Abstract
Bacteria inhabit enormously diverse niches and have a correspondingly large array of regulatory mechanisms to adapt to often inhospitable and variable environments. The stringent response allows bacteria to quickly reprogram transcription in response to changes in nutrient availability. Although the proteins controlling this response are conserved in almost all bacterial species, recent work has illuminated considerable diversity in the starvation cues and regulatory mechanisms that activate stringent signaling proteins in bacteria from different environments. In this review we describe the signals and genetic circuitries that control the stringent signaling systems of a copiotroph, a bacteriovore, an oligotroph and a mammalian pathogen – Escherichia coli, Myxococcus xanthus, Caulobacter crescentus and Mycobacterium tuberculosis, respectively – and discuss how control of the stringent response in these species is adapted to their particular lifestyles.
Keywords: stringent response, niche, ppGpp, RelA, SpoT, stress response
The stringent response mediates bacterial adaptation to environmental stresses
The stringent response (SR) is a broadly conserved bacterial stress response that controls adaptation to nutrient deprivation, and is activated by a number of different starvation and stress signals. The molecular hallmark of this response is synthesis of the small molecules guanosine 5’-diphosphate 3’-diphosphate (ppGpp) and guanosine 5’-triphosphate 3’-diphosphate (pppGpp) – together denoted (p)ppGpp - by RSH (Rel/Spo homolog) and SAS (small alarmone synthetase) proteins [1-3]. (p)ppGpp serves as a second messenger signal of the initial starvation or stress cue, and effects large-scale transcriptional change by binding directly to RNA polymerase in Gram-negative species [4, 5] and by altering the ratio of iNTPs in B. subtilis and likely other Gram-positive species [6, 7]. In general, (p)ppGpp downregulates transcription of genes encoding translational machinery and factors required for growth and division, and upregulates stress response genes [1, 2]. In addition to transcription, (p)ppGpp can also directly affect chromosome replication [8, 9] and alter the activity of certain enzymes that are involved in stress response [10, 11].
The SR was first described in Escherichia coli. However, RSH proteins exist in almost all species of bacteria, with the notable exception of some obligate intracellular pathogens and obligate symbionts [12, 13] which inhabit static microenvironments. Studies of SR in species from a range of environmental niches have demonstrated great diversity in the signals and conditions that activate (p)ppGpp synthesis (i.e. upstream signaling). There is also considerable variability in the means by which ppGpp alters cellular physiology in response to environmental change (i.e. downstream signaling), which is addressed in references [1, 2, 14-17].
RSH proteins are the most broadly conserved (p)ppGpp synthetases, and appear to be the primary enzymes responsible for (p)ppGpp accumulation during starvation. These proteins can be regulated on multiple levels: transcriptionally, e.g., see the section below on Mycobacterium tuberculosis (Mtb); post-translationally via sensing the levels of available amino acids; and through direct protein–protein interactions, e.g. see the section below on E. coli. In many species, the cytosolic concentration of (p)ppGpp is additionally controlled by small alarmone synthetase (SAS) proteins, which consist only of a (p)ppGpp synthetase domain, and small alarmone hydrolase (SAH) proteins which contain only a hydrolase domain [12] (Figure 1). While SAH proteins have not been studied in bacteria, SAS proteins have been characterized in Bacillus subtilis, Streptococcus mutans, Enterococcus faecalis and Vibrio cholerae [18-21]. The SAS proteins studied in Gram-positive species produce low levels of (p)ppGpp during log phase growth and higher levels during specific stress conditions such as alkaline stress [21] and inhibition of select amino acyl-tRNA synthetases [20]. The V. cholerae SAS protein produces (p)ppGpp during carbon and fatty acid starvation but not amino acid starvation [19]. SAS and SAH proteins are likely responsible for modulating (p)ppGpp levels under a variety of stress conditions and, like RSH proteins, are probably regulated transcriptionally [20] and post-translationally.
Figure 1. Domain structure of RSH proteins.
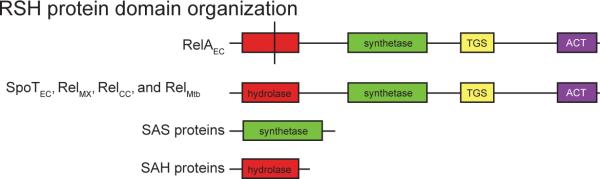
All RSH proteins contain – from N to C terminus – (p)ppGpp hydrolase, (p)ppGpp synthetase, TGS (conserved in Threonyl-tRNA synthetases, GTPases and SpoT ) and ACT domains (conserved in many proteins involved in small molecule metabolism; the ACT domain usually binds an amino acid or small molecule that allosterically regulates enzyme activity) [80]. The hydrolase domain of RelAEC contains sequence polymorphisms (indicated by vertical black line) that render it enzymatically inactive. We note there are several sites of sequence variation that distinguish RelAEC, SpoTEC and bifunctional Rel proteins besides the active sites of the catalytic domains; these are discussed in reference [12].
In this review, we describe the diversity in upstream sensory events and in the genetic circuitries that control (p)ppGpp levels in the cell. We further discuss how the environmental niche occupied by a bacterium shapes the structure and logic of this upstream SR signaling, thus ensuring a starvation response that is tailored to a particular niche. Our review focuses specifically on four environmentally-distinct bacterial species: (i) the enteric commensal, E. coli; (ii) the soil-dwelling bacteriovore, Myxococcus xanthus; (iii) the freshwater oligotroph, Caulobacter crescentus; and (iv) the mammalian pathogen, Mtb.
The copiotroph E. coli
The observation that stable RNA synthesis in E. coli is restricted upon amino acid starvation [22] was among the first molecular-level regulatory phenomena described in bacteria. Early genetic analyses [23] identified a specific chromosomal lesion in an E. coli mutant strain [24] that was known to accumulate RNA even when starved for amino acids. This mutation was reported to “relax the stringency” of amino acid control on RNA synthesis [23], and was later determined to disrupt the relA gene, whose product synthesizes (p)ppGpp [25, 26]. Decades of subsequent studies have shown that this regulatory system responds to diverse types of starvation, and functions to modulate DNA replication [8, 27, 28], translation [29], acid stress enzyme activity [11], and remodel the transcription of a large proportion of the genome [30-32].
E. coli encodes two paralogous RSH enzymes, RelAEC and SpoTEC (Figure 1 and Box 1). SpoT both synthesizes and hydrolyzes (p)ppGpp [33, 34]; RelAEC is only capable of synthesis. RelAEC associates with the 70S ribosome [35] in nutrient-replete conditions [36]. Upon amino acid starvation, uncharged tRNAs accumulate, enter the ribosomal A site and stall translation. This results in RelA dissociation from the ribosome [36] and initiation of (p)ppGpp production [37]. Thus, RelAEC is sensitive to starvation for any single amino acid. SpoTEC hydrolase activity is repressed when bound to free uncharged tRNAs (i.e. not associated with the ribosome) [38], and through association with the G-protein CgtA [39, 40]. Unlike RelAEC, SpoTEC does not synthesize (p)ppGpp in response to starvation for any single amino acid, but in response to shortages of multiple amino acids; the mechanism controlling this response is unknown [41]. Lipid starvation directly activates (p)ppGpp synthesis by SpoTEC [42]. Holo-acyl carrier protein (Acp-4′-phosphopantetheine), which shuttles growing lipid chains during fatty acid biosynthesis, binds to SpoTEC and induces (p)ppGpp synthesis [43]. The SpoTEC activating signal may be the lipid-free form of holo-Acp [43, 44]. SpoTEC also catalyzes (p)ppGpp accumulation in response to phosphate [45] and iron [46] starvation through unknown mechanisms (Figure 1A).
RelAEC, SpoTEC and other bacterial RSH enzymes contain (p)ppGpp synthetase and hydrolase domains within their N-terminal half. Two conserved domains, known as ACT and TGS, positioned C-terminal to the enzymatic domains, are important for regulation of (p)ppGpp levels in the cell [47, 48] (Figure 1). The TGS domain is required for interaction between holo-Acp and SpoTEC [43]. The ACT domain in RelAEC is required for detection of amino acid starvation via ribosomal protein L11 [48]. Between RelAEC and SpoTEC, E. coli upregulates production of (p)ppGpp in response to starvation for amino acids, carbon, nitrogen, fatty acids, phosphate and iron [2].
Studies on E. coli have determined the signaling inputs, transcriptional and other outputs, and many of the mechanistic details of stringent response activation and regulation. While some results from E. coli studies appear to be general, it is increasingly clear that the stringent response systems of many other species have divergent signaling inputs and mechanisms of regulation. It is probably best to consider the stringent response system in E. coli as an adaptation to its specific nutritional lifestyle, which may involve abrupt transitions from the mammalian gut to more nutrient-limited aquatic and soil environments [49] (Figure 2A). This system - in which (p)ppGpp accumulates quickly upon starvation for any single nutrient - causes rapid cessation of cell growth, activation of the stress response, and metabolic slowing [30-32]: adjustments for what is likely to be a life-long stay in a nutrient-limited environment [49].
Figure 2. Lifestyle and stringent response (SR) signaling in four bacteria.
Key events in SR activation signaling are diagrammed on the left; lifestyle transitions that affect SR signaling are diagrammed on the right. Arrowheads indicate positive regulation, perpendicular lines indicate negative regulation. A) In Escherichia coli, several starvation signals can individually activate the SR via either RelAEC or SpoTEC. RelAEC is activated at the ribosome when translation is halted due to the entry of an uncharged tRNA in the A site. SpoTEC is activated in lipid starvation through interactions with holo-acyl-carrier protein (Acp) and by several different starvation signals via unknown mechanisms. E. coli transitions from the nutrient-rich mammalian colon to nutrient-poor soil or aquatic environments; though it may also experience smaller variations in nutrient availability within these environments. In nutrient-rich conditions, cells are large and contain multiple replication forks, in nutrient-poor conditions cells are smaller and contain either one or zero replication forks. B) Myxococcus xanthus encodes a signal override system, involving both positive (CsgA) and negative (SocE) feedback loops, in its SR activation pathway that allows information about cell density and developmental progression to be integrated with information about nutrient availability. In the developmental progression of M. xanthus, vegetatively growing cells exhaust nutrients, activate the stringent response and A-signaling; cells then move in waves which coalesce into mounds that develop into mature, spore-forming fruiting bodies. (p)ppGpp levels remain high for the period indicated by the blue arrow, the green region indicates the time when the signal override is in effect. A-signaling is active during the period indicated by the purple arrow. C) The SR of Caulobacter crescentus has a higher threshold for activation than many other species, requiring both starvation for amino acids AND additional carbon or ammonium starvation signals. The amino acid starvation signal is transmitted through the ribosome, much like RelAEC. Information about the nutritional and developmental status of the cell are integrated at multiple levels: (p)ppGpp accumulation has a greater inhibitory effect on the swarmer-to-stalked transition than on the division of the stalked (purple) cell. In addition the swarmer cell (green) is more sensitive to starvation, producing higher levels of ppGpp than the stalked cell. Heavier lines indicate stronger regulation. D) Expression of Rel in mycobacteria is controlled through a complex signaling cascade that depends upon the amount of polyphosphate present in a cell, and results in populations of cells that have bistable expression of Rel. Cells that express high levels of Rel are thought to be biased toward a persister state (red), exhibiting slow growth and greater resistance to antibiotics and other stresses. Host-generated stresses that occur during infection likely fail to kill persister cells, which could then transition into vegetatively growing cells (blue) once stresses are alleviated.
The presence of two RSH genes, which are regulated by different starvation and stress signals, may simply function to expand the repertoire of signals to which this species can respond. This arrangement of RSH genes is found in most γ- and β-proteobacteria with diverse lifestyles, so it does not seem that having two RSH genes is necessarily an adaptation to the particular copiotrophic lifestyle of E. coli. We propose that γ- and β-proteobacteria in different niches likely respond to different subsets of starvation signals.
The bacteriovore M. xanthus
M. xanthus is a δ-protobacterium that lives in soil, feeds upon other bacteria and forms multicellular, spore-forming fruiting bodies upon nutrient exhaustion. M. xanthus gradually depletes available nutrients locally, rather than being abruptly excreted into a nutrient-limited environment. The RSH protein of M. xanthus (RelMX), which is predicted to be bifunctional [50] (Figure 1), synthesizes (p)ppGpp upon amino acid [51], glucose and ammonium starvation, and is tetracycline-sensitive and therefore presumed to be ribosome-dependent [52]. relMX mutants are unable to aggregate into fruiting bodies and to form spores upon nutrient exhaustion when grown in monoculture on simple growth medium [51]. The developmental process in M. xanthus involves two intercellular signaling programs that depend upon (p)ppGpp: A-signaling which responds to cell density, and C-signaling which coordinates aggregation later in development [53, 54].
There are two necessary conditions to initiate M. xanthus spore development: nutrient limitation and high cell density [55]. RelMX detects limitation of amino acids, carbon, nitrogen or phosphate [51, 52, 56]. (p)ppGpp activates the asg (A-signal generating) genes [51] that control expression or export of extracellular proteases that degrade extracellular proteins to release A-signal: a mixture of amino acids and peptides at concentrations above 10 μM [55, 57] (Figure 2B). At low cell density, A-signal remains below 10 μM, and the genes required for fruiting body development and sporulation are not expressed [55]. Once sufficient (p)ppGpp and A-signal have accumulated, the sporulation developmental program can continue (Figure 2B). However, (p)ppGpp signaling and A-signaling are ultimately in conflict: amino acids that accumulate as A-signal can serve as a nutrient source, which could suppress (p)ppGpp synthesis, halt the development of spores, and restore vegetative growth.
To resolve this signaling conflict, M. xanthus encodes a RelMX signal override system that maintains high (p)ppGpp levels throughout spore development despite the rise in available amino acids during A-signaling. (p)ppGpp synthesis is suppressed by SocE and activated by CsgA in the absence of SocE (Figure 2B). CsgA ensures elevated (p)ppGpp levels throughout development, so that available nutrients are used to complete sporulation and not encourage vegetative growth [58]. csgA transcription is activated by (p)ppGpp, forming a positive feedback loop that allows for continued (p)ppGpp synthesis even when nutrient levels temporarily rise [59] (Figure 2B). socE transcription is repressed by (p)ppGpp such that SocE levels are low during sporulation, permitting continued (p)ppGpp synthesis [58, 59].
It is not yet clear how SocE and CsgA regulate RelMX activity. These proteins may bind the RelMX protein itself and modulate its activity directly. Alternatively, SocE and CsgA may modulate expression of RelMX. (p)ppGpp levels in M. xanthus appear to be sensitive to the concentration of RelMX: overexpression of RelMX alone results in increased steady-state levels of (p)ppGpp [51], and a strain with reduced relMX transcription has lower levels of (p)ppGpp and corresponding defects in spore development [60].
Studies of the M. xanthus SR have revealed a complex override system in which the signals and signaling proteins that control (p)ppGpp synthesis by RelMX are regulated as a function of the developmental state of the bacterium. In this way, the SR signaling system has been ’wired’ to suit the developmentally-complex lifestyle of this spore-forming bacterium. We hope that future work will elucidate the molecular mechanism by which the M. xanthus regulatory proteins SocE and CsgA function to modulate SR signaling in this species.
The freshwater oligotoph C. crescentus
C. crescentus is an α-proteobacterium that inhabits oligotrophic (i.e., nutrient-poor) freshwater. This species experiences nearly constant nutrient deprivation, and has a number of adaptations that promote survival in its oligotrophic niche including (i) a membranous stalk and adhesive holdfast, which facilitate attachment to available nutrient sources; (ii) a dimorphic life cycle in which nutrient-seeking and cell growth functions are separated into different cell types; and (iii) a broad array of environmental sensing and regulatory systems. The sensory system that has the most profound effect on gene expression is the SR. We now know that the C. crescentus SR system is adapted to oligotrophy in both the signals it detects, and in the way it modulates cell development [28, 61-64].
C. crescentus encodes a single, bifunctional RSH protein [28] which we refer to as RelCC in this review. Although RelCC binds the ribosome and its activity is tetracycline-sensitive [61], it does not synthesize (p)ppGpp in response to starvation for any single amino acid or upon deprivation of multiple amino acids [61, 65]. However, (p)ppGpp synthesis by RelCC is activated upon general carbon or nitrogen starvation. Given that both carbon and nitrogen starvation result in amino acid limitation, and that ribosome poisons interfere with RelCC activity, it was predicted that amino acid starvation was a necessary, but insufficient signal for SR activation in C. crescentus. Analysis of (p)ppGpp accumulation in the presence of tetracycline and in a glucose auxotroph strain determined that full activation of the C. crescentus SR requires both amino acid starvation and an additional carbon or nitrogen starvation signal [61]. One activating signal is most likely an uncharged tRNA in the A site of the ribosome, as is the case for RelAEC. The second signal is one of two unknown RelCC activating signals elicited by either carbon or nitrogen starvation (Figure 2C). RelCC therefore functions as an AND logic gate that requires two activating signals. This stands in contrast to the stringent response of E. coli, which functions as an OR logic gate that can be activated by several single starvation signals.
The dimorphic cell cycle of C. crescentus is an adaptation to oligotrophy. This cell cycle program separates nutrient seeking and cell division functions into two distinct cell types. A chemotactic, but non-reproductive swarmer cell transitions irreversibly into an amotile and reproductive stalked cell. The amotile stalked cell then spawns a daughter swarmer cell (Figure 2C). Nutrient limitation biases the C. crescentus cell cycle to a longer swarmer cell lifetime [62, 64], which increases the probability of a swarmer cell finding a more optimal environment before differentiating into a reproductive stalked cell. This nutrient-dependent effect on developmental timing is mediated in part by the effect of (p)ppGpp on the expression and stability of regulatory proteins that control differentiation of the swarmer cell into a stalked cell [28, 62]. The stringent response and development are further integrated at the level of RelCC activity: swarmer cells accumulate higher levels of (p)ppGpp than stalked cells [62] (Figure 2C). The mechanism controlling this differential sensitivity to starvation is unknown, but it seems likely that bias in the rate of (p)ppGpp accumulation upon starvation is an important determinant of the different responses of the two C. crescentus cell types to nutrient limitation.
RelCC is capable of integrating information from a range of environmental signals including amino acid and carbon or nitrogen availability, as well as signals informing the developmental state of the cell. The logic of C. crescentus SR activation and the differential (p)ppGpp accumulation in swarmer versus stalked cells are likely adaptations to the oligotrophic niche of C. crescentus. This species grows and divides in environments that would likely not sustain prolonged growth of copiotrophs such as E. coli. AND-type control logic of the C. crescentus SR likely ensures that this species can continue slow growth even if one class of nutrient is temporarily limiting.
The mammalian pathogen Mtb
Mtb is a pathogenic actinomycete that causes tuberculosis. This species can remain dormant in granulomatous lesions in healthy lungs for years or decades, only to revive and cause disease in a subset of patients. The extraordinary ability of Mtb to survive for long periods in the oxygen-and nutrient-limited granuloma is facilitated by the SR [66, 67] and possibly also by entry of cells into what is known as a ‘persister’ state - wherein a subset of cells in a population transition into a phenotypically distinct state characterized by growth stasis and antibiotic tolerance.
The SR in Mtb is controlled by a single, bifunctional RSH enzyme, RelMtb. Mycobacteria accumulate (p)ppGpp in carbon and total starvation, in the presence of the respiration inhibitor sodium azide [68], and in response to the valine synthetase inhibitor D,L-norvaline [69]. Phosphate starvation [70], hypoxia, and activation of the alternative sigma factor, σE [71], increase relMtbtranscription, which is a means of upregulating (p)ppGpp synthesis. In addition, polyphosphate – long chains of inorganic orthophosphate – is involved in SR activation in mycobacteria. This stands in constrast to several other species, where polyphosphate levels are regulated downstream of (p)ppGpp [10, 61, 62].
In the related model mycobacterium, M. smegmatis, polyphosphate is synthesized by the enzyme polyphosphate kinase (Ppk); ppk null strains have reduced survival under starvation and hypoxia [71]. The survival defect of a Δppk strain is likely determined in part by an indirect effect of Ppk on relMS transcription. Specifically, polyphosphate functions as a phosphodonor to the stress-responsive sensor histidine kinase, MprB, which transfers its phosphoryl group to the response regulator, MprA. MprA~P activates transcription of sigE, which activates transcription of relMS (Fig. 2D) [71]. Thus, sigE and relMS expression are indirectly responsive to polyphosphate levels, which increase under stress conditions. Genetic disruption of the gene encoding exopolyphosphatase (ppx), which catalyzes the breakdown of polyphosphate, results in high levels of polyphosphate and elevated expression of sigE and relMS; this is correlated with slowed growth and increased tolerance to the antibiotic isoniazid [72], phenotypes that are hallmarks of persister cells. Thus, there is evidence that polyphosphate is an upstream determinant of mycobacterial persistence phenotypes that functions through Rel.
The transcriptional network that controls relMS expression via polyphosphate has positive feedback topology, in which MprA~P activates its own transcription (Figure 2D). This contributes to a bistable distribution in relMS expression among M. smegmatis cells, in which cells either exhibit high or low expression [73]. Stochasticity in ppk expression contributes to the maintenance of two populations of cells: those that express high levels of relMS and are thought to be more likely to exhibit a persister cell phenotype, and those with low relMS expression that are likely biased toward growth [73].
Mycobacteria have no flagella, limited motility and are apparently incapable of directed movement up or down nutrient gradients. Instead of spatially sampling their environment via chemotaxis, subpopulations of mycobacteria cells temporally sample their environment by entering a dormant state during stress. This strategy is aided when a population of cells can exist in at least two distinct physiological states. In one subpopulation cells are primed for growth; a second subpopulation exists in a growth-arrested and stress- and antibiotic-resistant state known as persistance. A complex signaling circuit causing bistable rel expression of may link the SR to the control of persister cells within the population, thereby facilitating Mtb adaptation to an inhospitable host niche.
Concluding remarks
While this review primarily focuses on variation in upstream control features of SR signaling between four species, it is important to note that we expect many aspects of the SR are broadly conserved among bacteria. The mechanism by which RelAEC detects amino acid starvation through ribosome stalling is apparently similar in most species with RSH proteins, though the signaling logic may vary, as described for C. crescentus. It also seems likely that, for example, holo-Acp will regulate RSH proteins in other bacteria, as lipid starvation is a general activator of (p)ppGpp synthesis in many genera.
Unusual upstream control features of SR activation have been documented in only a few species, most notably the ones described here. However, there is evidence that a number of other bacterial species have SR systems that differ from the archetypal E. coli system, both in terms of ‘activating’ environmental signals and in the regulatory logic that controls the SR. For example, a number of species are reported to be irresponsive to amino acid starvation, including Rhodobacter sphaeroides, Sinohizobium meliloti, andHelicobacter pylori [74-79]. R. sphaeroides, a photosynthetic α-proteobacterium, produces (p)ppGpp upon transition from light to dark [76]. The molecular details of SR regulation in these and other species remain largely undefined. While the genetic components of the SR regulatory system are broadly conserved, it is clear that the ecological niche and lifestyle of a bacterium has a profound influence on not only the regulatory output, but also on what signals and combinations of signals activate the SR.
There are several outstanding questions in the study of signal detection by SR regulatory systems. Perhaps most important is the question of how the enzyme activities of RSH proteins are regulated at the molecular level. The direct control of SpoTEC by holo-Acp is a nice example of post-translational control of an RSH protein, but the wide range of signals that activate (p)ppGpp synthesis by RSH proteins suggests there are multiple modes of post-translational regulation. The recently-discovered SAS and SAH proteins will likely prove to be important in modulating (p)ppGpp levels in diverse ways; how the synthetase and hydrolase activities of these proteins are regulated is yet another important question in the field.
Box 1. A note on nomenclature.
To avoid confusion we follow the suggested naming convention of Hauryliuk and colleagues [12] for this review. We use the name Rel followed by a genus and species subscript for the ancestral bifunctional enzymes (i.e. those possessing synthase and hydrolase activities). E. coli encodes two paralogous enzymes derived from Rel gene duplication: RelA (synthase activity only) and SpoT (synthase and hydrolase activities). Per convention for species encoding such paralogs [12], we use the names RelAEC and SpoTEC when discussing the enzymes of E. coli.
Literature Cited
- 1.Magnusson LU, et al. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Potrykus K, Cashel M. (p)ppGpp: Still Magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 3.Tozawa Y, Nomura Y. Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol (Stuttg) 2011;13:699–709. doi: 10.1111/j.1438-8677.2011.00484.x. [DOI] [PubMed] [Google Scholar]
- 4.Barker MM, et al. Mechanism of Regulation of Transcription Initiation by ppGpp. II. Models for Positive Control Based on Properties of RNAP Mutants and Competition for RNAP. J. Mol. Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 5.Barker MM, et al. Mechanishm of Regulation of Transcription Initiation by ppGpp. I. Effects of ppGpp of Trancription Initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 6.Krasny L, et al. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol. 2008;69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- 7.Lopez JM, et al. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol. 1981;146:605–613. doi: 10.1128/jb.146.2.605-613.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferullo DJ, Lovett ST. The Stringent Response and Cell Cycle Arrest in Escherchia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JD, et al. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda A, et al. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 11.Kanjee U, et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkinson GC, et al. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittenhuber G. Comparative Genomics and Evolution of Genes Encoding Bacterial (p)ppGpp Synthetases/Hydrolases (the Rel, RelA and SpoT Proteins). J Mol Microbiol Biotechnol. 2001;3:585–600. [PubMed] [Google Scholar]
- 14.Braeken K, et al. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Dalebroux ZD, et al. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol. 2010;76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain V, et al. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- 17.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 18.Abranches J, et al. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das B, et al. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol Microbiol. 2009;72:380–398. doi: 10.1111/j.1365-2958.2009.06653.x. [DOI] [PubMed] [Google Scholar]
- 20.Lemos JA, et al. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 21.Nanamiya H, et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- 22.Sands MK, Roberts RB. The effects of a truptophan-histidine deficiency in a mutant of Escherchia coli. J Bacteriol. 1952;63:505–511. doi: 10.1128/jb.63.4.505-511.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stent GS, Brenner S. A Genetic Locus for the Regulation of Ribonucleic Acid Synthesis. Proc. Natl. Acad. Sci. 1961;USA47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borek E, et al. Studies on a mutant of Escherichia coli with unbalanced ribonucleic acid synthesis. J Bacteriol. 1956;71:318–323. doi: 10.1128/jb.71.3.318-323.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cashel M, Gallant J. Two Compounds implicated in the Function of the RC Gene of Escherchia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 26.Cashel M, Kalbacher B. The Control of Ribonucleic Acid Synthesis in Escherchia coli. J. Biol. Chem. 1970;245:2309–2318. [PubMed] [Google Scholar]
- 27.Maciag M, et al. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Lesley JA, Shapiro L. SpoT Regulates DnaA Stability and Initiation of DNA Replication in Carbon-Starved Caulobacter crescentus. J. Bacteriol. 2008;190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milon P, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durfee T, et al. Transcription Profiling of the Stringent Response in Escherichia coli. J. Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Traxler MF, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sy J. In vitro degradation of guanosine 5′-diphosphate, 3′-diphosphate. Proc. Natl. Acad. Sci. USA. 1977;74:5529–5533. doi: 10.1073/pnas.74.12.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao H, et al. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoTnull mutations. J. Biol. Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 35.Ramagopal S, Davis BD. Localization of the Stringent Protein of Escherchia coli on the 50S Ribosomal Subunit. Proc Natl Acad Sci USA. 1974;71:820–824. doi: 10.1073/pnas.71.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.English BP, et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci USA. 2011;108:E365–373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseltine WA, Block R. Synthesis of Guanosine Tetra- and Pentaphosphate Requires the Presence of a Codon-Specific, Uncharged Transfer Ribonucleic Acid in the Acceptor Site of Ribosomes. Proc. Natl. Acad. Sci. USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An G, et al. Cloning the spoT Gene of Escherichia coli: Identification of the spoT Gene Product. J. Bacteriol. 1979;137:1100–1110. doi: 10.1128/jb.137.3.1100-1110.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang M, et al. G-Protein Control of the Ribosome-Associated Stress Response Protein SpoT. J. Bacteriol. 2007;189:6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wout P, et al. The Escherichia coli GTPase CgtAe Cofractionates with the 50S Ribosomal Subunity and Interacts with SpoT, a ppGpp Synthetase/Hydrolase. J. Bacteriol. 2004;186:5249–5257. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray KD, Bremer H. Control of spoT-dependent ppGpp Synthesis and Degradation in Escherchia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- 42.Seyfzadeh M, et al. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherchia coli. Proc Natl Acad Sci USA. 1993;90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 44.Angelini S, et al. Disrupting the Acyl Carrier Protein/SpoT interaction in vivo: identification of ACP residues involved in the interaction and consequence on growth. PLoS One. 2012;7:e36111. doi: 10.1371/journal.pone.0036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spira B, et al. Guanosine 3′,5′-Bispyrophosphate (ppGpp) Synthesis in Cells of Escherichia coli Starved for Pi. J. Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinella D, et al. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 47.Mechold U, et al. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiber G, et al. Overexpression of the relA Gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 49.Savageau MA. Escherichia coli Habitats, Cell Types, and Molecular Mechanisms of Gene Control. Am Nat. 1983;122:732–744. [Google Scholar]
- 50.Hogg T, et al. Conformational Antagonism between Opposing Active Sites in a Bifuctional RelA/SpoT Homolog Modulates (p)ppGpp Metabolism during the Stringent Response. Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 51.Harris BZ, et al. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthusduring starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konovalova A, et al. A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus. Mol Microbiol. 2012;84:260–275. doi: 10.1111/j.1365-2958.2012.08020.x. [DOI] [PubMed] [Google Scholar]
- 54.Konovalova A, et al. Two intercellular signals required for fruiting body formation in Myxococcus xanthus act sequentially but non-hierarchically. Mol Microbiol. 2012;86:65–81. doi: 10.1111/j.1365-2958.2012.08173.x. [DOI] [PubMed] [Google Scholar]
- 55.Kaplan HB, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139:89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 56.Manoil C, Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980;141:305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 58.Crawford EW, Jr., Shimkets LJ. The stringent response in Myxococcus xanthus is regulated by SocE and the CsgA C-signaling protein. Genes Dev. 2000;14:483–492. [PMC free article] [PubMed] [Google Scholar]
- 59.Crawford EW, Jr., Shimkets LJ. The Myxococcus xanthus socE and csgA genes are regulated by the stringent response. Mol Microbiol. 2000;37:788–799. doi: 10.1046/j.1365-2958.2000.02039.x. [DOI] [PubMed] [Google Scholar]
- 60.Ossa F, et al. The Myxococcus xanthus Nla4 protein is important for expression of stringent response-associated genes, ppGpp accumulation, and fruiting body development. J Bacteriol. 2007;189:8474–8483. doi: 10.1128/JB.00894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80:695–714. doi: 10.1111/j.1365-2958.2011.07602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boutte CC, et al. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J Bacteriol. 2012;194:28–35. doi: 10.1128/JB.05932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Britos L, et al. Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One. 2011;6:e18179. doi: 10.1371/journal.pone.0018179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.England JC, et al. Global Regulation of Gene Expression and Cell Differentiation in Caulobacter crescentus in Response to Nutrient Availability. J. Bacteriol. 2009;193:819–833. doi: 10.1128/JB.01240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiaverotti TA, et al. Conditions that Trigger Guanosine Tetraphosphate Accumulation in Caulobacter crescentus. J. Bacteriol. 1981;145:1463–1465. doi: 10.1128/jb.145.3.1463-1465.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahl JL, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. USA. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karakousis PC, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Primm TP, et al. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahl JL, et al. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol. 2005;187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rifat D, et al. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis. 2009;200:1126–1135. doi: 10.1086/605700. [DOI] [PubMed] [Google Scholar]
- 71.Sureka K, et al. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol. 2007;65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 72.Thayil SM, et al. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One. 2011;6:e28076. doi: 10.1371/journal.pone.0028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sureka K, et al. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One. 2008;3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Acosta R, Lueking D. Stringency in the Absence of ppGpp Accumulation in Rhodobacter sphaeroides. J Bacteriol. 1987;169:908–912. doi: 10.1128/jb.169.2.908-912.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belitsky B, Kari C. Absence of Accumulation of ppGpp and RNA during Amino Acid Starvation in Rhizobium meliloti. J. Biol. Chem. 1982;257:4677–4679. [PubMed] [Google Scholar]
- 76.Eccleston ED, Jr., Gray ED. Variations in ppGpp levels in Rhodopseudomonas spheroides during adaptation to decreased light intensity. Biochem. Biophys. Res. Comm. 1973;54:1370–1376. doi: 10.1016/0006-291x(73)91138-8. [DOI] [PubMed] [Google Scholar]
- 77.Howorth SM, England RR. Accumulation of ppGpp in symbiotic and free-living nitrogen-fixing bacteria following amino acid starvation. Arch. Microb. 1999;171:131–134. doi: 10.1007/s002030050689. [DOI] [PubMed] [Google Scholar]
- 78.Zhou YN, et al. Regulation of Cell Growth during Serum Starvation and Bacterial Survival in Macrophages by the Bifunctional Enzyme SpoT in Helicobacter pylori. J. Bacteriol. 2008;190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krol E, Becker A. ppGpp in Sinorhizobium meliloti: biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol Microbiol. 2011;81:1233–1254. doi: 10.1111/j.1365-2958.2011.07752.x. [DOI] [PubMed] [Google Scholar]
- 80.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]



