piRNAs are complementary to TEs, only this bulk of piRNA interactions against TEs is large enough to manifest direct target regulation. This mechanism depends upon piRNAs interacting with the nascent transcript, which can result in chromatin changes, but we do not rule out a role for PIWI in stimulating RNA turning over. Due to the limitations of plasmid transfections, we were unable to show that a Slicer-defective PIWI transgene could rescue reporter gene silencing after PIWI knockdown in OSS cells (data not shown). However, others have previously shown that Drosophila PIWI deficient in Slicer activity can rescue TE silencing after PIWI knockdown in OSCs (Saito et al. 2010) and rescue piwi mutant sterility (Darricarrere et al. 2013). This contrasts with mouse MIWI and MILI proteins that do require Slicer activity for TE silencing (De Fazio et al. 2011; Reuter et al. 2011).
With regard to other non-TE transcripts, we propose that limited interactions between PIWI and protein-coding transcripts, such as mismatches between transcripts and piRNAs, are insufficient to manifest strong silencing. Nevertheless, Piwi proteins may also have promiscuous RNA binding activity (Vourekas et al. 2012), including association to novel long noncoding RNAs expressed in gonadal cells (Sytnikova et al. 2014), which may allow Piwi proteins to accommodate the broad sequence diversity of piRNA precursors.
Our work adds a novel vantage point that complements the other recent studies: The PIWI/piRNA complex is versatile in mediating TGS on many loci in the nucleus. Future efforts with this system will enable better understanding of how the PIWI silencing mechanism operates on these transiently transfected piRNA reporter plasmids which are chromatinized but unlikely to be integrated. In 48 h after transfection, a TGS-like repression is established that includes H3K9me3 changes, but the decrease of RNA Pol II occupancy after PIWI knockdown is perplexing. What is unknown is whether PIWI-mediated silencing blocks transcription initiation or whether it may be similar to the nuclear RNAi (Nrde) pathway in Caenorhabditis elegans, which actually only impacts downstream RNA Pol II occupancy and transcriptional elongation (Guang et al. 2010; Burton et al. 2011). In this vein, our reporter system presents an excellent platform for future studies to examine the kinetics and mechanisms of de novo establishment of the TGS state by the PIWI/piRNA machinery. This next pursuit will allow us to address how the PIWI/piRNA complex directs chromatin repressive marks to target loci and how it affects RNA Polymerase II recruitment and elongation.
MATERIALS AND METHODS
OSS cells cultivation
OSS cells were cultured according to Niki (2009) in M3 media supplemented with 10% heat inactivated FBS, 1× fly extract, 10 µg/mL insulin, 0.6 mg/mL glutathione. For PIWI depletions, 500 pmol siRNAs were electroporated into a 10-cm plate of OSS cells using Amaxa kit V (program D013), and cells were cultured for 4 d before harvesting.
Luciferase reporter assays and reporter gene cloning
The siRNAs siGFP and siPiwi are according to Saito et al. (2009) and were nucleofected into OSS cells with the Amaxa and the V-kit with program D013. Reporter vectors were transfected 48 h after siRNA electroporation using FugeneHD (total 10 µg per 10 cm dish, equal amount of Firefly and Renilla reporter vectors). Seventy-two hours after reporter transfection cells were lysed, Renilla and Firefly luciferases activities were measured using the Dual Luciferase Assay Kit (Promega). The Renilla signal was normalized against Firefly. The Renilla and Firefly luciferase reporters were based on the reporter constructs used in Marr et al. (2012). The oligonucleotides used for amplifying the flamenco and tj 3′-UTR elements are listed in Supplemental Table S1 and genomic PCR fragments containing ApaI sites were cloned into pFREN-0 (the Renilla luciferase reporter driven by a mini-actin promoter).
RNA purification and Northern/RT-PCR analysis
Total RNA was extracted from OSS cells and fractions by TRI-reagent RT (MRC, Inc.). Remaining DNA was digested by 6U of DNaseI (NEB). To assess RNA expression level change reverse transcription was performed using 0.2 µg of RNA and M-MLV reverse transcriptase (Promega). For TE elements evaluation cDNA synthesis was performed by random primer and for genic sequences (dT)18 oligo was used. qPCR was performed using Go-Taq SybrGreen Master mix (Promega) on Bio-Rad C1000 machine. Oligonucleotides used for specific sequences amplification are listed in Supplemental Table S4. Relative changes in gene expression were calculated using the 2ΔΔCt method (Winer et al. 1999). RpL32/Rp49 mRNA was used for normalization of gene expression.
Western blots and antibodies
SDS-PAGE (SDS-polyacrylamide gel electrophoresis) and Western blotting were performed according to standard protocols. Rabbit polyclonal antibody to PIWI was raised to the synthetic peptide that corresponds to N-terminus (CADDQGRGRRRPLNEDD). Immunizations and bleedings were performed by Cocalico Biologicals Inc., antibody was purified using the SulfoLink kit (Pierce). Mouse anti-α-Tubulin (E7A) and anti-HA antibodies were purchased from the Developmental Studies Hybridoma Bank and Pierce, respectively.
Chromatin immunoprecipitation
ChIP was performed as previously described (Rozhkov et al. 2013) with the following modifications. Fully confluent 10-cm dishes of OSS cells were washed with 1× D-PBS, and crosslinked in 1.6% formaldehyde for 10 min at room temperature. Fixing was quenched for 5 min by addition of glycine to 125 mM, final concentration. Fixed cells were collected with rubber policeman, and pelleted. Cells were resuspended in lysis buffer (Rozhkov et al. 2013), and lysed on ice for 10 min. Cell lysates were then sonicated with Q125 Sonicator (QSonica) 30′′ ON/OFF cycle for 10 min to produce DNA fragments from 100 to 500 bp. Sheared chromatin was then cleared and stored at −20°C. For immunoprecipitation, chromatin aliquots were diluted 20-fold in chromatin dilution buffer (16.7 mM Tris–HCl at pH 8, 1.2 mM EDTA, 167 mM NaCl, 0.5% Triton X-100) and precleared with 10 μL of Protein A magnetic beads (Millipore #16-661) incubated in blocking buffer (0.5% Tween-20, 5 mg/mL BSA in PBS) for 1 h at 4°C. Precleared beads were removed, and replaced with antibody-conjugated beads. Reactions were incubated overnight at 4°C. Beads were then washed sequentially with the following: 6× with RIPA buffer (10 mM Tris–HCl at pH 8, 1 mM EDTA, 140 mM NaCl, 0.5% Triton X-100, 0.1% Deoxycholate), 2× with RIPA/500 buffer (RIPA supplemented with 500 mM NaCl), 2× with LiCl buffer (10 mM Tris–HCl at pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate), and 2× with TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA). To reverse crosslinking, beads were resuspended in Elution buffer (10 mM Tris–HCl at pH 8, 5 mM EDTA, 300 mM NaCl, 0.1% SDS), and incubated at 65°C overnight. Reactions were then incubated at 37°C sequentially with 10U RNase A and Proteinase K. Reactions were then phenol/chloroform extracted, ethanol precipitated and resuspended with dH2O. qPCR was then performed using primer pairs specified in Supplemental Table S1, including primers from Wang and Elgin (2011).
Mapping of piRNAs to TEs and mRNAs
The OSS cell piRNA reads from Lau et al. (2009b) were mapped with up to four mismatches using Bowtie2 (Langmead and Salzberg 2012) against the exons of a RefSeq transcript gene list from the Drosophila Dm3 assembly, and to the list of Drosophila consensus TE sequences obtained from the RepBase database (Kapitonov and Jurka 2008) and from FlyBase (Kaminker et al. 2002).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Arlo Greenleaf's and Karen Adelman's laboratory for providing antibodies to RNA Pol II and Michael Marr and Karen Adelman for advice on ChIP. We also thank Rachel Green's laboratory for the constructs for the tethered reporter assay, and the Hedstrom and Lovett laboratories for access to the luminometer and qPCR machine. We also thank Reazur Rahman for calculating the piRNA reads mapping to the Mec2 promoter, and Avital Rodal, Michael Marr, and Michael Rosbash for additional advice and comments on this manuscript. This work was supported by the National Institutes of Health (Core Facilities Grant P30NS045713 to the Brandeis Biology Department, T32GM007122 to J.P.C. and R00HD057298 to N.C.L.). Additional support was from the Searle Scholars program (N.C.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.046300.114.
REFERENCES
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol 24: 6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Sheridan R, Marks DS, Sander C 2007. Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol 3: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A 1995. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet 11: 349–353. [DOI] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S 2011. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci 108: 19683–19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A 2008. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci 105: 14964–14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrere N, Liu N, Watanabe T, Lin H 2013. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci 110: 1297–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Zinchenko MK, Hur JK, Nahvi A, Brunelle JL, Rogers EJ, Green R 2010. Allosteric regulation of Argonaute proteins by miRNAs. Nat Struct Mol Biol 17: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R 2011. A parsimonious model for gene regulation by miRNAs. Science 331: 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA 2004. Specificity of microRNA target selection in translational repression. Genes Dev 18: 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, Chambeyron S 2012. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res 22: 1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H 2006. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H 2011. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 45: 447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA 2005. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res 33: 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J 2008. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet 9: 411–412; author reply 414. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA 2007. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367. [DOI] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD 2009a. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J 28: 2945–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC 2009b. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr SK, Pennington KL, Marr MT 2012. Efficient metal-specific transcription activation by Drosophila MTF-1 requires conserved cysteine residues in the carboxy-terminal domain. Biochim Biophys Acta 1819: 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A 2007. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Okada TN, Siomi H, Siomi MC 2009. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y 2009. Culturing ovarian somatic and germline stem cells of Drosophila. Curr Protoc Stem Cell Biol Chapter 2: Unit 2E 1. [DOI] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K 2013. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Riu E, Chen ZY, Xu H, He CY, Kay MA 2007. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther 15: 1348–1355. [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC 2009. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr Biol 19: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kasai K, Saeki Y 2006. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol 80: 3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnikova YA, Rahman R, Chirn GW, Post C, Clark JP, Lau NC 2014. Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res 10.1101/gr.178129.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM 1999. Development and validation of real-time quantitative reverse transcriptase–polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270: 41–49. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





