Abstract
For proper development, cells must retain patterns of gene expression and repression through cell division. Repression via methylation of histone H3 on Lys27 (H3K27me) by Polycomb repressive complex 2 (PRC2) is conserved, but its transmission is not well understood. Our studies suggest that PRC2 represses the X chromosomes in Caenorhabditis elegans germ cells, and this repression is transmitted to embryos by both sperm and oocytes. By generating embryos containing some chromosomes with and some without H3K27me, we show that, without PRC2, H3K27me is transmitted to daughter chromatids through several rounds of cell division. In embryos with PRC2, a mosaic H3K27me pattern persists through embryogenesis. These results demonstrate that H3K27me and PRC2 each contribute to epigenetically transmitting the memory of repression across generations and during development.
Proper development depends on regulation of gene expression by packaging the genome into expressed and repressed chromatin domains. Our understanding of how that packaging is achieved and maintained is incomplete. Methylation of histone H3 on Lys27 (H3K27me) is a well-established mark of repressed chromatin that is generated by Polycomb repressive complex 2 (PRC2) in diverse phyla. In Drosophila, PRC2 and H3K27me maintain repression of important genes, including the Hox genes during somatic development and cell cycle genes during germline development (1–3). In mammals, H3K27me is also present on developmentally important genes in somatic and germ cells (4–6), and PRC2 serves numerous roles, including participation in X-chromosome inactivation and differentiation of embryonic stem cells (7, 8). In Caenorhabditis elegans, PRC2 is required only in germ cells where it participates in repression of the X chromosomes (9, 10). A critical question is how H3K27me-repressed chromatin states are passed from mother to daughter cells. One model is that H3K27-methylated histones are passed locally to the two daughter chromatids during DNA replication (11). Another model is that PRC2, but not methylated histones, is passed locally to daughter chromatids and newly establishes H3K27me after each round of DNA replication (12). We tested these models by examining cells containing or lacking PRC2 activity and with differentially H3K27-methylated chromosomes. We present evidence that H3K27-methylated histones transmit the memory of repression transgenerationally and short-term in embryos and that PRC2 promotes long-term memory during development.
C. elegans PRC2 is composed of MES-2 [ortholog of E(Z)/EZH2], MES-6 (ortholog of ESC/EED), and MES-3 (worm-specific subunit) and is essential for germline development but not somatic development (9, 13). PRC2 is maternally supplied to progeny and needed for the progeny’s primordial germ cells (PGCs) to survive and proliferate. In both naturally occurring sexes in C. elegans, hermaphrodites with two X chromosomes (XX) and males with one X (XO), germline development requires repression of the X chromosomes (14). PRC2-generated H3K27me3 participates in repressing the X chromosomes in XX germ cells: Repressive H3K27me3 is on all chromosomes but is concentrated on the X chromosomes in germ cells (13), and loss of PRC2 function causes germ lines to up-regulate numerous genes on the X (10). Whether XO germ lines require PRC2 repression has been unclear (15). We demonstrate that PRC2 is the main mode of X repression in XO worms, but that PRC2 function can be bypassed if the single X in XO worms experienced two other repressive conditions in the parental germ line: spermatogenesis and H3K9me.
The X chromosomes in germ cells are globally repressed during most stages of development except during late oogenesis (14, 16). Thus, a sperm-inherited X has a repressed transcriptional “history,” whereas an oocyte-inherited X lost repression before fertilization. We analyzed XO mes mutant males that inherited their X from these two different backgrounds (Fig. 1 and fig. S1). XO mes-3 mutants with an oocyte-inherited X (Xoo) had a severely underproliferated germ line, lacked sperm, and 0% were fertile. In striking contrast, XO mes-3 mutants with a sperm-inherited X (Xsp) generally had a well-proliferated germ line, and 73% were fertile. Similar results were observed for mes-2 and mes-6 mutants (fig. S1, A and D). We tested whether the gamete source of the X correlates with subsequent repression or expression of the X in male germ lines, using an X-linked lmn-1::GFP transgene that in wild-type worms is expressed in somatic cells and silenced in the germ line (17) (fig. S2). lmn-1::GFP was repressed in 91% of germ lines from XO (Xsp) mes-3 mutants but in only 5% of XO (Xoo) mes-3 mutants (Fig. 1 and fig. S2). These values are similar to the percentages of fertile XO mes-3 mutants (Fig. 1). Our findings suggest that fertility depends on continued X-chromosome repression in the germ line, which requires inheriting a repressed X. Although PRC2 likely modulates many aspects of gene expression in ways that are not essential for viability or fertility, our finding that XO worms with a sperm-inherited X do not require PRC2 suggests that the only essential role of PRC2 in worms is repression of the X chromosomes in germ cells.
Fig. 1. XO males with a sperm-inherited X do not require H3K27me and rely on H3K9me as an alternative mechanism of X repression.
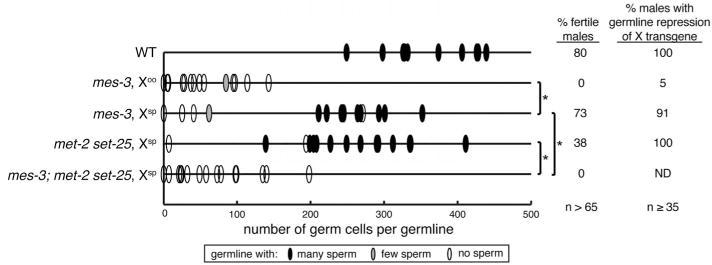
Analysis of germline proliferation, presence of sperm, fertility, and germline repression of an X-linked transgene in males with an oocyte-inherited X (Xoo) or sperm-inherited X (Xsp): wild type (WT), lacking H3K27me (mes-3), lacking H3K9me (met-2 set-25), or lacking both. Statistically significant differences between genotypes, *P < 0.01 Mann-Whitney U test. Also see figs. S1 and S2.
We reasoned that, in mutants lacking PRC2 and H3K27me, sperm may contribute to embryos an X chromosome repressed by an alternative mechanism, such as H3K9me. H3K27me is generally associated with developmentally regulated repression and H3K9me with repression via heterochromatin formation (18). To test if H3K9me is required for XO (Xsp) mes mutants to be fertile, we analyzed mes-3 mutants also mutant for met-2 and set-25 (lacking H3K9me) (19). With lack of both H3K27me and H3K9me, XO worms had severely underproliferated germ lines, and 0% were fertile (Fig. 1). Our findings show that H3K9 methylation provides an alternative mode of transmitting X repression to progeny. H3K9me likely enables Xsp to retain the heterochromatic state it experienced during spermatogenesis.
Our data suggest that the memory of X-chromosome repression can be inherited through sperm. As sperm mature in C. elegans, some histones are exchanged for sperm-specific histone variants and putative protamine-like proteins (20). H3K4me has been detected in C. elegans sperm (21). Our analysis revealed that H3K27me3 and H3K9me2 are also both present in mature sperm (fig. S3A) and transmitted to embryos via sperm (Fig. 2). To track the inheritance of sperm histone modifications in embryos, we analyzed embryos that could inherit histone modifications on paternally contributed chromosomes (P+) but not on maternally contributed chromosomes (M−) (Fig. 2A). In M−P+ one-cell embryos from oocytes lacking H3K27me3 (mes-3) fertilized by wild-type sperm with H3K27me3, we observed H3K27me3 on all sperm-contributed chromosomes but not on oocyte-contributed chromosomes (Fig. 2B). In M−P+ one-cell embryos from oocytes lacking H3K9me (met-2 set-25) fertilized by wild-type sperm, we observed H3K9me2 lightly on the sperm-contributed autosomes and heavily on the sperm-contributed X, similar to its pattern in the male germ line (22) (fig. S4C). These results demonstrate that the repressive histone marks H3K27me3 and H3K9me2 are transmitted to embryos by wild-type sperm.
Fig. 2. Repressive H3K27me3 is transmitted to embryos on sperm chromosomes and without PRC2 is transmitted through cell divisions.
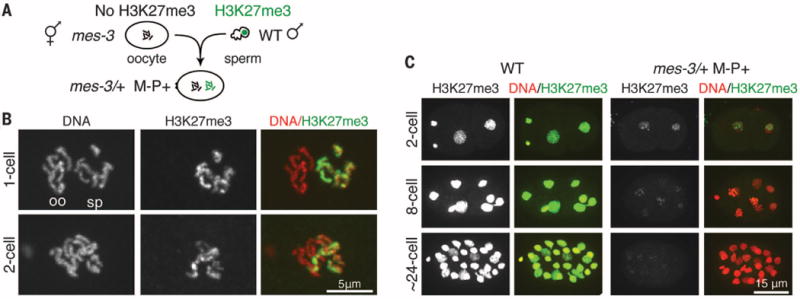
(A) Diagram of oocyte and sperm union to generate M−P+ embryos with H3K27me3 inherited from the sperm (P+) but not the oocyte (M−) and lacking maternal and paternal HMT. (B) Images of the two pronuclei in a one-cell embryo, and a diploid nucleus in a two-cell embryo. Merge panels show DNA (red) and H3K27me3 (green). The polar bodies [shown in (A)] identify the pronuclei as oocyte-derived (oo) or sperm-derived (sp). (C) Perdurance of paternally inherited H3K27me3 on a subset of chromosomes until the ~24-cell stage. Also see fig. S5, and fig. S6 for wild type.
mes M−P+ embryos lack a maternal load of PRC2 and do not inherit PRC2 by way of the sperm (fig. S3, B to D), which offers an opportunity to test if H3K27me3 on paternal chromosomes can be passed to daughter chromatids in the absence of histone methyltransferase (HMT) activity. In M−P+ embryos, H3K27me3 declined with age but persisted at easily visible levels on chromatin until the 16- to 24-cell stage, through at least four rounds of DNA replication (Fig. 2C and fig. S5C). H3K27me3 remained associated with a subset of chromosomes, likely sperm-derived chromosomes, and did not detectably spread to all chromosomes in each diploid nucleus (Fig. 2, B and C). We verified that two of the H3K27me3-stained chromosomes were sperm-derived (fig. S5, A and B). A similar pattern was observed for H3K9me2 in two-cell M−P+ embryos lacking MET-2 and SET-25 HMT activity (fig. S4C). Our results separate histone and HMT inheritance and demonstrate that modified histones can remain associated with DNA through several rounds of replication. These results also suggest that the memory of repression inherited on sperm chromosomes is transmitted through early embryo development.
To investigate propagation of repressive marks by ongoing methylation in the embryo, we generated embryos with HMT activity (maternally supplied in early embryos and transcribed from the embryonic genome in later-stage embryos) that contained histone marks on some but not all chromosomes. We analyzed M+P− embryos with H3K27me3 on oocyte-derived chromosomes but not on sperm-derived chromosomes (Fig. 3A). In such M+P− embryos, PRC2 maintained high levels of H3K27me3, and H3K27me3 remained restricted to one set of chromosomes throughout embryo development (Figs. 3 and 4). We verified H3K27me3-stained chromosomes to be oocyte-derived by using a maternally contributed III-X-IV fusion chromosome (Fig. 3). In the germ lines of larvae, we observed H3K27me3 gradually become detectable on all chromosomes as larval development progressed (Fig. 4). This spreading of H3K27me3 to all chromosomes is probably the result of germ cells turning on their transcriptional program and establishing repressed chromatin domains de novo. We speculate that H3K27me3-repressed chromatin is newly established during larval germline development each generation. In contrast to H3K27me3, maternally inherited H3K9me2 in the presence of H3K9 HMT activity spread to all chromosomes by the two-cell stage (fig. S4D). This difference in histone mark propagation may underlie C. elegans’ reliance on PRC2 for transgenerational regulation of repression. Both H3K27me and H3K9me provide short-term memory, but only PRC2 methylation of H3K27 provides long-term epigenetic memory to embryos.
Fig. 3. PRC2 maintains the memory of repression on gamete-of-origin chromosomes.
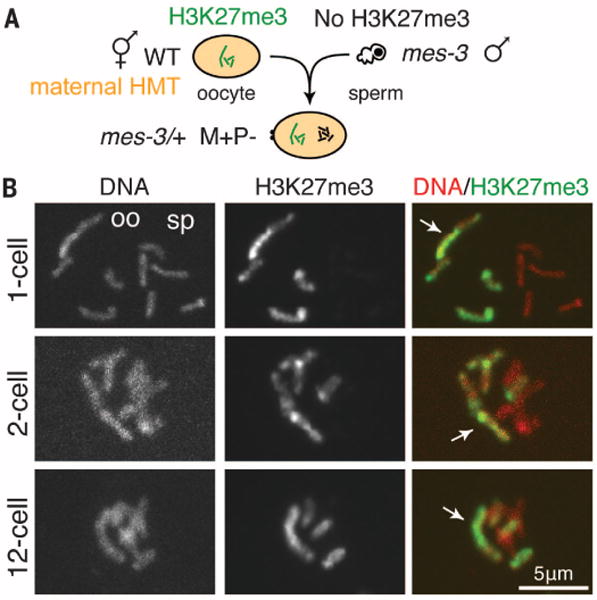
(A) Diagram of oocyte and sperm union to generate M+P− embryos with H3K27me3 inherited from the oocyte (M+) but not the sperm (P−) and containing maternal HMT. (B) Images (as described for Fig. 2B) of M+P− embryos with a III-X-IV fusion chromosome in the oocyte (M+) chromosome set (arrow). See fig. S6 for wild type.
Fig. 4. The memory of repression is maintained by PRC2 during embryogenesis and is newly established during germ cell development in larvae.
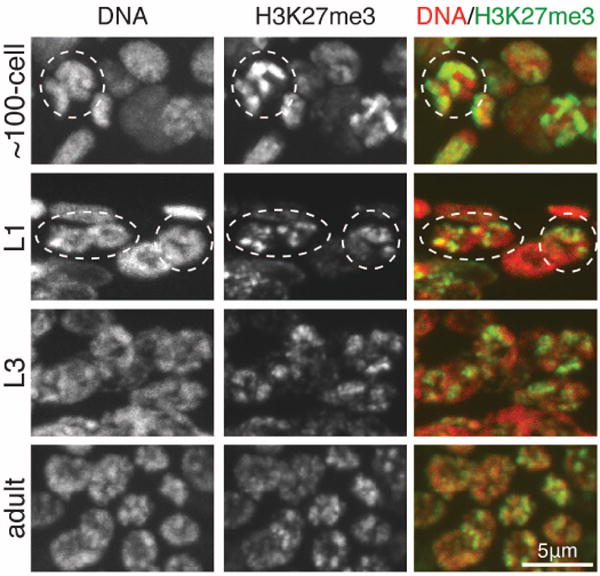
Images of worms generated as in Fig. 3. M+P− ~100-cell embryo (one nucleus circled), L1 larva (germ nuclei circled), and L3 and adult germ lines. See fig. S6 for wild type.
Our findings demonstrate that PRC2-generated H3K27 methylation is epigenetically transmitted across generations and, in C. elegans, is important for transmitting the memory of X-chromosome repression in the germ line. H3K27me3 is delivered to embryos on the chromosomes from both oocyte and sperm. In the absence of PRC2, H3K27me3 remains associated with gamete-of-origin chromosomes through several rounds of cell division (fig. S7), consistent with passage of modified histones in cis and locally during DNA replication (11, 23). In the presence of PRC2, H3K27me3 is propagated on gamete-of-origin chromosomes through embryogenesis (fig. S7). We speculate that worm PRC2 perpetuates paternally and maternally inherited patterns of repression through embryogenesis using a mechanism similar to mammalian PRC2: recruitment via its EED subunit to preexisting H3K27me3 and the resulting stimulation of HMT activity on neighboring unmethylated H3K27 (24). As in worms, other organisms may use a maternal supply of PRC2 to propagate inherited chromatin repression in embryos (25, 26); in worms, this transmits the memory of gene repression from parent germ lines to PGCs in progeny. As worm PGCs activate their transcription program in larvae, PRC2 likely catalyzes de novo H3K27me3 on all chromosomes in a pattern dictated by transcription factors, non-coding RNAs, nascent RNAs, and antagonism by marks of active chromatin (e.g., H3K4me and H3K36me) (10, 27–29).
Our studies in worms provide precedents for transmission of sperm H3K27me3 patterns to embryos and for important developmental consequences of paternal marking. Paternal inheritance could be similarly important in mammals, where H3K27me3 is present on sperm chromatin in a pattern that suggests involvement in embryogenesis (5, 30). Many of these H3K27me3 targets are also repressed in PGCs (30), where H3K27me3 increases just before epigenetic reorganization of the genome and may facilitate reacquisition of pluripotency by germ cells (31, 32). Thus, two common themes for PRC2 function in germ cells may be transmission of a memory of repression from parents to offspring and protection of the immortal and totipotent properties of germ cells.
Supplementary Material
Acknowledgments
We thank Strome laboratory members, N. Bhalla, and J. Tamkun for helpful discussions; H. Kimura for antibodies; and the Caenorhabditis Genetics Center for some strains (funded by NIH-ORIP-P40OD010440). This work was supported by NIH-T32GM008646, UCSC Dissertation-Year Fellowship, and Achievement Rewards for College Scientists Foundation Award to L.J.G. and NIH-R01GM034059 to S.S.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/345/6203/1515/suppl/DC1
Materials and Methods
References (33–40)
REFERENCES AND NOTES
- 1.Cao R, et al. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 2.Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 3.Iovino N, Ciabrelli F, Cavalli G. Dev Cell. 2013;26:431–439. doi: 10.1016/j.devcel.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki K, Tachibana M, Saitou M, Tokitake Y, Matsui Y. PLOS ONE. 2012;7:e46036. doi: 10.1371/journal.pone.0046036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammoud SS, et al. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein BE, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heard E. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Capowski EE, Martin P, Garvin C, Strome S. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Cell Reports. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzuolo C, Lo Sardo F, Diamantini A, Orlando V. PLOS Genet. 2011;7:e1002370. doi: 10.1371/journal.pgen.1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petruk S, et al. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender LB, Cao R, Zhang Y, Strome S. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- 14.Strome S, Kelly WG, Ercan S, Lieb JD. Cold Spring Harb Perspect Biol. 2014;6:a018366. doi: 10.1101/cshperspect.a018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garvin C, Holdeman R, Strome S. Genetics. 1998;148:167–185. doi: 10.1093/genetics/148.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly WG, et al. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beisel C, Paro R. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 19.Towbin BD, et al. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 20.Chu DS, et al. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arico JK, Katz DJ, van der Vlag J, Kelly WG. PLOS Genet. 2011;7:e1001391. doi: 10.1371/journal.pgen.1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bean CJ, Schaner CE, Kelly WG. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margueron R, Reinberg D. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margueron R, et al. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erhardt S, et al. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 26.Ohno K, McCabe D, Czermin B, Imhof A, Pirrotta V. Mech Dev. 2008;125:527–541. doi: 10.1016/j.mod.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko S, et al. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinn JL, et al. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitges FW, et al. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Brykczynska U, et al. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 31.Seki Y, et al. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 32.Saitou M, Yamaji M. Cold Spring Harb Perspect Biol. 2012;4:a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


