Abstract
Recently, extensive research has identified the non-invasive and cost-effective biomarker microRNA-106 (miR-106) in cancer detection. However, inconsistent results have prevented its usage in clinical. Therefore, we conducted this meta-analysis aimed to systematically determine diagnostic accuracy of miR-106 in distinguishing patients with cancer from cancer-free controls and further evaluate its value serving as a biomarker in clinical. We conducted a systematically literature search in databases (PubMed, web of science, Embase and the Cochrane Library) collecting relevant articles up to July 22th, 2014. The overall diagnostic accuracy of miR-106 was assessed by the following indexes: sensitivity, specificity, PLR, NLR and DOR. The SROC curve with AUC value was also generated for the assessment. Due to the significant heterogeneity, the random effects approach was chosen in our analysis and meta-regression was performed to explore the potential source of it. We also tested potential presence of publication bias using Deeks’ funnel plots test. Stata 12.0 statistical software was used for analysis in the present study. Overall, the 11 studies involving 756 cancer patients and 834 controls were considered eligible in our analysis. The results in our work showed that sensitivity of 0.57 (95% CI: 0.44-0.68) and specificity of 0.85 (95% CI: 0.72-0.92), with the under area AUC value of 0.75 (95% CI: 0.71-0.79) for miR-106 assay. Additionally, the combined PLR, NLR and DOR describing the discriminatory ability were 3.7 (95% CI: 2.2-6.2), 0.51 (95% CI: 0.42-0.62) and 7 (95% CI: 4-12) in the present analysis. The results in our meta-analysis showed that miR-106 had moderate accuracy in identifying cancer patients. Thus, further larger-scale prospective studies are needed to improve the diagnostic efficiency and explore the combination of miR-106 and other biomarkers with more pronounced accuracy.
Keywords: MicroRNA-106, cancer, meta-analysis, accuracy
Introduction
Cancer, known as one of the major public health challenge, is the leading cause of death worldwide, with about 12.7 million cancer new cases and 7.6 million cancer deaths occurred in 2008 [1]. Random biopsies are the most common used as well as the highly reliable method for diagnosis of cancer patients, but the potential sampling errors, invasive and expensive diagnostic procedures limit their usage in clinical [2,3]. The development of biomarkers, such as blood-based protein biomarkers (carcinoembryonic antigen CEA, carbohydrate antigen CA, or prostate specific antigen PSA), has enhanced early cancer detection, but the limited accuracy inhibits their use to distinguish aggressive from indolent tumors in early stage of cancer [4,5]. Due to the high mortality rate, low survival rate and lack of effective biological markers, more and more efforts have been made to find new means to supply existing detection methods.
Accumulating evidence has suggested the microRNAs (miRNAs, miRs) might serve as a novel and non-invasive biomarkers providing a natural pathway for controlling gene expression [6]. MiRNAs was small non-coding RNA molecules firstly founded in 1993 with size ranges from 19 to 25 [7]. MiRNAs may be involved in cancer progression as they have an influence on the degradation of mRNAs and the translation of many genes [8-12]. What’s more, numbers of studies have reported the potential association between abnormally miR-106 expression and the prevalence rate of cancer [13-16]. Further, the stability of miRNA in plasma, serum, feces and gastric juice is an outstanding goodness for potential biomarkers in diagnosis of cancer, which enables them to be detectable [17,18].
MicroRNA-106 (miR-106) of miRNA, has been reported to be abnormally expressed in various cancers, and thus has oncogenic activity in humans [19-21]. The diagnostic performance of miRNA-106 in cancer was first recognized when Tsujiura et al. demonstrated that the miR-106 was highly expressed in gastric cancer plasma with sensitivity of 0.79 and specificity of 0.63 [15]. Subsequent reports showed that the overexpression of miR-106 might also predict tumor stage, size, lymphatic and distant metastasis of the disease [22,23]. Additionally, Zhou et al. found that deregulated expression of miR-106a was associated significantly with survival in human colorectal cancer patients [24]. This evidence indicates that miR-106 might be strongly associated with cancer development, prediction and diagnosis, and may serving as a potential diagnostic tool in early cancer stage.
Though many studies showed that the miR-106 test could enhance the present cancer screening methods, disagreements still exist in identifying it as new biomarker of cancer as inconsistent diagnostic accuracy yielded by different single study. Considering the limits of the single study, different researchers adopted different study design, ascertained miR-106 expression using different specimen and explored the association of miR-106 with various cancers, we conduct this meta-analysis to evaluate the diagnostic efficacy of miR-106 and further discuss the possibility of it serving as diagnostic biomarker in clinical practice. As far as we know, there is no such work focusing on associations between miR-106 expression and cancers before.
Methods
Study selection
To identify relevant literature, we search several databases including PubMed, Web of science, Embase, and the Cochrane Library using the search strategy “microRNA-106” or “miR-106” or “miR-106a” or “miR-106b” and “cancer” or “tumor” or “carcinoma” or “neoplasms” and “sensitivity” or “specificity” or” accuracy” or “SROC curve” up to July 22th, 2014. Studies included should meet the criteria as follows: (1) evaluation of the miR-106 expression in diagnosing cancer patients; (2) case-control design with control group of benign disease patients or healthy people; (3) studies presenting sufficient data of sensitivity, specificity or enough information to calculate them. Additionally, studies were excluded if they were duplicate publications or conference report, editorials, letters or reviews or without sufficient data.
Data extraction and quality assessment
Two reviewers independently retrieved data from included studies. Data extracted including: study details (first author, published year and ethnicity), description of case and control group (sample size mean age and male ratio) and data for the final analysis [sensitivity, specificity, true positives (TP), false positives (FP), true negatives (TN) and false negatives (FN)].
The QUADAS-2, which is the standard in quality assessment for diagnostic accuracy studies, was used in the present meta-analysis to score the final quality of included studies [25].
Statistical analysis
Stata 12.0 statistical software was chosen for the analysis in present work [26]. The bivariate meta-analysis model was employed to summarize the sensitivity and specificity, so as to the parameters positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) [27]. The bivariate summary receiver operator characteristic (SROC) curve was also generated for the assessment. The AUC (area under the curve) of 1.0 indicates was recognized as a perfect performance indicating the discriminatory ability [28]. Sensitivity and specificity of miR-106 were presented as forest plots using the random effects approach. Due to the significant heterogeneity, meta-regression was performed to explore the potential source of it [29]. Sensitivity analysis was also performed to confirm the stability of the study. We also tested potential presence of publication bias using Deeks’ funnel plots test [30].
Results
Selection and characteristics of studies
Totally, 63 articles including 57 retrieved from the databases and 6 identified from a manual research were included based on the searching criteria, of which 13 articles duplication were excluded. After screening the titles, abstracts and key data for the rest 50 articles, 23 were excluded for 9 of them were reviews, 5 were not human studies and 9 were not relevant to cancer. The remaining 27 articles were for further full-text assessment, among which, 17 studies were excluded, including 7 studies not relevant to diagnosis, 4 not relevant to miRNAs and 5 without sufficient data. Finally, 10 articles including 11 studies were available in this meta-analysis [2,3,15,16,31-36]. The flow diagram of selection is shown in Figure 1.
Figure 1.
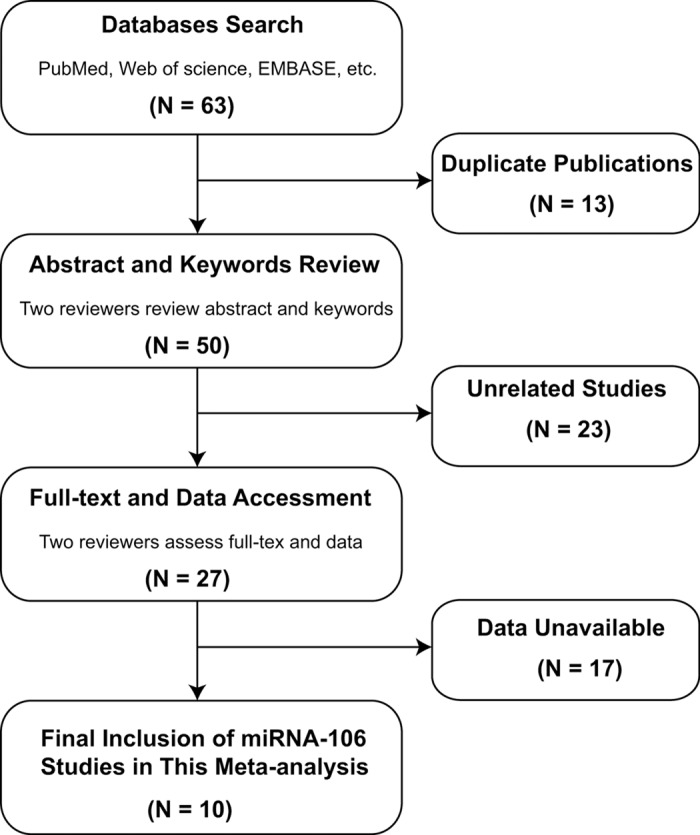
Flow chart of study selection.
Overall, the 11 studies published from 2010 to 2014 containing 756 cancer patients and 834 controls were available in our analysis. All the 11 diagnostic studies measured the miR-106 expression in serum (n = 3), plasma (n = 4), feces (n = 2), cerebrospinal fluid-CSF (n = 1) and gastric juice (n = 1) using real time quantitative polymerase chain reaction (qRT-PCR) methods. The cancer types analyzed in the studies included gastric cancer (n = 6), colorectal cancer (n = 4) and primary central nervous system lymphoma (n = 1). Among the 11 studies, 9 were conducted in Asian and 2 were conducted in Caucasian. We scored the 11 included studies at the guidance of QUADAS-2 assessment tool. All studies with score between 3 and 7 showed that majority of them got relativity high quality, which enhance the reliability of our analysis. The characteristics of and QUADAS-2 scores of each included study are listed in Table 1.
Table 1.
Characteristics of the included studies
| No. | First author | Year | Country | Ethnicity | Sample size | Male | Cancer | Specimen | Diagnostic power | QUADAS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||
| Case | Control | Case | Control | TP | FP | FN | TN | ||||||||
| 01 | Tsujiura et al [15] | 2010 | Japan | Asian | 69 | 30 | n.a. | n.a. | Gastric cancer | Plasma | 55 | 11 | 14 | 19 | 3 |
| 02 | Zhou et al [31] | 2010 | China | Asian | 90 | 27 | 62.3 | n.a. | Gastric cancer | Serum | 43 | 3 | 47 | 24 | 5 |
| 03 | Baraniskin et al [2] | 2011 | Germany | Caucasian | 23 | 30 | n.a. | n.a. | Lymphoma | CSF | 16 | 12 | 7 | 18 | 4 |
| 04 | Kuriyama et al [32] | 2012 | Japan | Asian | 69 | 126 | n.a. | n.a. | Colorectal cancer | Feces | 26 | 1 | 43 | 125 | 6 |
| 05 | Cai et al [3] | 2013 | China | Asian | 90 | 90 | 46.2 | 46.1 | Gastric cancer | Plasma | 59 | 18 | 31 | 72 | 6 |
| 06 | Cui et al [33] | 2013 | China | Asian | 42 | 99 | 64.2 | 53.2 | Gastric cancer | Gastric juice | 31 | 11 | 11 | 88 | 5 |
| 07 | Koga et al [34] | 2013 | Japan | Asian | 117 | 107 | 65 | 60 | Colorectal cancer | Feces | 40 | 3 | 77 | 104 | 5 |
| 08 | Luo et al [35] | 2013 | Germany | Caucasian | 80 | 144 | 68 | 62.5 | Colorectal cancer | Plasma | 15 | 7 | 65 | 137 | 6 |
| 09 | Shiotani et al [16] | 2013 | Japan | Asian | 64 | 64 | 67.9 | 68.4 | Gastric cancer | Serum | 36 | 19 | 28 | 45 | 6 |
| 62 | 70 | 67.8 | 66.7 | Gastric cancer | Serum | 47 | 34 | 15 | 36 | ||||||
| 10 | Zhang et al [36] | 2014 | China | Asian | 50 | 47 | 59 | 60 | Colorectal cancer | Plasma | 31 | 15 | 19 | 32 | 4 |
n.a.: Not available; QUADAS: Quality assessment of diagnostic accuracy studies; CSF: Cerebrospinal Fluid.
Diagnostic accuracy of miR-106
Sensitivity and specificity of 11 included studies presented in forests in Figure 2 indicated significant heterogeneity (I 2 = 91.07% for sensitivity and I 2 = 93.30% for specificity). Therefore, the random effects model was chosen in our analysis. Overall, the pooled sensitivity and specificity were 0.57 (95% CI: 0.44-0.68) and 0.85 (95% CI: 0.72-0.92) in our analysis. The SROC curve was shown in Figure 3A, with the AUC value of 0.75 (95% CI: 0.71-0.79), suggesting a moderate diagnostic accuracy of miR-106 in cancer detection. Additionally, the combined PLR was 3.7 (95% CI: 2.2-6.2), the NLR was 0.51 (95% CI: 0.42-0.62) and DOR was 7 (95% CI: 4-12) in the present analysis, suggesting that both the capacities of miR-106 differentiating patients with cancer from non-cancer individual were moderate. Since likelihood ratios are more valuable in clinical than sensitivity or specificity, and PLR > 10 or NLR < 0.1 is considered to be meaningful in clinical use, the results in our study indicated that miR-106 test have moderate ability to exclude or identify cancer but not powerful enough now in clinical use.
Figure 2.
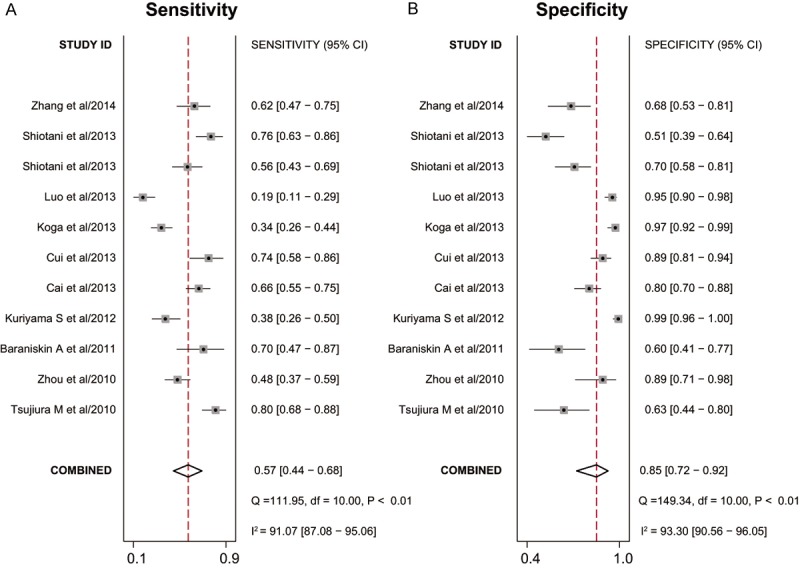
Forest plots of sensitivities and specificities from test accuracy studies of miR-106 in the diagnosis of cancer.
Figure 3.
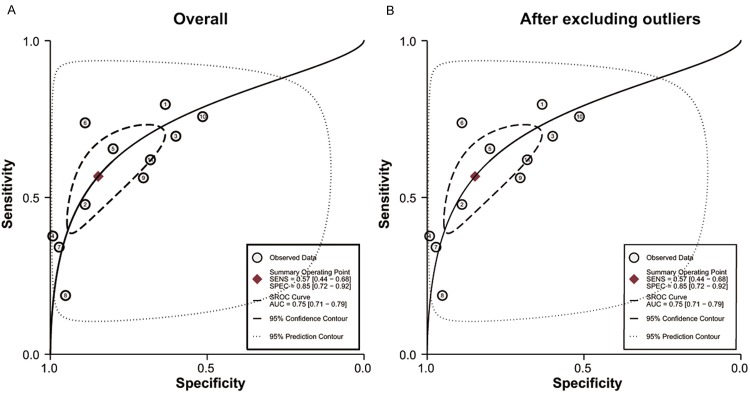
Summary receiver operating characteristic curves (SROC) of miR-106 describes the diagnostic performance (A: Before exclusion and B: After exclusion).
Meta-regression and sensitivity analysis
The heterogeneity is significant in our study. We considered 5 covariates (number of case, number of control, ethnicities, cancer types and specimen) may contribute to the heterogeneity and we performed meta-regression to assess their impact on sensitivity or specificity. The results in Figure 4 showed that neither number of case and control, nor ethnicities was the source of heterogeneity, but the cancer type and specimen have influence in specificity (P < 0.05). Sensitivity analysis (Figure 5) was conducted to find the impact of single individual to overall meta-analysis result if the data of outliners were removed. After two outliners [32,35] excluded from the test, the sensitivity increased from 0.57 to 0.63, specificity decreased from 0.85 to 0.79, PLR decreased from 3.7 to 3.1, NLR decreased from 0.51 to 0.47, DOR dropped from 7 to 6, and AUC had minimal change from 0.75 to 0.74 (Figure 3B). Although the exclusion changed sensitivity and specificity, the combined parameters remained stable, supporting the robustness of our work.
Figure 4.
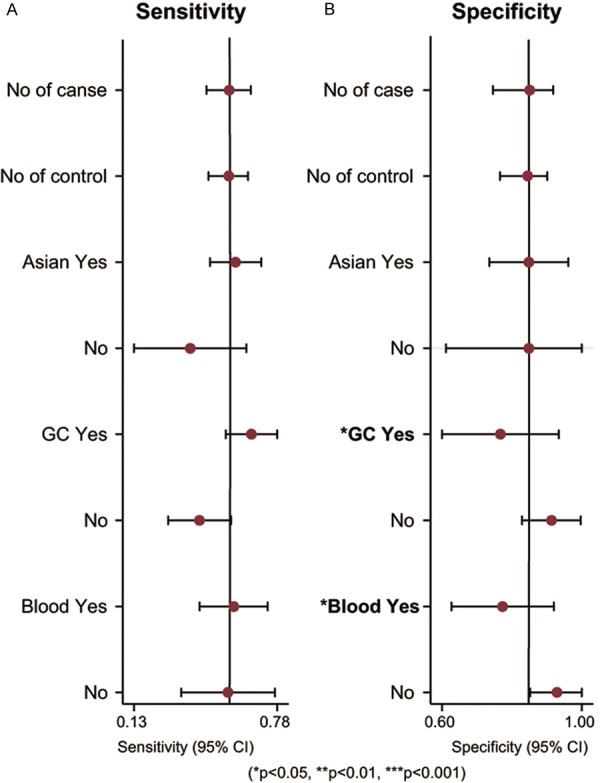
Meta-regression to explore the heterogeneity between studies.
Figure 5.
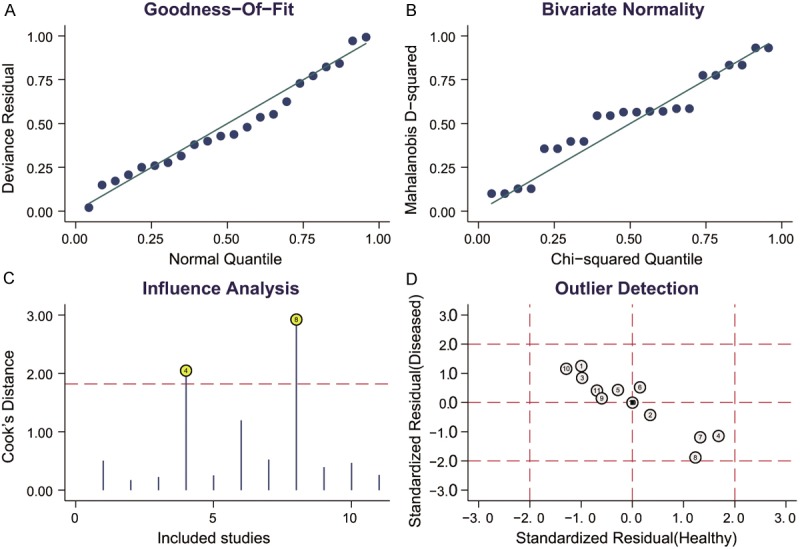
Sensitivity analysis. A: Goodness-of-fit; B: Bivariate normality; C: Influence analysis; D: Outlier detection.
Publication bias
Fagan’s nomogram in Figure 6 describes the possibility miR-106 assay to confirm or exclude cancer patients. In detail, for any people with a pre-test probability of 25% to have cancers, if the miR-106 test in cancer detection was positive, the post-test probability to have cancer would rise to 56%; while a negative result of miR-106 assay meaning the post-test probability would drop to 16% for the same people. Hence, miR-106 assay may play an important role as initial screening method for cancer. Deeks’ test was used in the meta-analysis to assess the publication bias. The P value of 0.28 suggested no publication bias exist.
Figure 6.
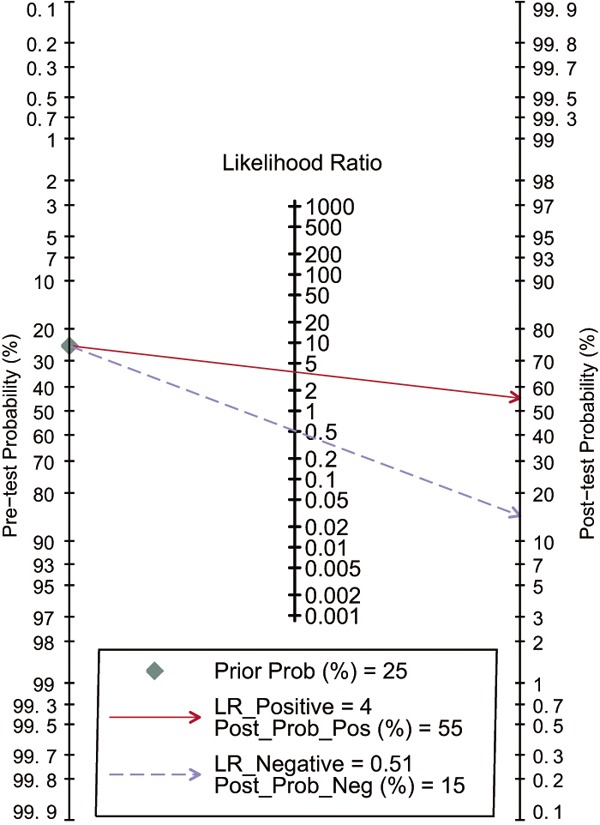
Fagan’s nomogram for assessment of post-test probabilities (PTPs).
Discussion
As cancer is still a major public health challenge worldwide, nowadays, early detection is the key point for patients to receive proper therapeutic treatment and thereby improve their health condition and prolong their life. Hence, more and more studies focus on novel strategies for early detection and prediction in cancer [37], and provide evidence that miRNAs may serve as promising biomarkers in early cancer diagnosis with high accuracy [37-39]. MiR-106 as a novel biomarker of early diagnosis has been reported to express abnormally in various cancers [2,3,31,32,36], but inconsistent conclusions about the diagnostic value of miR-106 for cancer patients exist. Since no such study systemically searches for the intersections of miR-106 expression and cancer, we conducted present meta-analysis collecting the results of miR-106 assay in cancer detection and further investigating candidate miRNA biomarkers in human cancers.
MiR-106b is located on chromosome 7q21 targeting genes p21 and E2F5, while miR-106a is a member of the miR-106a-92 cluster located at Xq26.2 [21,40,41]. Although miR-106 has found to be associated with cancer, its mechanism in caner development is still unclear. Yao et al. once explored the role of miR-106 on gastric cancer cell growth, proliferation, migration and invasion and further explained the possible effect of miR-106 in cancer [42]. They used flow cytometry analyses to test miR-106 expression on cell cycle and found that overexpressed miR-106b accelerated cells proliferation as a catalyst through the way shortened G0/G1 phase, subsequently facilitated their entry into the mitosis proliferation, thereby speeded up cell cycle progression. They also used trans well chamber assay to find the percentage of cell migration and invasion between miR-106 over expression group and the control group, but found the similar amount, indicating that miR-106 can’t affect migration and invasion of gastric cancer cell. Therefore, Yao et al. guessed that the over expression miR-106 speeded up cell cycle progression, and promote gastric cancer progress which may is possible mechanisms of miR-106b for cancers [42].
Our results showed that, miR-106 as a diagnostic biomarker in cancer detection yielded a pooled sensitivity of 0.57, specificity of 0.85, and AUC of 0.75, indicating a moderate overall accuracy. Ranging from 0 to infinity, DOR of 1 indicates that the assay can’t distinguish cancer and non-cancer individual. The discrimination ability of miR-106 assay for cancer diagnosis was not high enough as DOR was only 7 in our study. The PLR was 3.7 in the present meta-analysis, meaning that the probability of the patient having cancer is about 56% if the miR-106 test is positive according to Fagan’s nomogram. Actually, the PLR in our study was not high enough to confirm cancer patients and so as to NLR, was not low enough to excluded health individuals. Thus, there is a long way to go before the application of miR-106 into clinical.
Though difficulty still remains for miR-106 as biomarkers in cancer diagnosis, there are several points we can do to supply the miR-106 assay. Firstly, as our data suggested that with limited accuracy to confirm cancer patients or exclude healthy ones miR-106 may not be eligible to undertake early diagnosis. Many studies demonstrated that miRNA panel assays yielded higher accuracy than single one. Xavier et al. once demonstrated four miRNAs (miR-222, -328, -197, and -21) combined yielded 86% accuracy in differentiating malignant from benign indeterminate thyroid lesions compared the 78% accuracy of single miR-21 assay [43]. When tested the diagnostic performance of miR-106b in gastric cancer, Zhou et al. also investigated the combination of miR-106a and miR-17 and found that the combined group made an improvement in sensitivity from 0.48 to 0.63 [31]. As the sensitivity was relativity moderate in our analysis, the combination of multiple miRNAs may be the right way to solving the problem. Moreover, it has been reported that combination of protein-biomarkers and miRNAs may be a novel potential tool for cancer detection and prediction. MiR-141 and carcinoembryonic antigen (CEA) are both widely used biomarkers for CRC, Cheng et al. found that a combination of miR-141 and CEA was more sensitive than either marker used alone so that reduced the misdiagnosis rate of the CRC patients [44]. As far as we know, there are few researches focusing on the combination of miR-106 and other protein-biomarkers, since it may be an effective way to improve the diagnostic accuracy of miR-106, more attempts are needed in the further.
There are several drawbacks we should be mentioned in our study. Firstly, since the purpose of our analysis was to explore the possible role of miR-106 in cancer, the lack of cancer type in our analysis restricted the research extent as we only concluded gastric cancer, colorectal cancer and primary central nervous system lymphoma in the study. Future more fundamental research should investigate the association between miR-106 and other cancer types, which could promote the clinical application of miR-106. Secondly, as the standard process for miRNA detection has not been confirmed, different standard references, inconsistent cut-off value and different qRT-PCR methods were selected by our included studies, which may provide contradictory results. Thirdly, as the link between cancer and miR-106 has been noticed for recent year, limited studies evaluated the diagnostic value of miR-106 in cancer detection, resulted in sample size collected in our meta-analysis and the small-study effects inescapable. So further validate studies of miR-106 in large cohort are necessary to strengthen our conclusion.
Nevertheless, we also need to stress advantages in our meta-analysis. First, our meta-analysis is more reliable than single studies to some extent, as we systematically evaluated the overall diagnostic performance of miR-106 with cancers, and we performed meta-regression and sensitivity analysis to confirm the comparatively stability of our study. Second, although the current evidence suggests that the pooled accuracy of miR-106 was moderate with good specificity and relatively not so well sensitivity for cancer detection, we put forward the supplement using the combination of miR-106 and other miRNAs or protein-biomarkers to improve the accuracy of miR-106 detection. Finally, this is the first available meta-analysis providing valuable data of association between miR-106 and various cancers, which will be helpful in the clinical application of it.
In conclusion, the sensitivity, specificity of miR-106 in cancer is moderate, larger-scale prospective studies are needed to improve the diagnostic efficiency and explore the biomarkers combination with more pronounced accuracy in the further.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zollner H, Reinacher-Schick A, Schmiegel W, Hahn SA, Schroers R. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 3.Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol. 2013;30:452. doi: 10.1007/s12032-012-0452-0. [DOI] [PubMed] [Google Scholar]
- 4.Lumachi F, Brandes AA, Ermani M, Bruno G, Boccagni P. Sensitivity of serum tumor markers CEA and CA 15-3 in breast cancer recurrences and correlation with different prognostic factors. Anticancer Res. 2000;20:4751–4755. [PubMed] [Google Scholar]
- 5.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers--blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 6.Bartels CL, Tsongalis GJ. [MicroRNAs: novel biomarkers for human cancer] . Ann Biol Clin (Paris) 2010;68:263–272. doi: 10.1684/abc.2010.0429. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, Zhang MY, Sun Y, Huang BJ, Chen M, He QM, Jiang N, Chen L, Cho WC, Yun JP, Zeng J, Liu LZ, Li L, Guo Y, Wang HY, Ma J. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–641. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 13.Horner PJ, Harris JR. A herpes simplex skin ulcer in a patient with AIDS--an unusual presentation. Int J STD AIDS. 1990;1:288–289. doi: 10.1177/095646249000100413. [DOI] [PubMed] [Google Scholar]
- 14.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, Inoue K, Uedo N, Iishi H, Haruma K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer. 2013;109:2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 20.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 21.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 22.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 23.Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y, Li D, Zhong J. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009;400:97–102. doi: 10.1016/j.cca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Diaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Pena C, Rodriguez R, Munoz C, Garcia F, Bonilla F, Dominguez G. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 25.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 26.Deville WL, Buntinx F, Bouter LM, Montori VM, de Vet HC, van der Windt DA, Bezemer PD. Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol. 2002;2:9. doi: 10.1186/1471-2288-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell AJ, Vaze A, Rao S. Clinical diagnosis of depression in primary care: a meta-analysis. Lancet. 2009;374:609–619. doi: 10.1016/S0140-6736(09)60879-5. [DOI] [PubMed] [Google Scholar]
- 28.Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Hansen ES, Tauris P. Methicillin-induced nephropathy. A case with linear deposition of IgG and C3 on the tubular-basement-membrane. Acta Pathol Microbiol Scand A. 1976;84:440–442. [PubMed] [Google Scholar]
- 30.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 2012;88:709–717. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- 32.Kuriyama S, Hamaya Y, Yamada T, Sugimoto M, Osawa S, Sugimoto K, Miyajima H, Kanaoka S. Fecal microrna assays as a marker for colorectal cancer screening. Gastroenterology. 2012;142:S770. [Google Scholar]
- 33.Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, Xiao B, Xia T, Yu X, Le Y, Guo J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119:1618–1626. doi: 10.1002/cncr.27903. [DOI] [PubMed] [Google Scholar]
- 34.Koga Y, Yamazaki N, Yamamoto Y, Yamamoto S, Saito N, Kakugawa Y, Otake Y, Matsumoto M, Matsumura Y. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013;22:1844–1852. doi: 10.1158/1055-9965.EPI-13-0512. [DOI] [PubMed] [Google Scholar]
- 35.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Meng L, Fan Z, Liu B, Pei Y, Zhao Z. [Expression of plasma miR-106a in colorectal cancer and its clinical significance] . Nan Fang Yi Ke Da Xue Xue Bao. 2014;34:354–357. [PubMed] [Google Scholar]
- 37.Fan C, Chen C, Wu D. The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013;40:3389–3394. doi: 10.1007/s11033-012-2416-z. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, Hernando E, Wei JJ. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol. 2012;228:204–215. doi: 10.1002/path.4000. [DOI] [PubMed] [Google Scholar]
- 39.Vosa U, Vooder T, Kolde R, Vilo J, Metspalu A, Annilo T. Meta-analysis of microRNA expression in lung cancer. Int J Cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Shi XB, Nori D, Chao CK, Chen AM, Valicenti R, White Rde V. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate. 2011;71:567–574. doi: 10.1002/pros.21272. [DOI] [PubMed] [Google Scholar]
- 41.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Schira J, Muller HW, Wernet P. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 2011;6:e16138. doi: 10.1371/journal.pone.0016138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao YL, Wu XY, Wu JH, Gu T, Chen L, Gu JH, Liu Y, Zhang QH. Effects of microRNA-106 on proliferation of gastric cancer cell through regulating p21 and E2F5. Asian Pac J Cancer Prev. 2013;14:2839–2843. doi: 10.7314/apjcp.2013.14.5.2839. [DOI] [PubMed] [Google Scholar]
- 43.Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, Buitrago D, Cooper D, Zeiger MA, Zarnegar R, Elemento O, Fahey TJ 3rd. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]


