Abstract
Aim
We investigated genetic variation of Irish pike populations and their relationship with European outgroups, in order to elucidate the origin of this species to the island, which is largely assumed to have occurred as a human-mediated introduction over the past few hundred years. We aimed thereby to provide new insights into population structure to improve fisheries and biodiversity management in Irish freshwaters.
Location
Ireland, Britain and continental Europe.
Methods
A total of 752 pike (Esox lucius) were sampled from 15 locations around Ireland, and 9 continental European sites, and genotyped at six polymorphic microsatellite loci. Patterns and mechanisms of population genetic structure were assessed through a diverse array of methods, including Bayesian clustering, hierarchical analysis of molecular variance, and approximate Bayesian computation.
Results
Varying levels of genetic diversity and a high degree of population genetic differentiation were detected. Clear substructure within Ireland was identified, with two main groups being evident. One of the Irish populations showed high similarity with British populations. The other, more widespread, Irish strain did not group with any European population examined. Approximate Bayesian computation suggested that this widespread Irish strain is older, and may have colonized Ireland independently of humans.
Main conclusions
Population genetic substructure in Irish pike is high and comparable to the levels observed elsewhere in Europe. A comparison of evolutionary scenarios upholds the possibility that pike may have colonized Ireland in two ‘waves’, the first of which, being independent of human colonization, would represent the first evidence for natural colonization of a non-anadromous freshwater fish to the island of Ireland. Although further investigations using comprehensive genomic techniques will be necessary to confirm this, the present results warrant a reappraisal of current management strategies for this species.
Keywords: Conservation biogeography, dispersal, management, molecular markers, non-anadromous freshwater fish, phylogeography, population genetics, post-glacial biota
Introduction
The faunal assemblage of islands depend upon a complex interplay of both extrinsic (e.g. area, distance to nearest neighbours, latitude) and intrinsic factors (e.g. life histories, migration and adaptation), which determine the success of natural colonization (Heaney, 2001; Dennis et al., 2012). As a large island on the north-western fringe of Europe, isolated from the rest of Europe swiftly after the retreat of the Pleistocene ice sheets (Edwards & Brooks, 2008), Ireland represents a very suitable scenario for investigating colonization patterns and potential barriers to dispersal. Historically, much of the Irish colonization debate has centred on mammalian fauna and the presence or absence of potential land bridges (Davenport et al., 2008). Little focus has been directed to alternative potential colonization routes of freshwater fish species, with the assumption that they could only have been anthropogenically introduced prevailing (Igoe 2004; King et al., 2011).
The isolation of Ireland by sea since the last glaciation has resulted in a depauperate freshwater fauna (Griffiths, 1997; Maitland, 2004; King et al., 2011), consisting exclusively of diadromous species [e.g. brown/sea trout (Salmo trutta), salmon (Salmo salar), European eel (Anguilla anguilla) and lampreys (Petromyzonidae)], which were able to naturally recolonize Ireland at the end of the last glaciation (Wheeler, 1977; Maitland, 2004; King et al., 2011), and stenohaline species introduced during the last few hundred years (Fitzmaurice, 1984; Griffiths, 1997). Today the few freshwater natives (11 species) are outnumbered by non-natives (13 species), which create increasing pressures primarily through competition for resources (e.g. roach, Rutilus rutilus) (Stokes et al., 2004; King et al., 2011). Although the introduction rate of alien species has greatly increased in recent decades in line with globalization (Cambray, 2003; Minchin, 2007; Gozlan et al., 2010), almost half of the introduced fish species now present in Ireland have been here for many hundreds of years (Fitzmaurice, 1984), and have no known date nor source of introduction (King et al., 2011).
Northern pike (Esox lucius Linnaeus, 1758) (Esocidae) is a freshwater fish with a circumpolar distribution in the Northern Hemisphere (Maes et al., 2003; Aguilar et al., 2005). Throughout its range, pike is of particular interest owing to its socio-economic value through recreational and commercial fishing (Casselman & Lewis, 1996; Laikre et al., 2005; Launey et al., 2006; Lucentini et al., 2009). Pike are almost ubiquitous in Ireland; however, they have long been thought to be non-native (Kennedy, 1969; Fitzmaurice, 1984; O’Grady & Delanty, 2008), based almost exclusively on the seminal paper by Went (1957). Went (1957) attempted to trace the earliest evidence of pike in Ireland, concluding that there were no references to pike prior to the 16th century, and that where references did exist they pertained to its absence, leading many to interpret Went’s paper as a suggested introduction date of the 16th century. This has led the status of pike in Ireland to become a contentious issue within stakeholder groups (e.g. Barbe & Garrett, 2013). Controversial policies, such as culling and transfer of pike during predator control operations aimed at protecting the native brown trout (Fitzmaurice, 1984; O’Grady & Delanty, 2008), have been common in the management of this species during recent decades, potentially compromising the integrity of genetic stocks [Inland Fisheries Trust (IFT) annual reports, e.g. IFT (1966–67, 1979–80); Minchin, 2007].
Pike is noted for its pronounced low levels of genetic variability when compared to other freshwater fish (Seeb et al., 1987; Senanan & Kapuscinski, 2000), including the closely related ‘muskellunge’, Esox masquinongy (Desjardins, 1996; Miller & Senanan, 2003). It has been suggested that severe post-glacial bottlenecks as a result of northward expansion from restricted refugia have been responsible for such reduced genetic variability (Maes et al., 2003; Jacobsen et al., 2005; Launey et al., 2006); however, the same patterns are not observed in other freshwater fish species that must have been subject to similar conditions (Miller & Senanan, 2003). The status of pike as a top predator may in part explain its unusually low polymorphism level, as predator population sizes depend upon suitable prey densities (Maes et al., 2003; Jacobsen et al., 2005). The only study to date that has investigated nuclear genetic variation in Ireland found monomorphism at all microsatellite loci examined (Jacobsen et al., 2005). Examination of mtDNA also showed very low variability and lacked power for inferring post-glacial dispersal patterns (Maes et al., 2003; Nicod et al., 2004), as did investigations of allozymes (Healy & Mulcahy, 1980).
The number, extent and source of introductions of pike into Irish waterways are currently unknown. Elucidation of patterns of genetic structure in Irish pike may lead to important discoveries about the origin of populations, their current connectivity, and the impact that indiscriminate transfer and mixing of individuals has had on populations, through the potential introgression of maladapted genes, threatening the genetic integrity of natural populations (Tallmon et al., 2004; Launey et al., 2006). Increasing local (e.g. resource extraction) and regional (e.g. climate, floods) disturbances make resilience within systems of paramount importance, and effective management strategies necessary (Folke et al., 2004; Venturelli & Tonn, 2006). Molecular data are vital not only to establish the origins of populations, but also in order to determine their viability, demographics and to distinguish discrete stocks for management purposes. Here we present the first Ireland-wide population genetic investigation using a suite of polymorphic microsatellite markers to illustrate the nature of population connectivity in Irish pike and their relationship to British and continental European populations. Specifically, we aim to test whether (1) there is identifiable population structure in Irish pike, (2) the timing of introduction is consistent with the historical periods so far hypothesized, and (3) potential sources for Irish pike populations can be identified.
Materials and methods
Sampling
Pike were sampled from 15 locations around Ireland using a combination of electrofishing, gill-netting and angling, between August 2010 and November 2011. Gill-netting and electrofishing were carried out opportunistically in collaboration with Inland Fisheries Ireland (the state agency responsible for the protection, management and conservation of Ireland’s inland fisheries) during their routine surveys (Table 1). Samples were also obtained while attending angling competitions (Table 1), and occasionally through organized trips to sample areas of particular interest (Lee & Bane). Sampling locations were chosen to give as broad a representation and coverage of Irish pike populations as possible (Fig. 1). Fin clips were taken and preserved in 100% ethanol before storage at −20 °C until analysis. Tissue or scale samples were also obtained from England, France, Sweden, Germany and Romania (Table 1). European samples were selected to cover hypothesized European lineages [i.e. previously identified genetically distinct northern European and Balkan (Danubian) populations; Maes et al., 2003; Jacobsen et al., 2005; Launey et al., 2006] including the most likely sources of introduction/natural colonization (Britain, north-western France).
Table 1.
Pike samples across Ireland and Europe. Study sites, indicating site name, region, GPS location (DD), sample size (n) and method employed (gill = gill nets, elec = electrofishing, ang = angling). German and Swedish samples were shipped as tissue by colleagues. The following overall genetic variability measures are also reported: number of alleles per locus (A), allelic richness (R), expected (He) and observed (Ho) heterozygosity. Multilocus estimates of FIS and P-values for multilocus Hardy–Weinberg equilibrium tests are provided (significant values in bold). Irish samples were collected between August 2010 to November 2011; European and British samples were collected between March 2002 and September 2012.
| Mean over all loci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Habitat | Lat. (DD) | Long. (DD) | Method | n | A | R | He | Ho | FIS | P |
| Europe | |||||||||||
| Somova | Lake | 45.1835 | 28.6832 | Gill | 10 | 8.83 | 7.11 | 0.90 | 0.67 | 0.26 | < 0.001 |
| Baltic | Sea | 58.5 | 17.7 | 20 | 9.00 | 5.38 | 0.73 | 0.73 | 0.00 | 0.6131 | |
| Wittensee | Lake | 54.3860 | 9.7564 | 39 | 8.17 | 4.57 | 0.67 | 0.62 | 0.07 | 0.4328 | |
| Dollnsee | Lake | 52.9945 | 13.5820 | 44 | 5.50 | 3.87 | 0.60 | 0.60 | 0.00 | 0.4390 | |
| Loire | River | 47.4133 | 0.9844 | Elec | 24 | 7.50 | 4.87 | 0.67 | 0.63 | 0.06 | 0.0293 |
| Britain | |||||||||||
| Frome | River | 50.6836 | −2.1087 | Elec | 32 | 3.17 | 2.41 | 0.39 | 0.37 | 0.04 | 0.0001 |
| Thames | River | 51.6383 | −1.1792 | Elec | 30 | 4.83 | 3.06 | 0.50 | 0.47 | 0.06 | 0.0170 |
| Winderemere | Lake | 54.3760 | −2.9351 | Gill | 29 | 4.00 | 2.72 | 0.36 | 0.39 | −0.10 | < 0.001 |
| Leven | Canal | 53.8896 | −0.3564 | Elec | 30 | 4.17 | 2.93 | 0.47 | 0.42 | 0.10 | 0.3275 |
| Ireland | |||||||||||
| Bane | Lake | 54.0300 | −6.9130 | Ang | 12 | 1.33 | 1.25 | 0.05 | 0.03 | 0.49 | 0.1809 |
| Barrow | River | 52.6648 | −6.9842 | Elec | 48 | 2.67 | 1.96 | 0.27 | 0.29 | −0.05 | 0.9335 |
| Carra | Lake | 53.7166 | −9.2560 | Gill | 20 | 1.67 | 1.47 | 0.13 | 0.13 | 0.07 | 0.7099 |
| Conn | Lake | 54.0253 | −9.2519 | Gill/Ang | 30 | 1.67 | 1.37 | 0.10 | 0.09 | 0.07 | |
| Corrib | Lake | 53.4913 | −9.3137 | Gill | 39 | 2.17 | 1.57 | 0.17 | 0.18 | −0.09 | 0.8526 |
| Deel | River | 53.5866 | −7.1277 | Elec | 35 | 2.00 | 1.47 | 0.12 | 0.14 | −0.11 | 0.0897 |
| Derg | Lake | 52.8407 | −8.4582 | Ang | 40 | 2.33 | 1.69 | 0.19 | 0.19 | 0.03 | 0.6009 |
| Dromore | River | 54.0926 | −7.0126 | Ang | 25 | 2.00 | 1.55 | 0.14 | 0.14 | 0.03 | 0.5529 |
| Grand | Canal | 53.2463 | −7.8936 | Elec | 44 | 3.17 | 2.20 | 0.31 | 0.28 | 0.11 | 0.0058 |
| Inny | River | 53.6511 | −7.4191 | Elec | 34 | 4.00 | 2.17 | 0.27 | 0.29 | −0.11 | 0.6698 |
| Lee | Various | 53.6511 | −8.9623 | Ang | 52 | 2.50 | 1.99 | 0.32 | 0.34 | −0.07 | 0.8482 |
| Royal | Canal | 53.3734 | −6.4697 | Elec | 50 | 2.50 | 1.66 | 0.19 | 0.18 | 0.04 | 0.7889 |
| Scur | Lake | 54.0257 | −7.9522 | Gill | 27 | 2.17 | 1.45 | 0.13 | 0.14 | −0.12 | 0.9893 |
| Shannon | River | 52.7125 | −8.5086 | Ang | 8 | 2.00 | 1.89 | 0.22 | 0.23 | −0.04 | 0.2007 |
| Sheelin | Lake | 53.8031 | −7.3114 | Gill | 40 | 2.83 | 1.99 | 0.25 | 0.28 | −0.14 | 0.0843 |
Figure 1.

Locations of pike sampling sites in Ireland and Europe. The Shannon, Derg, Inny, Sheelin and Scur are all directly part of the Shannon system. Dromore River and Lough Bane are a part of the Erne system, which connects to the Shannon via the Shannon-Erne waterway (16 locks) at Upper Lough Erne in County Fermanagh. The Grand Canal connects the River Liffey in Dublin to the River Shannon at Shannon Harbour, Co. Offaly, via 44 locks, and connects with the River Barrow via the Barrowline Canal (9 locks). The Royal Canal also connects Dublin’s River Liffey with the Shannon, at the more northerly Abbeyshrule in County Longford, meeting the River Deel along its way.
Genetic analysis
Genomic DNA was isolated from tissue samples using a modified salt extraction protocol for DNA (Miller et al., 1988). Launey et al. (2003) suggested that many loci would be required to identify genetic segregation between pike populations owing to the low levels of variability at a global scale in this species; thus 30 loci were selected from the literature for testing, based on the number of alleles observed and the geographical range previously examined (see Appendix S1 in Supporting Information). These loci were examined in individuals selected from geographically distinct regions of Ireland (Grand, Corrib, Carra, Barrow, Sheelin, Shannon, Lee). Six of these loci proved to be variable during screening. A total of 752 individuals were successfully amplified and genotyped at the six microsatellite loci: Elu19 (Miller & Kapuscinski, 1997), Eluc004 and Eluc045 (Aguilar et al., 2005), B118INRA (Launey et al., 2003), B24 and B451 (Wang et al., 2011). Loci were amplified in a single 10 μL multiplex polymerase chain reaction (PCR) containing 1 μL of DNA (25 ng/μL), 5 μL Multiplex PCR Mastermix (Qiagen, Crawley, UK) and labelled primers (FAM, VIC, NED and PET) with the following concentrations: Elu 19 (0.25 μm), Eluc004 (0.4 μm), Eluc045 (0.4 μm), B118INRA (0.25 μm), B24 (0.4 μm) and B451 (0.25 μm). Amplification conditions were as follows: 95 °C for 15 min; 35 cycles of 94 °C for 45 s, 58 °C for 45s, 72 °C for 45s and a final extension at 72 °C for 45 min. All PCR products were run on a 16-capillary system ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), with an internal size standard (600 LIZ, Applied Biosystems) using the program GeneMapper 4.0 (Applied Biosystems). Ten per cent of samples were randomly selected and re-amplified and scored at all six loci.
Data analysis
micro-checker 2.2.3 (van Oosterhout et al., 2004) was used to check for scoring errors, large allele dropout and possible scoring errors for each population (1000 randomizations). Allele frequencies, number of alleles, allelic richness, expected and observed heterozygosity values (He and Ho), linkage disquilibrium, FIS and FST values were computed using fstat 2.9.3.2 (Goudet, 2001) with default settings. Loci were tested for departures from selective neutrality using the LOSITAN (Antao et al., 2008) FST outlier method, under default settings for both the infinite allele model (IAM) and stepwise mutation model (SMM) (Beaumont & Nichols, 1996). genepop 4.1.4 (Rousset, 2008) was used to test for departure from Hardy–Weinberg equilibrium (HWE). bottleneck 1.2 (Piry et al., 1999) was used to detect recent population reductions. The IAM, SMM and a two-phase mutation model (TPM; 20% and 70% SMM) were all tested and assessed with a Wilcoxon sign-rank test.
Pairwise FST values from fstat were visualized and compared using a non-metric multidimensional scaling analysis (NMDS) plot, as implemented in past 2.17c (Hammer et al., 2001).
Population substructure was assessed using the Markov chain Monte Carlo Bayesian clustering method of the software structure 2.3.4 (Pritchard et al., 2000), which infers the most likely number of population clusters (K) by minimizing Hardy–Weinberg departures and linkage disequilibrium within groups. Individuals are assigned to clusters based on probability of membership (Q-statistic). structure analysis was carried out at two levels, firstly with all populations, and separately with only Irish populations. Five independent runs were performed for each K value (1–28) using a burn-in period of 100,000 and followed by 400,000 iterations. Assignment tests were run under the default settings with the admixture model and correlated allelic frequencies. The program structure harvester 0.6.93 (Earl & vonHoldt, 2012) was used to assess and visualize likelihood values across the multiple values of K, to detect the K that best fit the data, using both the mean posterior probability of the data [L(K)] and the Evanno et al. (2005) method (ΔK). The software clumpp 1.1.2 (Jakobsson & Rosenberg, 2007) was used to assign clusters to which each run corresponded (search options: fullsearch) and visualized with the software distruct 1.1 (Rosenberg, 2004).
Hierarchical analysis of molecular variance (AMOVA) was performed using arlequin 3.5.1.3 (default settings; Excoffier & Lischer, 2010). Multiple groups were tested based initially upon geographical location (e.g. Europe, Britain and Ireland), and secondly informed by structure plots and NMDS plots of pairwise FST values. The optimal groupings were selected based upon largest FCT (between groups/regions) in relation to FSC (between populations within groups/regions).
Approximate Bayesian computation (using the program diyabc 1.0.4; Cornuet et al., 2008) was used to estimate the relative likelihood of alternative scenarios for the initial introduction of pike into Ireland. The program uses reference tables (containing parameters based on known or estimated values) to establish scenarios from which simulated data sets could be compared to the observed values (see Appendix S2 for details). Baltic and Danubian samples were excluded as they were unlikely to be the direct source for Irish pike populations: Baltic pike reached the enclosed sea (and probably adapted to mildly brackish conditions) as it became inhabitable after the end of the last glaciation. Pike from the Danube (Romania) belong to a separate lineage (Maes et al., 2003) at the most south-eastern edge of Europe. The English ‘Leven’ was not included in the ‘Britain’ group, based on FST values, which show that it groups more with European than other British samples, and so would add noise when testing hypotheses on Irish colonization. Seventeen scenarios, covering all likely colonization avenues were explored (Appendix S2). The effective population sizes (Ne) were set from 10 to 10,000; bottleneck sizes (d) were assigned an effective population size of 10, and each competing scenario was given equal prior probability. Mutation model prior distributions were taken as default and each scenario was simulated 500,000 times. The relative likelihoods of the scenarios were compared by both logistic regression and direct approach on 1% and 0.2% of the closest simulated data sets, respectively, and the fit of the model to the data were visualized using principal components analysis (PCA), as implemented in diyabc. To increase computational efficiency, the 17 test scenarios were split into five groups and the four best supported scenarios were then re-run together and used to estimate posterior parameters such as time since event and effective population sizes. Estimates for time since coalescent events are given in generations; assuming this relates to the age at first spawning (Martin & Palumbi, 1993), we converted this to years, by taking a value of 2 years as average age at first spawning, as reported for Ireland (Healy, 1956; Roche et al., 1999; O’Grady & Delanty, 2008) and Europe (Raat, 1988; Arlinghaus et al., 2009).
Mantel tests were used to assess the degree of association between matrices of genetic divergence (FST) and geographic distance (km overland). Tests were conducted in past (Hammer et al., 2001) and P-values were obtained through randomization (10,000).
Results
No consistent linkage disequilibrium between locus pairs was observed; Elu19 × EluB118INRA appeared linked in the Windermere population only. There was no evidence of systematic allelic dropout, null alleles or possible scoring errors. LOSITAN indicated that loci B24 and Eluc045 were possible candidates for balancing selection. The total number of alleles per locus ranged from 10 (Elu19) to 30 (B451) across the study area, and allelic richness ranged from 1.6 to 5.2 per locus. Loci varied in their degree of information content, ranging from monomorphism in some samples, up to 17 alleles in others. The average number of alleles within populations ranged from 1.33 to 9 (Table 1), and no consistent departures from Hardy–Weinberg equilibrium were detected. Danube, Loire, Frome, Thames, Windermere and Grand all significantly departed from HWE – generally due to heterozygote deficiency at 1–4 loci (not always the same loci), possibly as a result of the fact that sampling was spread over multiple areas for these samples, and so are small samples representative of large populations over large areas, and possibly bear the signature of Wahlund effect, reflected in the lack of heterozygotes. A pattern of decreasing genetic diversity was observed when moving from Europe towards Ireland.
The River Inny (Ireland; P = 0.03), Lake Windermere (Britain; P = 0.03) and Lake Wittensee (Germany; P = 0.02) all appeared to have undergone bottlenecks under SMM, and Lake Somova (Romania) appeared bottlenecked under the IAM (P = 0.03) and TPM, tested with both 20% and 70% SMM (P = 0.03).
Genetic differentiation among all samples was evident, with global multilocus FST = 0.328 [95% confidence interval (CI): 0.264–0.419]. For the Irish samples alone, global multilocus FST was of the same order of magnitude, at 0.27 (95% CI: 0.161–0.304), with some loci in some Irish samples being fixed at one allele (e.g. locus Elu19 in Bane, Conn, Carra, Deel, Dromore, Inny, Lee, Royal, Scur, Shannon, Sheelin). Overall, pairwise comparisons indicated strong differentiation of the European samples from the Irish, with Britain giving intermediate values. Within Ireland, high pairwise sample FST values were also observed (Table 2).
Table 2.
Pairwise FST values for all pike populations (upper) and pairwise significance (*P > 0.05) after sequential Bonferroni corrections (***) (lower). Site names have been shortened to three letters from Table 1. Irish pairwise values are highlighted in bold.
| DAN | BAL | WIT | KLE | LOI | FRO | THA | WIN | LEV | BAN | BAR | CAR | CON | COR | DEE | DER | DRO | GRA | INN | LEE | ROY | SCU | SHA | SHE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAN | 0.12 | 0.15 | 0.19 | 0.15 | 0.33 | 0.27 | 0.36 | 0.30 | 0.51 | 0.46 | 0.51 | 0.59 | 0.56 | 0.58 | 0.54 | 0.54 | 0.44 | 0.45 | 0.42 | 0.57 | 0.55 | 0.35 | 0.49 | |
| BAL | *** | 0.10 | 0.17 | 0.06 | 0.31 | 0.21 | 0.28 | 0.23 | 0.39 | 0.33 | 0.37 | 0.45 | 0.41 | 0.43 | 0.39 | 0.39 | 0.30 | 0.32 | 0.34 | 0.44 | 0.41 | 0.26 | 0.35 | |
| WIT | *** | *** | 0.11 | 0.10 | 0.31 | 0.24 | 0.31 | 0.18 | 0.42 | 0.37 | 0.39 | 0.44 | 0.42 | 0.44 | 0.42 | 0.42 | 0.35 | 0.34 | 0.35 | 0.46 | 0.42 | 0.31 | 0.38 | |
| KLE | *** | *** | *** | 0.11 | 0.26 | 0.22 | 0.33 | 0.28 | 0.43 | 0.37 | 0.40 | 0.45 | 0.43 | 0.44 | 0.42 | 0.42 | 0.36 | 0.35 | 0.33 | 0.44 | 0.43 | 0.32 | 0.39 | |
| LOI | *** | *** | *** | *** | 0.23 | 0.11 | 0.20 | 0.18 | 0.33 | 0.20 | 0.28 | 0.33 | 0.31 | 0.32 | 0.30 | 0.31 | 0.20 | 0.21 | 0.21 | 0.34 | 0.33 | 0.17 | 0.24 | |
| FRO | *** | *** | *** | *** | *** | 0.29 | 0.42 | 0.39 | 0.59 | 0.42 | 0.54 | 0.60 | 0.56 | 0.57 | 0.55 | 0.57 | 0.45 | 0.47 | 0.38 | 0.59 | 0.57 | 0.46 | 0.51 | |
| THA | *** | *** | *** | *** | *** | *** | 0.24 | 0.20 | 0.32 | 0.23 | 0.27 | 0.35 | 0.28 | 0.32 | 0.26 | 0.28 | 0.18 | 0.23 | 0.22 | 0.29 | 0.31 | 0.16 | 0.21 | |
| WIN | *** | *** | *** | *** | *** | *** | *** | 0.27 | 0.55 | 0.21 | 0.41 | 0.42 | 0.42 | 0.44 | 0.44 | 0.49 | 0.28 | 0.36 | 0.26 | 0.53 | 0.49 | 0.34 | 0.35 | |
| LEV | *** | *** | *** | *** | *** | *** | *** | *** | 0.44 | 0.40 | 0.42 | 0.49 | 0.43 | 0.47 | 0.41 | 0.43 | 0.33 | 0.36 | 0.39 | 0.43 | 0.45 | 0.32 | 0.36 | |
| BAN | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.50 | 0.39 | 0.71 | 0.31 | 0.43 | 0.12 | 0.05 | 0.19 | 0.16 | 0.52 | 0.12 | 0.21 | 0.27 | 0.21 | |
| BAR | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.28 | 0.30 | 0.28 | 0.29 | 0.33 | 0.41 | 0.18 | 0.26 | 0.12 | 0.47 | 0.37 | 0.25 | 0.24 | |
| CAR | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.35 | 0.02 | −0.01 | 0.06 | 0.16 | 0.05 | 0.04 | 0.38 | 0.31 | 0.07 | 0.02 | 0.05 | |
| CON | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.30 | 0.34 | 0.39 | 0.53 | 0.23 | 0.32 | 0.38 | 0.48 | 0.51 | 0.41 | 0.23 | |
| COR | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.04 | 0.07 | 0.15 | 0.05 | 0.05 | 0.37 | 0.28 | 0.09 | 0.03 | 0.05 | |
| DEE | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.17 | *** | *** | 0.10 | 0.21 | 0.07 | 0.06 | 0.41 | 0.36 | 0.11 | 0.06 | 0.08 | |
| DER | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.05 | *** | *** | *** | 0.01 | 0.04 | 0.02 | 0.42 | 0.16 | 0.03 | 0.00 | 0.03 | |
| DRO | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.16 | *** | * | *** | *** | *** | 0.52 | 0.09 | 0.06 | 0.47 | 0.13 | 0.05 | 0.05 | 0.09 | |
| GRA | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.03 | 0.29 | 0.19 | 0.09 | −0.01 | 0.01 | |
| INN | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.35 | 0.20 | 0.04 | 0.00 | 0.04 | |
| LEE | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.48 | 0.45 | 0.35 | 0.33 | |
| ROY | *** | *** | *** | *** | *** | *** | *** | *** | *** | * | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.25 | 0.22 | 0.16 | |
| SCU | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | * | *** | *** | *** | *** | 0.05 | 0.10 | |
| SHA | *** | *** | *** | *** | *** | *** | *** | *** | *** | * | *** | 0.28 | *** | 0.20 | 0.17 | 0.32 | 0.38 | 0.28 | 0.43 | *** | *** | 0.15 | 0.02 | |
| SHE | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | * | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | 0.16 |
Private alleles were rare in Ireland, with only four alleles (10%) found private to Ireland across all sites sampled (found in the Barrow, Sheelin, Inny and Grand). A large number of alleles were shared by the mainland European samples but not observed in either Britain or Ireland. 25% of British alleles were shared with Europe, but not Ireland. 22% of the main Irish group alleles were shared with Europe but not found in Britain; the Lee–Barrow group only had one allele shared with Europe that was not present in Britain. 24% of all alleles present in the Danubian sample and 11% of Baltic alleles were private.
structure harvester illustrated that K = 2 and K = 3 were the most likely scenarios for the ‘all samples’ test (Fig. 1, Appendix S1). Graphs of these scenarios were then examined to investigate groupings, as illustrated in Fig. 2a. The K = 2 scenario highlights the divergence of the Irish group (orange) in relation to mainland European and British samples (blue), and the K = 3 scenario depicts the separation of the British samples (yellow). Two Irish samples (Lee and Barrow) strongly group with Britain, with the other Irish samples grouping separately into the orange group. Also of note is the Leven Canal (East Yorkshire, along the east coast of England) which groups with mainland Europe.
Figure 2.
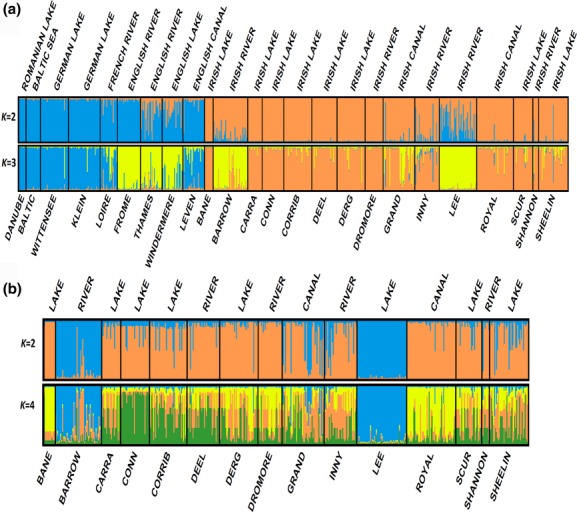
Results from structure indicating individual assignment and population clustering of pike for (a) 752 individuals from all 24 locations sampled (Table 1), and (b) 504 individuals from 15 locations in Ireland. Individual sampling location is listed below the figure, with its site type above. Each vertical bar represents an individual’s assignment into K clusters (colours). Results of Evanno’s ΔK and L(K) (Appendix S1: Fig. S1) indicate that the best supported K values for all populations are 2 and 3, respectively. The best supported values for Ireland only are K = 2 and K = 4. For ‘Ireland only’, the distinction of the Lee & Barrow populations is evident at K = 2, and maintained at K = 4, where the distinction of Lough Conn, Lough Bane and the Royal Canal can now be seen.
structure analysis of just the Irish samples enabled the elucidation of finer-scale structure within Ireland (Fig. 2b). Here K = 2 and K = 4 were the best supported scenarios (Fig. 1b, Appendix S1). Overall, K = 2 completely supports the above findings of two highly divergent strains within Ireland (the blue group, related to the British samples, and a second distinct Irish group, shown in orange). Examination of the K = 4 graph demonstrates that divergent groups exist within Ireland, such as Lough Conn (homogeneously ‘green’), and the Royal Canal (yellow), whereas the majority of the weakly assigned individuals (multiple colours) relate to individuals from samples connected to the River Shannon.
The NMDS plot of pairwise FST values also supports the above groupings (Fig. 3) and illustrates a close relationship between the French Loire and British Thames river samples, and a grouping of the Irish Lee, Barrow and British Windermere samples. Within the ‘Shannon’ type genotypes, the samples not directly connected with the main river system (Bane, Carra, Corrib and Deel) appear on the outskirts of this cluster in the NMDS plot, as does the Royal Canal. Lough Conn appears highly distinct, further reflecting the groups found with structure.
Figure 3.

Non-metric multidimensional scaling (NMDS) plot of population pairwise FST distances (Table 2) enabling visualization of evolutionary relationships between pike populations.
AMOVA was carried out using multiple grouping designs in order to investigate potential explanations for the observed variance (Table 3). The best designs (2 and 4) were selected as those whose ‘among groups’ factor explained the majority of the variance observed (i.e. the largest FCT), whilst also minimizing the ‘within samples’ variation (FSC). Design 2 reflects the separation of Lee & Barrow, grouping them with Lough Windermere (Fig. 3). The rest of Ireland remains distinct, and Britain groups with the European samples. Design 4 supports the further separation of the divergent Lough Conn and River Frome samples into separate groups.
Table 3.
Analyses of molecular variance (AMOVA) for pike populations. Four grouping scenarios are reported, the highest support is found for those displaying the largest FCT in relation to FSC, i.e. the largest percentage of variation accounted for by the grouping design, which minimizes the variation within these groups.
| Grouping | Source of variation | d.f. | % variation | F-index | P |
|---|---|---|---|---|---|
| 1. Ireland | Among groups | 2 | 23.23 | 0.232 (FCT) | < 0.001 |
| 2. Britain | Among populations within groups | 21 | 17.08 | 0.223 (FSC) | < 0.001 |
| 3. Europe | |||||
| 1. Main Ireland | Among groups | 2 | 25.15 | 0.252 (FCT) | < 0.001 |
| 2. Britain & Europe | Among populations within groups | 21 | 14.06 | 0.188 (FSC) | < 0.001 |
| 3. Barrow, Lee, Windermere | |||||
| 1. Main Ireland | Among groups | 2 | 24.69 | 0.247 (FCT) | < 0.001 |
| 2. Europe & Leven | Among populations within groups | 21 | 14.27 | 0.190 (FSC) | < 0.001 |
| 3. Britain, Barrow & Lee | |||||
| 1. Main Ireland | Among groups | 4 | 26.95 | 0.270 (FCT) | < 0.001 |
| 2. Europe, Leven & Thames | Among populations with groups | 19 | 11.57 | 0.158 (FSC) | < 0.001 |
| 3. Barrow, Lee, Windermere | |||||
| 4. Frome | |||||
| 5. Conn |
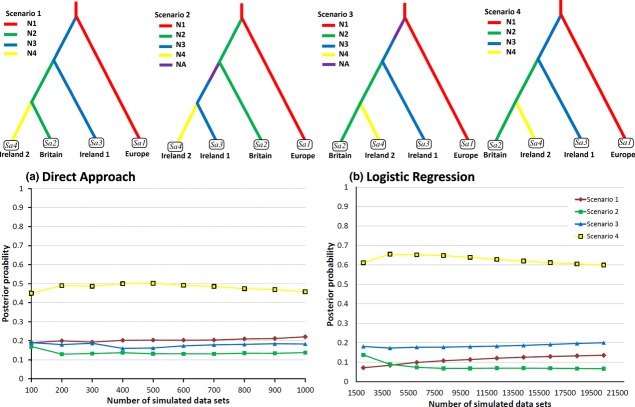
Despite the wide range of scenarios tested with diyabc, one in particular stood out for its best fit to the observed data, producing the highest support values with both the direct estimate and logistic regression (Fig. 4). This scenario was consistently the strongest supported when tested against differing groups of competing scenarios (Appendix S2). The first split in this scenario suggests colonization of Ireland and Britain (Ne = 2300, 95% CI: 1190–3990) from Europe (Ne = 9130, 95% CI: 6980–9940) c. 8000 years ago (t3 median = 4200 generations, 95% CI: 1280–9090). The second split appears to indicate a split between the Irish and British (Ne = 4450, 95% CI: 2220–7490) populations some 4000 years ago (t2 = 1720, 95% CI: 644–4560). The third and final split illustrates a more recent introduction from Britain into Ireland (Ne = 744, 95% CI: 263–1550) c. 1000 years ago (t1 = 615; 95% CI: 152–2160).
Figure 4.
Approximate Bayesian computation (diyabc 1.0.4; Cornuet et al., 2008) was used to estimate the relative likelihood of alternative scenarios for the initial introduction of pike into Ireland. Above, the diyabc graphs illustrate the four final best supported scenarios tested together. Ireland 1 refers to the main group of Irish genotypes, Ireland 2 refers to the Barrow and Lee populations, which group with the British Windermere in Fig. 3. For each group the scenario is illustrated (colours indicate different population sizes, Ne), and graphs indicate the relative likelihoods of the four best scenarios compared by (a) direct approach, and (b) logistic regression on the 1% (20,000) and 0.005% (1000) of the closest simulated data sets, respectively. The graphs clearly illustrate that Scenario 4 is the scenario with the best support. See Appendix S2 for more details on approximate Bayesian computation analyses.
Within Ireland, isolation-by-distance was observed (r = 0.48; P = 0.002), which was maintained when only the main older strain was examined, albeit slightly less strongly (r = 0.26; P = 0.03). Taking rivers only (r = 0.11; P = 0.36) and lakes only (r = 0.45; P = 0.059) indicated – despite the decreased power as a result of reduced samples – that it is the lakes that are responsible for the majority of isolation effect, as may be expected from rivers acting as corridors, and within which there may be more movement and hence gene flow.
Discussion
The present study unveils for the first time the genetic diversity within and among pike populations inhabiting Ireland’s water bodies, and clarifies their relationships with populations from European locations. We found evidence for strong spatial structure, with FST values within Ireland being of the same order of magnitude as across Europe (Jacobsen et al., 2005; Launey et al., 2006), and the existence of distinct populations, probably corresponding to multiple colonization dates, which indicates that pike may have first colonized Ireland naturally. This information is significant for the reappraisal of current management strategies in this economically (angling) and ecologically (top-predator) important species, and will contribute new perspectives to the long-standing debate on the mechanisms and timing of colonization dynamics of Britain and Ireland (Lynch, 1996; Griffiths, 1997; Woodman et al., 1997; Carden et al., 2012).
Phylogeography and colonization history
Ireland’s fauna is emblematic for its extremely complex series of colonization events and introductions, the patterns of which are still largely unknown and vigorously debated (McCormick, 1999; Davenport et al., 2008; McDevitt et al., 2011). The once popular ‘land-bridge hypothesis’ – which proposed land corridor connections between Ireland and Britain or north-western France – has been debunked as recent research has shown sea level to have risen much faster than previously thought (Brooks et al., 2007; Edwards & Brooks, 2008). As more case studies become available, the story becomes increasingly complex, hindering generalization and identification of common patterns (e.g. pygmy shrew; McDevitt et al., 2011), and even revealing multiple colonization events (e.g. red deer; Carden et al., 2012).
structure (Fig. 2) and the NMDS plot (Fig. 3) highlighted some degrees of similarity that were unexpected based on their geographical location, e.g. British pike group with some Irish samples (Lee and Barrow), and the Leven Canal (north-eastern England) is more similar to northern European populations than to British ones. The Lee–Barrow–Windermere group signal is observed to a lesser degree in some other samples (e.g. Grand Canal), which can be explained by the connection existing between the Barrowline Canal and the Grand Canal. Pike from the River Thames group with the River Loire from north-western France, perhaps reflecting some historical connection, prior to inundation of Doggerland which removed any remaining connections between Britain and Europe around 7000–8000 years ago (Wheeler, 1977; Weninger et al., 2008). It is at this point that pike populations in mainland Europe and in the British Isles became demographically independent. The results of the diyabc analysis provide stark support for this time frame, with the posterior probability distribution for the first Europe versus British–Irish split agreeing with an 8000 year timeline.
diyabc analysis rejected the seemingly more obvious, ‘simple’ explanation according to which Ireland would be colonized from Britain and any more population subdivision would have resulted from more recent processes within the island. Instead, analyses indicate that around 3500–4000 years ago, Irish and British pike populations became isolated; this may have corresponded to the Irish Sea assuming its contemporary fully marine nature and becoming an impassable barrier for freshwater fish. Finally, a second pike contingent appears to have entered the island around 1000 years ago and is currently distributed in the south of the island.
Overall, ABC, structure and hierarchical AMOVA analyses, each one based on independent methods, concur to indicate a strong separation between two distinct Irish units, whose introduction to Ireland may have followed rather different paths: the first more widespread group appears to have reached Ireland and Britain shortly after the retreat of the ice sheets; the second, mainly present in the southern river catchments of the Lee and the Barrow, was likely introduced by humans during the Middle Ages. This is further supported by the alleles Ireland and Britain both share with Europe, but not with each other. If all Irish pike had colonized from Britain, Ireland should consist of a subset of the British alleles. It is also worthy of note that a similar geographical division between the north and west (Boreal race) and south-eastern (Celtic race) Ireland has been previously observed in Atlantic salmon, Salmo salar (Child et al., 1976).
The greater level of admixture observed from the populations connected to the Shannon is not surprising as the Shannon system has been a major focus of pike management works since the 1960s, involving both culling and transfer of pike among areas of the system (IFT annual reports 1952–1980). However, further spatial subdivision is detectable within this ‘older’ Irish group. Some divergence might have occurred in very recent times due to human activities – such as the closure of the Royal Canal in 1961, provoking the subsequent isolation and drift of the population for almost 50 years until the canal was reopened in 2010. Similar processes may have been at work in Lough Bane, which is a very small, somewhat isolated waterbody (approximately 200 m × 400 m). Other patterns are more difficult to reconstruct; for instance, the lack of divergence of Lough Corrib and Lough Carra from the main ‘Shannon’ group may be linked to recurrent management operations on these waterbodies. The divergence of the Lough Conn population, which lacks unique alleles, probably reflects a recent founding event.
History of pike in Ireland and management implications
Northern pike are thought to have been anthropogenically introduced to Ireland around the 16th century (Went, 1957). However, our results refute this simplistic view. One strain has indeed probably been introduced from Britain, perhaps from populations related to the Windermere pike; however, a much earlier introduction has been found to be incompatible with anthropogenic transfers. Albeit widespread in the island, this putatively older Irish strain is both significantly genetically depauperate and considerably divergent from the British and European sites examined here. Interestingly, the two main Irish groups seem to exhibit little geographical overlap (Fig. 1).
The more recent introduction to Ireland may have been facilitated in the 12th century by the Normans, who are responsible for many of the introductions to this island [e.g. hedgehogs (Erinaceus europaeus), fallow deer (Dama dama), black rat (Rattus rattus), rabbit (Oryctolagus cuniculus); McCormick, 1999]. In support of this date of introduction is the very rare finding of two pike cleithra bones found during the excavation of the Anglo-Norman castle at Trim, Co. Meath, dating to the late 13th–early 14th century (Hamilton-Dyer, 2011). The fact that they are cleithra (head bones) indicates that the pike may have been present alive, as the usual method of shipping fish at that time was beheaded and dried (Hoffmann, 2009). Furthermore, Longfield (1929) states that pike were likely to have been introduced by the 14th century, and that by the 16th century they were thoroughly at home in Ireland. At this time, pike exports from the south of Ireland (Youghal, Dungarvan, Cork and Kinsale) to southern English towns (Longfield, 1929) greatly exceeded those of brown trout. In one year alone, 1507, Dartmouth imported 3850 pike from Ireland.
Went (1957) stated that there was no old Irish name for pike, and that the modern name is ‘gailliasc’ which literally translates into ‘strange or foreign fish’, thus suggesting an introduction (Fitzmaurice, 1984). However, Farran’s (1946) paper on the local names of Irish fish contains over 10 variations of names for pike, which included liús, lús, lusaigh and lusc – all of which are similar to both the old English name for pike (luce) and the Latin ‘lucius’ or ‘lupus’.
This study has revealed greater population structure than was previously hypothesized to be present based on the expectations of 16th century introduction and the only previous account of genotypic variation (or more aptly the lack thereof) in Irish pike (Jacobsen et al., 2005). Collective evidence indicates that pike spatial structure within Ireland is meaningful, and warrants thoughtful consideration and examination of current habitats and populations. Management practices should remain precautionary and avoid breaching population barriers such as through translocations (Miller & Senanan, 2003; Tallmon et al., 2004), especially between to the two putative Irish strains. This is particularly significant when observing the structure assignments in the Grand Canal and the River Barrow (Fig. 2), which seem to indicate some mixing of the strains, probably as a result of the Barrowline Canal connection. Careful consideration should be given to assessing life history and ecological interactions, particularly between these units, and monitoring should continue using molecular genetic approaches, which may lead to the identification of further divergent populations. Furthermore, as lakes have been shown to maintain isolation-by-distance despite translocation practices, it may indicate that translocated individuals do not adapt well in the new habitat, which would be an important finding to take into consideration for minimizing inefficiencies of management strategies. Finally, recently developed genomic approaches should be used to monitor and investigate possible adaptive divergence in different environmental contexts (e.g. genomic scans or transcriptomic approaches), and add support to the evolutionary history and colonization pathway of the species.
As Irish systems come under increasing pressure, particularly from invasive species [e.g. curly waterweed (Lagarosiphon major), zebra mussel (Dreissena polymorpha), the freshwater clam Corbicula fluminea], attention must be paid to these newly discovered Irish pike population units. This research provides the first piece of evidence to help achieve that goal, and highlights the complexity inherent in natural systems, and the need for empirical knowledge as a basis for appropriate biodiversity management.
Acknowledgments
This study was fully funded through grant assistance generously provided by Inland Fisheries Ireland. The authors are grateful to all the IFI staff that contributed and facilitated the collection of samples for this project, particularly Paul McCloone and Will Corcoran. Thanks also go to the many anglers at the Cork and District Pike Angling Club, and the Irish Federation of Pike anglers, particularly John Chambers, for allowing us to sample their catch. Thanks to our many colleagues and collaborators, Robert Arlinghaus, Catherine Boisneau, Bill Brazier, Gareth Davies, Erik Eschbach, Janice Fletcher, Matthias Geiger, Rudy Gozlan, Kevin Grimes, Bernd Haenfling, Emilie Hardouin, Fran Igoe, Ross Macklin, Lovisa Wennerström and Ian Winfield, who contributed samples for the study. Thanks to Jens Carlsson, Andy Halpin, Shelia Hamilton-Dyer, Richard Hoffman, Evelyn Keaveney, Criostoir MacCarthaigh, Liam MacMathuna and Ken Whelan for their inputs on historical aspects of pike in Ireland. Sincere thanks to Chiara Benvenuto, Ilaria Coscia, Carlotta Sacchi, Maria Sala-Bozano and Alexia Massa-Gallucci for technical support, and to Jens Carlsson, Bernd Haenfling, Allan McDevitt, Albert Phillimore and two anonymous referees for helpful and constructive feedback on the manuscript. The authors confirm that no conflict of interest exists.
Biosketch
Debbi Pedreschi conducted this work as part of her PhD studies at the School of Biology & Environmental Sciences at University College Dublin. Her interests span ecology, genetics and conservation biology throughout the aquatic sciences.
All co-authors have a long-standing interest in fisheries biology and management, with particular foci on the mechanisms of population substructure, colonization and the history of Irish freshwater fish fauna.
Author contributions: M.K.Q., S.M., J.C. & M.O’G. conceived the project and coordinated sampling; D.P. collected and analysed the data and wrote the manuscript, with S.M. advising on data analyses and contributing to the writing.
Editor: Albert Phillimore
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Appendix S1 List of microsatellite loci tested.
Appendix S2 List and results of all diyabc scenarios tested.
References
- Aguilar A, Banks JD, Levine KF. Wayne RK. Population genetics of northern pike (Esox lucius) introduced into Lake Davis, California. Canadian Journal of Fisheries and Aquatic Sciences. 2005;62:1589–1599. [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A. Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlinghaus R, Matsumura S. Dieckmann U. Quantifying selection differentials caused by recreational fishing: development of modelling framework and application to reproductive investment in pike (Esox lucius. Evolutionary Applications. 2009;2:335–355. doi: 10.1111/j.1752-4571.2009.00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe F. Garrett S. The pike in Ireland: a (necessary) review. 2013. Available at: http://homepage.eircom.net/˜sheelin/IPSPikeInIre.html (accessed 6 March 2013) [Google Scholar]
- Beaumont MA. Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society B: Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Brooks AJ, Bradley SL, Edwards RJ, Milne GA, Horton B. Shennan I. Postglacial relative sea-level observations from Ireland and their role in glacial rebound modelling. Journal of Quaternary Science. 2007;23:175–192. [Google Scholar]
- Cambray JA. Impact on indigenous species biodiversity caused by the globalisation of alien recreational freshwater fisheries. Hydrobiologia. 2003;500:217–230. [Google Scholar]
- Carden RF, McDevitt AD, Zachos FE, Woodman PC, O’Toole P, Rose H, Monaghan NT, Campana MG, Bradley DG. Edwards CJ. Phylogeographic, ancient DNA, fossil and morphometric analyses reveal ancient and modern introductions of a large mammal: the complex case of red deer (Cervus elaphus) in Ireland. Quaternary Science Reviews. 2012;42:74–84. [Google Scholar]
- Casselman JM. Lewis CA. Habitat requirements of northern pike (Esox lucius. Canadian Journal of Fisheries and Aquatic Sciences. 1996;53(Supplement 1):161–174. [Google Scholar]
- Child AR, Burnell AM. Wilkins NP. The existence of two races of Atlantic salmon (Salmo salar L.) in the British Isles. Journal of Fish Biology. 1976;8:35–43. [Google Scholar]
- Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, Guillemaud T. Estoup A. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24:2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport JL, Sleeman DP. Woodman PC. Mind the gap: postglacial colonization of Ireland. Dublin: Special Supplement to the Irish Naturalists’ Journal; 2008. [Google Scholar]
- Dennis RL, Hardy PB. Dapporto L. Nestedness in island faunas: novel insights into island biogeography through butterfly community profiles of colonization ability and migration capacity. Journal of Biogeography. 2012;39:1412–1426. [Google Scholar]
- Desjardins MD. Molecular genetic variation in Muskellunge (Esox masquinongy) and northern pike (Esox lucius) from watersheds in the upper Midwest. IL: University of Illinois at Urbana-Champaign; 1996. [Google Scholar]
- Earl DA. vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Edwards RJ. Brooks AJ. The island of Ireland: drowning the myth of an Irish land-bridge? In: Davenport JJ, Sleeman DP, Woodman PC, editors; Mind the gap: postglacial colonisation of Ireland. Dublin: Special Supplement to the Irish Naturalists’ Journal; 2008. pp. 19–34. [Google Scholar]
- Evanno G, Regnaut S. Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Farran GP. Local names of Irish fishes (concluded) The Irish Naturalists’ Journal. 1946;13:370–376. [Google Scholar]
- Fitzmaurice P. The effects of freshwater fish introductions into Ireland. Rome: Food and Agriculture Organization of the United Nations; 1984. EIFAC Technical Paper, Vol. 42 (Supplement 1–2) [Google Scholar]
- Folke C, Carpenter S, Walker B, Scheffer M, Elmqvist T, Gunderson L. Holling CS. Regime shifts, resilience, and biodiversity in ecosystem management. Annual Review of Ecology, Evolution, and Systematics. 2004;35:557–581. [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001. Available at: http://www2.unil.ch/popgen/softwares/fstat.htm. [Google Scholar]
- Gozlan RE, Britton JR, Cowx I. Copp GH. Current knowledge on non-native freshwater fish introductions. Journal of Fish Biology. 2010;76:751–786. [Google Scholar]
- Griffiths D. The status of the Irish freshwater fish fauna: a review. Journal of Applied Ichthyology. 1997;13:9–13. [Google Scholar]
- Hamilton-Dyer S. Fish and bird bones. In: Hayden A, editor. Trim Castle, Co. Meath: excavations 1995–1998. Dublin: Stationery Office; 2011. pp. 411–418. Archaeological Monograph Series 6. [Google Scholar]
- Hammer Ø, Harper DAT. Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:4–9. [Google Scholar]
- Healy A. Pike (Esox lucius L.) in three Irish lakes. Dublin: Scientific Proceedings, Royal Dublin Society; 1956. [Google Scholar]
- Healy JA. Mulcahy MF. A biochemical genetic analysis of populations of the northern pike, Esox lucius L., from Europe and North America. Journal of Fish Biology. 1980;17:317–324. [Google Scholar]
- Heaney LR. Dynamic disequilibrium: a long-term, large-scale perspective on the equilibrium model of island biogeography. Global Ecology and Biogeography. 2001;9:59–74. [Google Scholar]
- Hoffmann RC. Strekfusz: a fish dish links Jagiellonian Krakow to distant waters. In: Górecki P, van Dusen N, editors. Central and Eastern Europe in the Middle Ages: a cultural history. London: Tauris; 2009. pp. 116–124. [Google Scholar]
- IFT. Inland Fisheries Trust (IFT) annual report. Dublin: Inland Fisheries Trust; 1966. [Google Scholar]
- IFT. Inland Fisheries Trust (IFT) annual report. Dublin: Inland Fisheries Trust; 1979. [Google Scholar]
- Igoe F. Threatened Irish freshwater fishes. Biology & Environment: Proceedings of the Royal Irish Academy. 2004;104:1–3. [Google Scholar]
- Jacobsen BH, Hansen MM. Loeschcke V. Microsatellite DNA analysis of northern pike (Esox lucius) populations: insights into the genetic structure and demographic history of a genetically depauperate species. Biological Journal of the Linnean Society. 2005;84:91–101. [Google Scholar]
- Jakobsson M. Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kennedy M. Irish pike investigations: 1. Spawning and early life history. Irish Fisheries Investigations Series A (Freshwater) 1969;5:4–33. [Google Scholar]
- King JL, Marnell F, Kingston N, Rosell R, Boylan P, Caffrey JM, FitzPatrick Ú, Gargan PG, Kelly FL, O’Grady MF, Poole R, Roche WK. Cassidy D. Ireland Red List No. 5: Amphibians, reptiles & freshwater fish. Dublin, Ireland: National Parks and Wildlife Service, Department of Arts, Heritage and the Gaeltacht; 2011. [Google Scholar]
- Laikre L, Miller LM, Palme A, Palm S, Kapuscinski AR, Thoresson G. Ryman N. Spatial genetic structure of northern pike (Esox lucius) in the Baltic Sea. Molecular Ecology. 2005;14:1955–1964. doi: 10.1111/j.1365-294X.2005.02570.x. [DOI] [PubMed] [Google Scholar]
- Launey S, Krieg F, Morin J. LaRoche J. Five new microsatellite markers for Northern pike (Esox lucius. Molecular Ecology Notes. 2003;3:366–368. [Google Scholar]
- Launey S, Morin J, Minery S. LaRoche J. Microsatellite genetic variation reveals extensive introgression between wild and introduced stocks, and a new evolutionary unit in French pike Esox lucius L. Journal of Fish Biology, 68 (Supplement B) 2006;19:3–216. [Google Scholar]
- Longfield AK. Anglo-Irish trade in the sixteenth century. London: G. Routledge; 1929. [Google Scholar]
- Lucentini L, Palomba A, Gigliarelli L, Sgaravizzi G, Lancioni H, Lanfaloni L, Natali M. Panara F. Temporal changes and effective population size of an Italian isolated and supportive-breeding managed northern pike (Esox lucius) population. Fisheries Research. 2009;96:139–147. [Google Scholar]
- Lynch J. Postglacial colonization of Ireland by mustelids, with particular reference to the badger (Meles meles L.) Journal of Biogeography. 1996;23:179–185. [Google Scholar]
- Maes GE, Van Houdt JKJ, De Charleroy D. Volckaert FAM. Indications for a recent Holarctic expansion of pike based on a preliminary study of mtDNA variation. Journal of Fish Biology. 2003;63:254–259. [Google Scholar]
- Maitland PS. Ireland’s most threatened and rare freshwater fish: an international perspective on fish conservation. Biology and Environment: Proceedings of the Royal Irish Academy. 2004;104:5–16. [Google Scholar]
- Martin AP. Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proceedings of the National Academy of Sciences USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Early evidence for wild animals in Ireland. Archäologie in Eurasien. 1999;6:355–371. [Google Scholar]
- McDevitt AD, Vega R, Rambau RV, Yannic G, Herman JS, Hayden TJ. Searle JB. Colonization of Ireland: revisiting ‘the pygmy shrew syndrome’ using mitochondrial, Y chromosomal and microsatellite markers. Heredity. 2011;107:548–557. doi: 10.1038/hdy.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LM. Kapuscinski AR. Historical analysis of genetic variation reveals low effective population size in a northern pike (Esox lucius. Population Genetics. 1997;147:1249–1258. doi: 10.1093/genetics/147.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LM. Senanan W. A review of northern pike population genetics research and its implications for management. North American Journal of Fisheries Management. 2003;23:297–306. [Google Scholar]
- Miller SA, Dykes DD. Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin D. A checklist of alien and cryptogenic aquatic species in Ireland. Aquatic Invasions. 2007;2:341–366. [Google Scholar]
- Nicod JC, Wang YZ, Excoffier L. Largiader CR. Low levels of mitochondrial DNA variation among central and southern European Esox lucius populations. Journal of Fish Biology. 2004;64:1442–1449. [Google Scholar]
- O’Grady MF. Delanty K. The ecology, biology and management of pike in Irish waters with particular reference to wild brown trout lake fisheries. Central Fisheries Board, Dublin: Internal Position Paper; 2008. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DP. Shipley P. microchecker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Piry S, Luikart G. Cornuet JM. Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M. Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raat AJ. Synopsis of biological data on the northern pike, Esox lucius Linnaeus, 1758. Rome: Food and Agriculture Organization of the United Nations; 1988. FAO Fisheries Synopsis, No. 30. [Google Scholar]
- Roche W, O’Grady M. Bracken JJ. Some characteristics of a pike Esox lucius L. population in an Irish reservoir. Hydrobiologia. 1999. pp. 217–223.
- Rosenberg NA. distruct: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rousset F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Seeb JE, Seeb LW, Oates DW. Utter FM. Genetic variation and postglacial dispersal of populations of northern pike (Esox lucius) in North America. Canadian Journal of Fisheries and Aquatic Sciences. 1987;44:556–561. [Google Scholar]
- Senanan W. Kapuscinski AR. Genetic relationships among populations of northern pike (Esox lucius. Canadian Journal of Fisheries and Aquatic Sciences. 2000;57:391–404. [Google Scholar]
- Stokes K, O’Neill K. McDonald RA. Invasive species in Ireland. Belfast: Unpublished report to Environment & Heritage Service and National Parks & Wildlife Service. Queen’s University Belfast; 2004. [Google Scholar]
- Tallmon DA, Luikart G. Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends in Ecology and Evolution. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Venturelli PA. Tonn WM. Diet and growth of northern pike in the absence of prey fishes: initial consequences for persisting in disturbance-prone lakes. Transactions of the American Fisheries Society. 2006;135:1512–1522. [Google Scholar]
- Wang J, Wang C, Qian L, Ma Y, Yang X, Jeney Z. Li S. Genetic characterization of 18 novel microsatellite loci in northern pike (Esox lucius L.) Genetics and Molecular Biology. 2011;34:169–172. doi: 10.1590/S1415-47572010005000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger B, Schulting R, Bradtmöller M, Clare L, Collard M, Edinborough K, Hilpert J, Jöris O, Niekus M, Rohling EJ. Wagner B. The catastrophic final flooding of Doggerland by the Storegga Slide tsunami. Documenta Praehistorica. 2008;35:1–24. [Google Scholar]
- Went AEJ. The pike in Ireland. The Irish Naturalists’ Journal. 1957;12:177–182. [Google Scholar]
- Wheeler A. The origin and distribution of the freshwater fishes of the British Isles. Journal of Biogeography. 1977;4:1–24. [Google Scholar]
- Woodman P, McCarthy M. Monaghan N. The Irish Quaternary fauna project. Quaternary Science Reviews. 1997;16:129–159. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 List of microsatellite loci tested.
Appendix S2 List and results of all diyabc scenarios tested.