Abstract
Background: The aim of the current study was to evaluate the association of PAI-1 4G/5G polymorphism with coronary artery disease (CAD) risk using a meta-analysis. Methods: All eligible studies were identified through a search of PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), Database of Chinese Scientific and Technical Periodicals, and China Biology Medical literature database (CBM) before June 2014. The association between the PAI-1 4G/5G polymorphism and CAD risk was estimated by odds ratio (OR) and 95% confidence interval (CI). Results: A total of 72 studies including 23557 cases and 21526 controls were eventually collected. The PAI-1 4G/5G polymorphism was significant associated with CAD risk in overall population (OR=1.19, 95% CI 1.10-1.28, P < 0.00001). The combination of adjusted ORs for CAD was 1.20 (95% CI 1.03-1.40, P=0.02). This polymorphism was associated with CAD risk in Caucasians (OR=1.10, 95% CI 1.02-1.19, P=0.01) and Asians (OR=1.46, 95% CI 1.21-1.75, P < 0.0001). This polymorphism significantly increased MI risk (OR=1.15, 95% CI 1.06-1.25, P=0.001). In the subgroup analysis by age, this polymorphism was significantly associated with early-onset CAD risk (OR=1.21, 95% CI 1.02-1.43, P=0.03). In the gender subgroup analyses, a statistically significant association was found in male CAD patients (OR=1.10, 95% CI 1.01-1.20, P=0.04). Both T2DM patients and non-T2DM patients carrying 4G allele showed increased CAD risks (OR=2.23, 95% CI 1.27-3.92, P=0.005 and OR=1.64, 95% CI 1.19-2.25, P=0.002, respectively). Conclusions: This meta-analysis suggested that PAI-1 4G/5G polymorphism was a risk factor for CAD.
Keywords: Coronary artery disease, plasminogen activator inhibitor-1, meta-analysis, genetic
Introduction
Cardiovascular diseases, the first cause of death in the Western countries are a real common health problem. Despite the high responsibility of factors such as high level of total cholesterol, systemic hypertension, smoking, type 2 diabetes (T2DM) in coronary artery disease (CAD), evidence from family studies show that genetic factors contribute to the predisposition to CAD.
The plasminogen activator inhibitor-1 (PAI-1), a 52 kDa glycoprotein belong to the serine proteinase inhibitor super family, is a multifaceted proteolytic factor. It is the principal inhibitor of tissue and urinary plasminogen activators, and therefore constitutes an important regulatory protein in fibrinolysis [1]. Impaired fibrinolysis due to high PAI-1 activity has been shown to be associated with an increased risk of thrombotic events [2]. PAI-1 overexpression may also promote development of weak plaques with thin fibrous caps by inhibiting both u-PA receptor- and integrin-mediated cell adhesion and migration [3]. In addition, increased plasma PAI-1 levels have been reported in survivors of myocardial infarction (MI) compared with the general population [4]. Therefore, PAI-1 might play an important role in the pathogenesis of CAD.
The PAI-1 gene, located in 7q21.3-22, spans 12.3 kb and contains 9 exons and 8 introns. The polymorphism of the 4G/5G gene is located in the PAI-1 gene promoter region. The most commonly studied functional variant in the PAI-1 gene is the guanine deletion polymorphism at position -675 nucleotides relative to the transcription start site (rs1799889). The PAI-1 -675 4G allele has higher transcriptional activity than the PAI-1 -675 5G allele and homozygous possession of -675 4G is associated with higher plasma PAI-1 levels [5]. A number of papers investigated the association between this polymorphism and CAD risk. However, the results remained inconclusive [6-73]. Metaanalysis is a useful method for investigating associations between genetic factors and diseases, because a quantitative approach is used to combine the results from different studies on the same topic, thereby providing more reliable conclusions. Thus, we performed a meta-analysis to clarify the association of PAI-1 4G/5G polymorphism with CAD.
Methods
Publication search
A computerized literature search was performed to identify the relevant studies from five electronic databases including PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), Database of Chinese Scientific and Technical Periodicals, and China Biology Medical literature database (CBM). The search terms were used as follows: (coronary artery disease or coronary heart disease or atherosclerosis) and (polymorphism or variant or mutation) and (plasminogen activator inhibitor-1 or PAI-1). All searched studies were retrieved and the bibliographies were checked for other relevant publications.
Inclusion and exclusion criteria
The following criteria were used for the literature selection: first, studies should concern the association of PAI-1 4G/5G polymorphism with CAD risk; second, studies must be observational studies (case-control or cohort); third, papers must offer the size of the sample, odds ratios (ORs) and their 95% confidence intervals (CIs), the genetic distribution or the information that can help infer the results. Studies were excluded if one of the following existed: first, studies were not relevant to PAI-1 or CAD; second, the design based on family or sibling pairs; third, sample size or OR and 95% CI were not reported; fourth, reviews and abstracts. As for the studies from the same institution, only the one with the largest sample size was included. No language restrictions were imposed.
Data extraction
Data were extracted by two authors independently. If encountered the conflicting evaluations, an agreement was reached following a discussion; if could not reached agreement, another author was consulted to resolve the debate. The following information was extracted from each study: first author, year of publication, original country, ethnicity, endpoint, age, gender, sample size, covariates.
Statistical analysis
OR and 95% CI were employed to evaluate the strength of the association between 4G/5G polymorphism and the risk of CAD in dominant model. Departure from Hardy-Weinberg equilibrium (HWE) in controls was tested by the chi-square test. The Q statistic and the I2 statistic were used to assess the degree of heterogeneity among the studies included in the meta-analysis. The random-effects model was used to estimate the pooled OR (the DerSimonian and Laird method). Subgroup analyses were carried out by ethnicity, endpoint, age, gender and T2DM. We defined the early-onset CAD was the first event before 50 years old. Sensitivity analysis was performed through sequentially excluded individual studies to assess the stability of the results. The potential publication bias was examined visually in a funnel plot of log [OR] against its standard error (SE), and the degree of asymmetry was tested using Egger’s test [74].
All statistical tests were performed using Revman 5.1 software (Nordic Cochrane Center, Copenhagen, Denmark) and STATA 11.0 software (Stata Corporation, College Station, TX, USA). A P value < 0.05 was considered statistically significant, except for tests of heterogeneity where a level of 0.10 was used.
Results
Study characteristics
The flow chart in Figure 1 summarizes this literature review process. In this current study, a total of 68 eligible studies met the inclusion criteria [6-73]. Four articles reported two cohorts, and each cohort was considered as a case-control study. Finally, a total of 72 studies involving 23557 cases and 21526 controls were included in this meta-analysis. There were 24 studies performed using Asians, 45 studies using Caucasians, and 2 studies using Africans. Thirteen studies included only male CAD patients, and four studies included female CAD patients. Ten studies reported adjusted ORs and CIs and four studies reported the information of T2DM. Three studies were not in HWE. The characteristics of each study included in this meta-analysis are presented in Table 1. Genotype frequencies and HWE examination results are listed in Table 2.
Figure 1.
Flow of study identification, inclusion, and exclusion.
Table 1.
Characteristics of the included studies
| First author | Year | Country | Ethnicity | Endpoint | Age of patients | Female (%) | Case (n) | Control (n) | Adjustment for covariates |
|---|---|---|---|---|---|---|---|---|---|
| Dawson | 1993 | Sweden | Caucasian | MI | < 45 (39.9±0.4) | 14 | 107 | 73 | NA |
| Eriksson | 1995 | Sweden | Caucasian | MI | < 45 | 0 | 93 | 100 | NA |
| Ye | 1995 | France/UK | Caucasian | MI | 25-64 | 0 | 476 | 601 | NA |
| Mansfield | 1995 | UK | Caucasian | CAD | 61-70 | 0 | 38 | 122 | NA |
| Burzotta | 1997 | Italy | Caucasian | MI | > 45 (59±7) | 25 | 108 | 175 | NA |
| Ridker | 1997 | USA | Caucasian | MI | 62.9±8.8 | 0 | 374 | 495 | NA |
| Ossei-Gerning | 1997 | UK | Caucasian | MI | 59.8 | NA | 158 | 150 | NA |
| Iwai | 1998 | Japan | Asian | MI | 59.3±10.3 | 22.5 | 204 | 148 | NA |
| Kohler | 1998 | Finland | Caucasian | MI | 57-59 | 27.7 | 181 | 188 | NA |
| Margaglione | 1998 | Italy | Caucasian | MI | 22-65 | 23 | 198 | 981 | NA |
| Pastinen | 1998 | Finland | Caucasian | MI | 58.1±4.9 | 19.2 | 151 | 150 | NA |
| Junker | 1998 | Germany | Caucasian | MI | 38.6±4.4 | 0 | 241 | 179 | NA |
| Sugano | 1998 | Japan | Asian | MI | 63.1±9.2 | 12.1 | 66 | 62 | NA |
| Ardissino | 1999 | Italy | Caucasian | MI | 40.7±4.1 | 7.5 | 200 | 200 | NA |
| Anderson 1 | 1999 | USA | Caucasian | MI | 63.7±11.6 | 23 | 375 | 978 | NA |
| Anderson 2 | 1999 | USA | Caucasian | CAD | 62.5±10.9 | 20 | 898 | 329 | NA |
| Doggen | 1999 | Netherlands | Caucasian | MI | 56.1±9.0 | 0 | 331 | 302 | NA |
| Gardemann 1 | 1999 | Germany | Caucasian | CAD | 62.7 | 0 | 1791 | 594 | NA |
| Gardemann 2 | 1999 | Germany | Caucasian | MI | 62.2 | 0 | 1214 | 1351 | NA |
| Grancha | 1999 | Spain | Caucasian | CAD | 56±5 | 100 | 41 | 62 | NA |
| Beneš | 2000 | Czech | Caucasian | CAD | 49.5±4.5 | 0 | 175 | 222 | NA |
| Canavy | 2000 | France | Caucasian | CAD/MI | 55 | 22 | 244 | 244 | NA |
| Hooper | 2000 | USA | African | MI | 60.7±9.2 | 53 | 110 | 185 | NA |
| Mikkelsson | 2000 | Finland | Caucasian | MI | 47.9±9 | 0 | 68 | 164 | NA |
| Song | 2000 | Korea | Asian | CAD | 60.7±9.2 | 37.3 | 158 | 139 | NA |
| Fu | 2000 | China | Asian | MI | 51.3±6.7 | 42.5 | 87 | 92 | NA |
| Viitanen | 2001 | Finland | Caucasian | CAD | 56±1 | 40.7 | 118 | 110 | NA |
| Dai | 2001 | China | Asian | CAD/MI | 57±9 | NA | 250 | 95 | NA |
| Fu | 2001 | China | Asian | CAD | 66±10 | 50 | 123 | 172 | NA |
| Shang | 2001 | China | Asian | CAD | NA | NA | 38 | 80 | NA |
| Ortlepp | 2002 | Germany | Caucasian | CAD | 58±12.8 | 68 | 100 | 100 | NA |
| Yamada | 2002 | Japan | Asian | MI | 62.5±10.8 | 100 | 589 | 704 | NA |
| Guan | 2002 | China | Asian | CAD | 34-90 | 38.1 | 126 | 121 | NA |
| Li | 2002 | China | Asian | CAD | 60 ± 8 | 33.3 | 36 | 16 | NA |
| ATVBISG | 2003 | Italy | Caucasian | MI | < 45 | 12.3 | 1210 | 1210 | Smoking, diabetes, hypertension, family history, |
| body mass index, hypercholesterolemia, alcohol, | |||||||||
| cocaine, physical exercise | |||||||||
| Crainich | 2003 | USA | Caucasian | MI | 73.5±5.5 | 40.2 | 264 | 753 | NA |
| Juhan-Vague | 2003 | Europe | Caucasian | MI | < 60 | 0 | 483 | 507 | NA |
| Leander 1 | 2003 | Sweden | Caucasian | MI | 58.3±7.1 | 0 | 851 | 1051 | Age, residential area |
| Leander 2 | 2003 | Sweden | Caucasian | MI | 61.5±6.8 | 100 | 361 | 505 | Age, residential area |
| Petrovič | 2003 | Slovenia | Caucasian | MI | 58.3±11.3 | 33.8 | 154 | 194 | NA |
| Zhan | 2003 | China | Asian | MI | 67.1±10.4 | 21.4 | 56 | 83 | NA |
| Ding 1 | 2003 | China | Asian | CAD | NA | NA | 60 | 109 | Age, body mass index, family history |
| Ding 2 | 2003 | China | Asian | CAD | NA | NA | 49 | 63 | Age, body mass index, family history |
| Wang | 2003 | China | Asian | CAD | 59±12 | 24 | 67 | 30 | NA |
| Zhai | 2003 | China | Asian | CAD | 62.8±9 | 32 | 122 | 172 | NA |
| Tobin | 2004 | UK | Caucasian | MI | 61.9±9.2 | 32 | 547 | 505 | NA |
| Pegoraro | 2005 | Indian | Asian | MI | < 45 | NA | 195 | 300 | NA |
| Whiting | 2005 | USA | Caucasian | CAD | NA | NA | 881 | 261 | Diabetes, family history |
| Zak | 2005 | Poland | Caucasian | CAD | 45.9±6 | 34.9 | 146 | 121 | NA |
| Agirbasli | 2006 | Turkey | Caucasian | CAD | < 55 | 20 | 100 | 100 | NA |
| Su | 2006 | China | Asian | CAD | 54.5±8.9 | 21.6 | 812 | 931 | Age, sex, BMI, HDL-C, LDL-C, hypertension, diabetes, and smoking |
| Xia | 2006 | China | Asian | CAD | 57.7±8.1 | 28.6 | 166 | 63 | NA |
| Morange | 2007 | France | Caucasian | MI | 51.91±5.44 | 0 | 510 | 543 | NA |
| Sampaio | 2007 | Brazil | Caucasian | MI | 34.4±4.9 | 38.1 | 115 | 104 | Age, gender, ethnic background, hypertension, diabetes, hypercholesterolemia, obesity, smoking, stress, and sedentary lifestyle |
| Taymaz | 2007 | Turkey | Caucasian | CAD | NA | NA | 115 | 41 | NA |
| Onalan | 2008 | Turkey | Caucasian | MI | 59±11 | 19.9 | 156 | 281 | NA |
| Saely | 2008 | Austria | Caucasian | CAD | NA | NA | 406 | 266 | Age, gender, BMI, smoking, hypertension, LDL cholesterol, HDL cholesterol, triglycerides, and use of aspirin, statins, angiotensin converting enzyme inhibitors and beta adrenoreceptor blocking agents |
| Sarecka | 2008 | Poland | Caucasian | CAD | 43.8±6.1 | 32.6 | 178 | 202 | Smoking, elevated level of total cholesterol, LDL-cholesterol, triacylglycerols, overweight or obesity |
| Zhang | 2008 | China | Asian | CAD | 63.6±4.9 | 27 | 155 | 190 | NA |
| Isordia-Salas | 2009 | Mexico | Caucasian | MI | 40±4.6 | 16.5 | 127 | 127 | NA |
| Tàssies | 2009 | Spain | Caucasian | CAD | 60±13 | 23 | 248 | 200 | NA |
| Var | 2009 | Turkey | Caucasian | CAD | 55.3±11.3 | 35 | 86 | 90 | Age, sex, smoking and hypertension |
| Chen | 2009 | China | Asian | CAD | 60±11 | 51 | 293 | 178 | NA |
| Abboud | 2010 | Tunisia | African | MI | 59.0±12.0 | 19 | 305 | 328 | NA |
| Cao | 2010 | China | Asian | MI | 64.62 | 33.6 | 116 | 60 | NA |
| Koch | 2010 | Germany | Caucasian | MI | 64±12 | 24.2 | 3657 | 1211 | Age, gender, history of arterial hypertension, history of hypercholesterolaemia, current cigarette smoking, and diabetes mellitus |
| Agirbasli | 2011 | Turkey | Caucasian | CAD | 45.4±7 | 43.3 | 90 | 90 | NA |
| Ahmed | 2011 | Pakistan | Caucasian | MI | 52.1±11.3 | 19.7 | 229 | 217 | NA |
| Ashavaid | 2011 | India | Asian | CAD | 58.6±10.4 | 19.7 | 446 | 473 | NA |
| Lima | 2011 | Brazil | Caucasian | CAD | 60 | 50 | 123 | 38 | NA |
| Zhao | 2012 | China | Asian | CAD | 40-82 | 32.9 | 146 | 113 | NA |
| Lin | 2012 | China | Asian | CAD | 43±14 | 38 | 65 | 132 | NA |
MI, myocardium infarction; HDL, high-density lipoprotein; LDL, low-density lipoprotein; BMI, body mass index; NA, not available.
Table 2.
Distribution of PAI-1 -675 4G/5G polymorphism among patients and controls
| Case | Control | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Study | 4G/4G | 4G/5G | 5G/5G | 4G/4G | 4G/5G | 5G/5G | HWE |
| Dawson | 29 | 51 | 27 | 23 | 24 | 26 | No |
| Eriksson | 40 | 38 | 15 | 26 | 54 | 20 | Yes |
| Ye | 148 | 230 | 98 | 189 | 271 | 141 | No |
| Mansfield | 20 | 15 | 3 | 37 | 67 | 18 | Yes |
| Burzotta | 32 | 46 | 30 | 52 | 86 | 37 | Yes |
| Ridker | 101 | 191 | 82 | 133 | 247 | 115 | Yes |
| Ossei-Gerning | 59 | 73 | 26 | 36 | 65 | 49 | Yes |
| Iwai | 83 | 99 | 22 | 53 | 76 | 19 | Yes |
| Kohler | 66 | 91 | 27 | 54 | 86 | 48 | Yes |
| Margaglione | 68 | 85 | 45 | 239 | 493 | 249 | Yes |
| Pastinen | 46 | 74 | 31 | 30 | 80 | 40 | Yes |
| Junker | 86 | 112 | 43 | 52 | 93 | 34 | Yes |
| Sugano | 5 | 28 | 33 | 6 | 27 | 29 | Yes |
| Ardissino | 38 | 93 | 69 | 32 | 102 | 66 | Yes |
| Anderson 1 | 105 | 193 | 77 | 303 | 457 | 218 | Yes |
| Anderson 2 | 267 | 433 | 198 | 97 | 155 | 77 | Yes |
| Doggen | 88 | 170 | 73 | 84 | 150 | 68 | Yes |
| Gardemann 1 | 624 | 985 | 362 | 167 | 305 | 122 | Yes |
| Gardemann 2 | 382 | 606 | 226 | 409 | 684 | 258 | Yes |
| Grancha | 6 | 23 | 12 | 11 | 30 | 21 | Yes |
| Beneš | 53 | 91 | 31 | 77 | 103 | 42 | Yes |
| Canavy | 48 | 97 | 56 | 64 | 121 | 59 | Yes |
| Hooper | 7 | 42 | 59 | 11 | 79 | 104 | Yes |
| Mikkelsson | 18 | 38 | 12 | 29 | 78 | 57 | Yes |
| Song | 62 | 64 | 32 | 54 | 60 | 25 | Yes |
| Fu | 39 | 29 | 19 | 25 | 45 | 22 | Yes |
| Viitanen | 29 | 65 | 24 | 28 | 51 | 31 | Yes |
| Dai | 85 | 110 | 55 | 12 | 48 | 35 | Yes |
| Fu | 58 | 49 | 16 | 38 | 85 | 49 | Yes |
| Shang | 13 | 18 | 7 | 20 | 37 | 23 | Yes |
| Ortlepp | 36 | 48 | 16 | 24 | 54 | 22 | Yes |
| Yamada | 215 | 300 | 75 | 315 | 316 | 73 | Yes |
| Guan | 50 | 52 | 24 | 23 | 70 | 28 | Yes |
| Li | 13 | 18 | 5 | 5 | 11 | 0 | Yes |
| ATVBISG | 335 | 589 | 286 | 342 | 588 | 280 | Yes |
| Crainich | 70 | 136 | 58 | 200 | 387 | 166 | Yes |
| Juhan-Vague | 125 | 249 | 109 | 133 | 269 | 105 | Yes |
| Leander 1 | 256 | 415 | 153 | 283 | 542 | 203 | Yes |
| Leander 2 | 103 | 180 | 61 | 153 | 226 | 110 | Yes |
| Petrovič | 45 | 74 | 35 | 68 | 89 | 37 | Yes |
| Zhan | 40 | 14 | 2 | 25 | 52 | 6 | No |
| Ding 1 | 8 | 26 | 26 | 15 | 39 | 55 | Yes |
| Ding 2 | 15 | 23 | 11 | 10 | 25 | 28 | Yes |
| Wang | 8 | 35 | 24 | 2 | 7 | 21 | Yes |
| Zhai | 58 | 49 | 16 | 38 | 85 | 49 | Yes |
| Tobin | 159 | 280 | 108 | 162 | 237 | 106 | Yes |
| Pegoraro | 42 | 99 | 54 | 65 | 132 | 103 | Yes |
| Whiting | 263 | 427 | 191 | 78 | 121 | 62 | Yes |
| Zak | 34 | 74 | 38 | 44 | 58 | 19 | Yes |
| Agirbasli | 28 | 46 | 26 | 23 | 60 | 17 | Yes |
| Su | 272 | 390 | 150 | 275 | 446 | 210 | Yes |
| Xia | 79 | 67 | 20 | 18 | 28 | 17 | Yes |
| Morange | 105 | 236 | 120 | 96 | 254 | 124 | Yes |
| Sampaio | 23 | 47 | 45 | 16 | 45 | 43 | Yes |
| Taymaz | 31 | 58 | 26 | 15 | 20 | 6 | Yes |
| Onalan | 51 | 75 | 30 | 73 | 112 | 96 | Yes |
| Saely | NA | NA | NA | NA | NA | NA | Yes |
| Sarecka | 38 | 94 | 46 | 69 | 103 | 30 | Yes |
| Zhang | 58 | 62 | 35 | 52 | 87 | 51 | Yes |
| Isordia-Salas | 9 | 64 | 54 | 17 | 38 | 72 | Yes |
| Tàssies | 56 | 121 | 71 | 48 | 92 | 60 | Yes |
| Var | 43 | 24 | 19 | 24 | 36 | 30 | Yes |
| Chen | 100 | 140 | 53 | 47 | 99 | 32 | Yes |
| Abboud | 88 | 156 | 61 | 42 | 180 | 106 | Yes |
| Cao | 61 | 41 | 14 | 15 | 27 | 18 | Yes |
| Koch | 1091 | 1787 | 779 | 360 | 590 | 261 | Yes |
| Agirbasli | 36 | 35 | 19 | 24 | 43 | 23 | Yes |
| Ahmed | 64 | 86 | 79 | 52 | 89 | 76 | Yes |
| Ashavaid | 112 | 218 | 116 | 113 | 247 | 113 | Yes |
| Lima | 46 | 34 | 43 | 12 | 12 | 14 | Yes |
| Zhao | 46 | 68 | 32 | 23 | 57 | 33 | Yes |
| Lin | 29 | 28 | 8 | 34 | 63 | 35 | Yes |
HWE, Hardy-Weinberg equilibrium.
Quantitative data synthesis
The results of this meta-analysis are shown in Table 3. We found that PAI-1 4G/5G polymorphism was significant associated with CAD risk in overall population (OR=1.19, 95% CI 1.10-1.28, P < 0.00001, Figure 2). The combination of adjusted ORs for CAD was 1.20 (95% CI 1.03-1.40, P=0.02). In the subgroup analysis according to ethnicity, the results suggested that PAI-1 4G/5G polymorphism was associated with CAD risk in Caucasians (OR=1.10, 95% CI 1.02-1.19, P=0.01) and Asians (OR=1.46, 95% CI 1.21-1.75, P < 0.0001). However, no significant association was observed in Africans (OR=1.38, 95% CI 0.70-2.70, P=0.35). In terms of subgroup analyses by endpoint, the PAI-1 4G/5G polymorphism significantly increased MI risk (OR=1.15, 95% CI 1.06-1.25, P=0.001). In the subgroup analysis by age, the PAI-1 4G/5G polymorphism was significantly associated with early-onset CAD risk (OR=1.21, 95% CI 1.02-1.43, P=0.03) but not with late-onset CAD risk (OR=0.90, 95% CI 0.72-1.13, P=0.37). In the gender subgroup analyses, a statistically significant association was found in male CAD patients (OR=1.10, 95% CI 1.01-1.20, P=0.04) but not with female CAD patients (OR=1.03, 95% CI 0.89-1.19, P=0.73). Stratification by T2DM status showed that both T2DM patients and non-T2DM patients carrying 4G allele were associated with increased CAD risks (OR=2.23, 95% CI 1.27-3.92, P=0.005 and OR=1.64, 95% CI 1.19-2.25, P=0.002, respectively).
Table 3.
The effect of PAI-1 -675 4G/5G polymorphism on CAD risk
| Comparison | Study | No. ofstudies | Test of association | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| OR (95% CI) | Z | P Value | χ 2 | P Value | I 2 (%) | |||
| 4G/4G + 4G/5G vs. 5G/5G | Overall | 72 | 1.19 (1.10-1.28) | 4.54 | < 0.00001 | 144.13 | < 0.00001 | 51 |
| 4G/4G + 4G/5G vs. 5G/5G | Adjusted | 10 | 1.20 (1.03-1.40) | 2.29 | 0.02 | 13.95 | 0.12 | 36 |
| 4G/4G + 4G/5G vs. 5G/5G | Caucasian | 45 | 1.10 (1.02-1.19) | 2.54 | 0.01 | 70.61 | 0.007 | 38 |
| 4G/4G + 4G/5G vs. 5G/5G | Asian | 24 | 1.46 (1.21-1.75) | 4.03 | < 0.0001 | 55.19 | 0.0002 | 58 |
| 4G/4G + 4G/5G vs. 5G/5G | African | 2 | 1.38 (0.70-2.70) | 0.93 | 0.35 | 5.12 | 0.02 | 80 |
| 4G/4G + 4G/5G vs. 5G/5G | MI | 39 | 1.15 (1.06-1.25) | 3.27 | 0.001 | 68.35 | 0.002 | 44 |
| 4G/4G + 4G/5G vs. 5G/5G | Early-onset | 12 | 1.21 (1.02-1.43) | 2.20 | 0.03 | 6.31 | 0.71 | 0 |
| 4G/4G + 4G/5G vs. 5G/5G | Late-onset | 4 | 0.90 (0.72-1.13) | 0.89 | 0.37 | 1.83 | 0.61 | 0 |
| 4G/4G + 4G/5G vs. 5G/5G | Male | 13 | 1.10 (1.01-1.20) | 2.10 | 0.04 | 7.23 | 0.84 | 0 |
| 4G/4G + 4G/5G vs. 5G/5G | Female | 4 | 1.03 (0.89-1.19) | 0.34 | 0.73 | 4.54 | 0.21 | 34 |
| 4G/4G + 4G/5G vs. 5G/5G | T2DM | 4 | 2.23 (1.27-3.92) | 2.80 | 0.005 | 0.48 | 0.79 | 0 |
| 4G/4G + 4G/5G vs. 5G/5G | Non-T2DM | 3 | 1.64 (1.19-2.25) | 3.03 | 0.002 | 0.57 | 0.75 | 0 |
MI, myocardium infarction; T2DM, type 2 diabetes.
Figure 2.
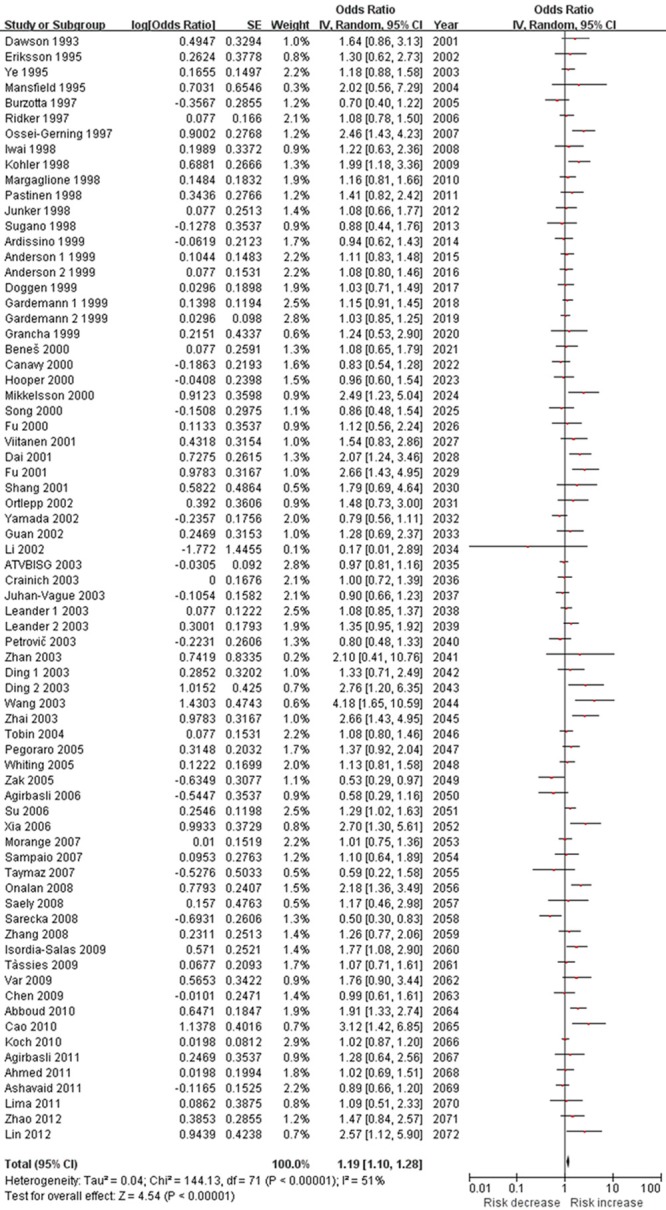
Meta-analysis of the association between the PAI-1 4G/5G polymorphism and CAD risk.
Sensitivity analysis was used to evaluate the stability of the overall results by sequential omission of individual studies. In this meta-analysis, the results of sensitive analysis showed that any single study did not influence the overall results qualitatively (data not shown).
Funnel plots and the Egger’s test were used to assess publication bias. In the funnel plot analysis, the shape of the funnel plot seemed symmetrical (Figure 3). Furthermore, Egger’s test did not detect any publication bias (P=0.239). Therefore, there was no significant publication bias in the studies included in current analyses.
Figure 3.
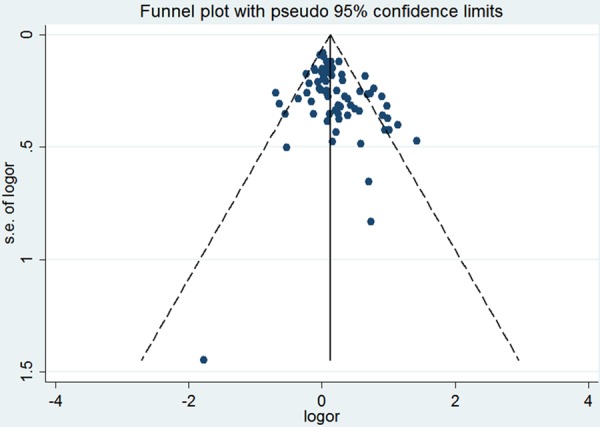
Funnel plot of the association between the PAI-1 4G/5G polymorphism and CAD risk.
Discussion
This present meta-analysis investigating the relationship between PAI-1 4G/5G polymorphism and risk of CAD. Seventy-two studies with a total of 45083 subjects were eligible. At the overall analysis, the PAI-1 4G/5G polymorphism was significantly associated with CAD risk. Even the studies reporting adjusted ORs were included, the result was still significant. We also found that this polymorphism increased MI risk significantly. In the subgroup analysis by ethnicity, we noted that Asians and Caucasians carrying the 4G allel had an increased CAD risk. Only two studies investigated the association between PAI-1 4G/5G polymorphism and risk of CAD in Africans. Therefore, more studies are still needed. In the stratified analysis by age, we found PAI-1 4G/5G polymorphism showed increased early-onset CAD risk but not late-onset CAD risk. There were only four studies about late-onset CAD risk, the positive association between PAI-1 4G/5G polymorphism and late-onset CAD risk could not be ruled out, because studies with small sample size may have insufficient statistical power to detect a slight effect. The subgroup analysis based on gender found that this polymorphism showed increased CAD risk in male patients but not in female patients. Since the number of studies included in female subgroup analysis was small, the results lacked sufficient reliability to confirm or refute an association in a definitive manner. In the future, more studies should be designed to analyze these associations. When subgroup analysis was performed according to T2DM status, significant associations were showed in T2DM patients and non-T2DM patients. This result suggested that T2DM did not change the effect of PAI-1 4G/5G polymorphism on CAD. Previous meta-analysis has assessed the association between PAI-1 4G/5G polymorphism and risk of CAD. For example, Koch and coworkers found that the risk of MI in 4G allele carriers was found to be significantly elevated [67]. Li suggested that PAI-1 4G/5G polymorphism was associated with increased CAD risk in Chinese Han population [75]. Nikolopoulos et al. also indicated that PAI-1 4G allele slightly increased the risk for MI [76]. These results were all in line with our results. However, our study had some advantages. First, it was the first time studying T2DM and PAI-1 4G/5G polymorphism interactions. Second, we sought to find as many publications as we could by means of various searching approaches. Third, the main result remained statistically signifiant when the adjusted ORs were combined.
PAI-1 is a glycoprotein that belongs to the serine protease inhibitor superfamily. It is equimolecularly combined with the tissue plasminogen activator (tPA) single chain, double chains, and double chain urokinase plasminogen activator (uPA). Consequently, tPA and uPA activities are rapidly inhibited by PAI-1. Mice in which PAI-1 gene was invalidated were protected from thrombotic risk after vascular injury [77]. Case-control studies in humans have shown that high PAI-1 plasma levels were associated with an increased risk of CAD and that plasma levels of PAI-1 were higher in patients with MI than in control individuals [78]. Therefore, PAI-1 might be involved in the development of CAD. PAI-1 4G/5G polymorphism is one of the DNA sequence variations that plays a key rolein regulating PAI-1 gene expression. Studies have shown that the PAI-1 activity of the 4G allele promoter is higher than that of 5G in a cytokine-stimulated state. Unlike the 5G allele that binds a transcription repressor protein, resulting in low PAI-1 expression, the 4G allele does not bind a transcription repressor, thus conferring a high PAI-1 expressor nature to the allele [5].
Some limitations should be addressed. First, there was only two case-control study investigated the association of PAI-1 4G/5G polymorphism and risk of CAD in Africans. Therefore, more studies with large sample sizes are needed to further identify the association among Africans. Second, because small negative studies are less likely to be published, the possibility of publication bias cannot be ruled out completely, even though the Egger’s test and funnel plots did not provide the evidence of publication bias in this meta-analysis. Third, a lack of original data from the eligible studies limited evaluation of the effects of the gene-gene and gene-environment interactions during CAD development.
In conclusion, this meta-analysis suggested that PAI-1 4G/5G polymorphism was associated with increased CAD risk. Further studies with large sample size were needed to confirm our findings.
Disclosure of conflict of interest
None.
References
- 1.Pepper MS. Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler Thromb Vasc Biol. 2001;21:1104–17. doi: 10.1161/hq0701.093685. [DOI] [PubMed] [Google Scholar]
- 2.Cortellaro M, Cofrancesco E, Boschetti C, Mussoni L, Donati MB, Cardillo M Catalano M, Gabrielli L, Lombardi B, Specchia G. Increased fibrin turnover and high PAI-1 activity as predictors of ischemic events in atherosclerotic patients. A case-control study. The PLAT Group. Arterioscler Thromb. 1993;13:1412–7. doi: 10.1161/01.atv.13.10.1412. [DOI] [PubMed] [Google Scholar]
- 3.Sobel BE. Increased plasminogen activator inhibitor-1 and vasculopathy. A reconcilable paradox. Circulation. 1999;99:2496–8. doi: 10.1161/01.cir.99.19.2496. [DOI] [PubMed] [Google Scholar]
- 4.Zorio E, Gilabert-Estellés J, España F, Ramón LA, Cosín R, Estellés A. Fibrinolysis: the key to new pathogenetic mechanisms. Curr Med Chem. 2008;15:923–9. doi: 10.2174/092986708783955455. [DOI] [PubMed] [Google Scholar]
- 5.Dawson S, Hamsten A, Wiman B, Henney A, Humphries S. Genetic variation at the plasminogen activator inhibitor-1 locus is associated with altered levels of plasma plasminogen activator inhibitor-1 activity. Arterioscler Thromb. 1991;11:183–90. doi: 10.1161/01.atv.11.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Dawson SJ, Wiman B, Hamsten A, Green F, Hu- mphries S, Henney AM. The two allele seuences of a common polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene respond differently to interleukin-1 in HepG2 cells. J Biol Chem. 1993;268:10739–45. [PubMed] [Google Scholar]
- 7.Eriksson P, Kallin B, van’t Hooft FM, Bavenholm P, Hamsten A. Allele-specific increase in basal transcription of the plasminogen-activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci U S A. 1995;92:1851–5. doi: 10.1073/pnas.92.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye S, Green FR, Scarabin PY, Nicaud V, Bara L, Dawson SJ, Humphries SE, Evans A, Luc G, Cambou JP, et al. The 4G/5G genetic polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene is associated with differences in plasma PAI-1 activity but not with risk of myocardial infarction in the ECTIM study. Etude CasTemoins de I’nfarctus du Mycocarde. Thromb Haemost. 1995;74:837–41. [PubMed] [Google Scholar]
- 9.Mansfield MW, Stickland MH, Grant PJ. Plasminogen activator inhibitor-1 (PAI-1) promoter polymorphism and coronary artery disease in non-insulin-dependent diabetes. Thromb Haemost. 1995;74:1032–1034. [PubMed] [Google Scholar]
- 10.Burzotta F, Di Castelnuovo A, Amore C, D’Orazio A, Donati MB, Iacoviello L. 4G/5G polymorphism in the promoter region of the PAI-1 gene is not a risk factor for familial myocardial infarction in subjects over 45 years. Thromb Haemost. 1997;78:1294–5. [PubMed] [Google Scholar]
- 11.Ridker PM, Hennekens CH, Lindpaintner K, Stampfer MJ, Miletich JP. Arterial and venous thrombosis is not associated with the 4G/5G polymorphism in the promoter of the plasminogen activator inhibitor gene in a large cohort of US men. Circulation. 1997;95:59–62. doi: 10.1161/01.cir.95.1.59. [DOI] [PubMed] [Google Scholar]
- 12.Ossei-Gerning N, Mansfield MW, Stickland MH, Wilson IJ, Grant PJ. Plasminogen activator inhibitor-1 promoter 4G/5G genotype and plasma levels in relation to a history of myocardial infarction in patients characterized by coronary angiography. Arterioscler Thromb Vasc Biol. 1997;17:33–7. doi: 10.1161/01.atv.17.1.33. [DOI] [PubMed] [Google Scholar]
- 13.Iwai N, Shimoike H, Nakamura Y, Tamaki S, Kinoshita M. The 4G/5G polymorphism of the plasminogen activator inhibitor gene is associated with the time course of progression to acute coronary syndromes. Atherosclerosis. 1998;136:109–14. doi: 10.1016/s0021-9150(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 14.Kohler HP, Stickland MH, Ossei-Gerning N, Carter A, Mikkola H, Grant PJ. Association of a common polymorphism in the factor XIII gene with myocardial infarction. Thromb Haemost. 1998;79:8–13. [PubMed] [Google Scholar]
- 15.Margaglione M, Cappucci G, Colaizzo D, Giuliani N, Vecchione G, Grandone E, Pennelli O, Di Minno G. The PAI-1 gene locus 4G/5G polymorphism is associated with a family history of coronary artery disease. Arterioscler Thromb Vasc Biol. 1998;18:152–6. doi: 10.1161/01.atv.18.2.152. [DOI] [PubMed] [Google Scholar]
- 16.Pastinen T, Perola M, Niini P, Terwilliger J, Salomaa V, Vartiainen E, Peltonen L, Syvänen A. Array-based multiplex analysis of candidate genes reveals two independent and additive genetic risk factors for myocardial infarction in the Finnish population. Hum Mol Genet. 1998;7:1453–62. doi: 10.1093/hmg/7.9.1453. [DOI] [PubMed] [Google Scholar]
- 17.Junker R, Heinrich J, Schulte H, Tataru M, Köhler E, Schönfeld R, Nowak-Göttl U, Assmann G. Plasminogen activator inhibitor-1 4G/5G-polymorphism and factor V Q506 mutation are not associated with myocardial infarction in young men. Blood Coagul Fibrinolysis. 1998;9:597–602. doi: 10.1097/00001721-199810000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Sugano T, Tsuji H, Masuda H, Nakagawa K, Nishimura H, Kasahara T, Yoshizumi M, Nakahara Y, Kitamura H, Yamada K, Yoneda M, Maki K, Tatsumi T, Azuma A, Nakagawa M. Plasminogen activator inhibitor-1 promoter 4G/5G genotype is not a risk factor for myocardial infarction in a Japanese population. Blood Coagul Fibrinolysis. 1998;9:201–4. doi: 10.1097/00001721-199803000-00013. [DOI] [PubMed] [Google Scholar]
- 19.Ardissino D, Mannucci PM, Merlini PA, Duca F, Fetiveau R, Tagliabue L, Tubaro M, Galvani M, Ottani F, Ferrario M, Corral J, Margaglione M. Prothrombotic genetic risk factors in young survivors of myocardial infarction. Blood. 1999;94:46–51. [PubMed] [Google Scholar]
- 20.Anderson JL, Muhlestein JB, Habashi J, Carlquist JF, Bair TL, Elmer SP, Davis BP. Lack of association of a common polymorphism of the plasminogen activator inhibitor-1 gene with coronary artery disease and myocardial infarction. J Am Coll Cardiol. 1999;34:1778–83. doi: 10.1016/s0735-1097(99)00424-6. [DOI] [PubMed] [Google Scholar]
- 21.Doggen CJ, Bertina RM, Cats VM, Reitsma PH, Rosendaal FR. The 4G/5G polymorphism in the plasminogen activator inhibitor-1 gene is not associated with myocardial infarction. Thromb Haemost. 1999;82:115–20. [PubMed] [Google Scholar]
- 22.Gardemann A, Lohre J, Katz N, Tillmanns H, Hehrlein FW, Haberbosch W. The 4G4G genotype of the plasminogen activator inhibitor 4G/5G gene polymorphism is associated with coronary atherosclerosis in patients at high risk for this disease. Thromb Haemost. 1999;82:1121–6. [PubMed] [Google Scholar]
- 23.Grancha S, Estellés A, Tormo G, Falco C, Gilabert J, España F, Cano A, Segui R, Aznar J. Plasminogen activator inhibitor-1 (PAI-1) promoter 4G/5G genotype and increased PAI-1 circulating levels in postmenopausal women with coronary artery disease. Thromb Haemost. 1999;81:516–21. [PubMed] [Google Scholar]
- 24.Benes P, Muzík J, Benedík J, Frélich M, Elbl L, Vasku A, Znojil V, Vácha J. Single effects of apolipoprotein B, (a), and E polymorphisms and interaction between plasminogen activator inhibitor-1 and apolipoprotein (a) genotypes and the risk of coronary artery disease in Czech male caucasians. Mol Genet Metab. 2000;69:137–43. doi: 10.1006/mgme.1999.2957. [DOI] [PubMed] [Google Scholar]
- 25.Canavy I, Henry M, Morange PE, Tiret L, Poirier O, Ebagosti A, Bory M, Juhan-Vague I. Genetic polymorphisms and coronary artery disease in the south of France. Thromb Haemost. 2000;83:212–6. [PubMed] [Google Scholar]
- 26.Hooper WC, Lally C, Austin H, Renshaw M, Dilley A, Wenger NK, Phillips DJ, Whitsett C, Rawlins P, Evatt BL. The role of the t-PA I/D and PAI-1 4G/5G polymorphisms in African-American adults with a diagnosis of myocardial infarction or venous thromboembolism. Thromb Res. 2000;99:223–30. doi: 10.1016/s0049-3848(00)00236-x. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsson J, Perola M, Wartiovaara U, Peltonen L, Palotie A, Penttila A, Penttilä A, Karhunen PJ. Plasminogen activator inhibitor-1 (PAI-1) 4G/5G polymorphism, coronary thrombosis, and myocardial infarction in middle-aged Finnish men who died suddenly. Thromb Haemost. 2000;84:78–82. [PubMed] [Google Scholar]
- 28.Song J, Yoon YM, Jung HJ, Hong SH, Park H, Kim JQ. Plasminogen activator inhibitor-1 4G/5G promoter polymorphism and coagulation factor VII Arg353- > Gln polymorphism in Korean patients with coronary artery disease. J Korean Med Sci. 2000;15:146–52. doi: 10.3346/jkms.2000.15.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu L, Jin H, Song K, Zhang C, Shen J, Huang Y. Relationship between gene polymorphism of the PAI-1 promoter and myocardial infarction. Chin Med J (Engl) 2001;114:266–9. [PubMed] [Google Scholar]
- 30.Viitanen L, Pihlajamäki J, Halonen P, Lehtonen M, Kareinen A, Lehto S, Laakso M. Association of angiotensin converting enzyme and plasminogen activator inhibitor-1 promoter gene polymorphisms with features of the insulin resistance syndrome in patients with premature coronary heart disease. Atherosclerosis. 2001;157:57–64. doi: 10.1016/s0021-9150(00)00705-x. [DOI] [PubMed] [Google Scholar]
- 31.Dai Y, Gao RL, Ye Y, Wu YJ, Chen JL, et al. The 4G/5G genetic polymorphism of the plasminogen activator inhibitor-1 (PAI-1) gene is not associated with plasma PAI-1 antigen level and the risk of coronary artery disease in Chinese. Chin Circ J. 2001;16:275–8. [Google Scholar]
- 32.Fu Y, Wang XD, Zhai YL, Fan Z, Yang L, et al. The 4G/5G polymorphism of the plasminogen activator inhibitor-1 gene in patients with coronary heart disease. J Capital Univers Med Sci. 2001;22:119–22. [Google Scholar]
- 33.Shang SH, Wang C, Yu YC, Gu Y, Wu GT, Wu ZC. Plasminogen activator inhibitor-1 (PAI-1) promoter polymorphism and coronary artery disease in type 2 diabetes. J Tongji Univers (Med Sci) 2001;4:16–8. [Google Scholar]
- 34.Ortlepp JR, Lauscher J, Janssens U, Minkenberg R, Hanrath P, Hoffmann R. Analysis of several hundred genetic polymorphisms may improve assessment of the individual genetic burden for coronary artery disease. Eur J Intern Med. 2002;13:485–92. doi: 10.1016/s0953-6205(02)00182-6. [DOI] [PubMed] [Google Scholar]
- 35.Yamada Y, Izawa H, Ichihara S, Takatsu F, Ishihara H, Hirayama H, Sone T, Tanaka M, Yokota M. Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med. 2002;347:1916–23. doi: 10.1056/NEJMoa021445. [DOI] [PubMed] [Google Scholar]
- 36.Guan L, Ji X, Wang J, Zhang A, Zhang Y, Zhao l. Association of plasminogen activator inhibitor-1 gene 4G/5G polymorphism and coronary heart disease in Chinese patients. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:393–6. [PubMed] [Google Scholar]
- 37.Li XS, Xian SX, Huang HQ. Relationship between plasminogen activator inhibitor-1 gene and coronary heart diseasewith blood-stagnation syndrome. J Guangzhou Univers Tradition Chin Med. 2002;19:261–4. [Google Scholar]
- 38.Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group. No evidence of association between prothrombotic gene polymorphisms and the development of acute myocardial infarction at a young age. Circulation. 2003;107:1117–22. doi: 10.1161/01.cir.0000051465.94572.d0. [DOI] [PubMed] [Google Scholar]
- 39.Crainich P, Jenny NS, Tang Z, Arnold AM, Kuller LH, Manolio T, Sharrett AR, Tracy RP. Lack of association of the plasminogen activator inhibitor-1 4G/5G promoter polymorphism with cardiovascular disease in the elderly. J Thromb Haemost. 2003;1:1799–804. doi: 10.1046/j.1538-7836.2003.00255.x. [DOI] [PubMed] [Google Scholar]
- 40.Juhan-Vague I, Morange PE, Frere C, Aillaud MF, Alessi MC, Hawe E, Boquist S, Tornvall P, Yudkin JS, Tremoli E, Margaglione M, Di Minno G, Hamsten A, Humphries SE. The plasminogen activator inhibitor-1-675 4G/5G genotype influences the risk of myocardial infarction associated with elevated plasma proinsulin and insulin concentrations in men from Europe: the HIFMECH study. J Thromb Haemost. 2003;1:2322–9. doi: 10.1046/j.1538-7836.2003.00458.x. [DOI] [PubMed] [Google Scholar]
- 41.Leander K, Wiman B, Hallqvist J, Sten-Linder M, de Faire U. PAI-1 level and the PAI-1 4G/5G polymorphism in relation to risk of non-fatal myocardial infarction: results from the Stockholm Heart Epidemiology Program (SHEEP) Thromb Haemost. 2003;89:1064–71. [PubMed] [Google Scholar]
- 42.Petrovic D, Globocnik-Petrovic M, Peterlin B. 4G4G genotype of PAI-1 gene promoter polymorphism is not associated with myocardial infarction in Caucasians with type-2 diabetes. Cardiology. 2003;100:157–8. doi: 10.1159/000073935. [DOI] [PubMed] [Google Scholar]
- 43.Zhan M, Zhou Y, Han Z. Plasminogen activator inhibitor-1 4G/5G gene polymorphism in patients with myocardial or cerebrovascular infarction in Tianjin, China. Chin Med J (Engl) 2003;116:1707–10. [PubMed] [Google Scholar]
- 44.Ding GX, Shen J, Chen JW. The relationship among polymorphism of PAI-1 gene, type 2 diabetes mellitus, hypertension and coronary heart disease. Acta Univers Med Nanjing (Nat Sci) 2003;1:4–7. [Google Scholar]
- 45.Wang YS, Wang SY, Zhang M, Bai CX, Li SQ. Polymorphisms of PAI-1 4G/5G in patients with coronary heart disease complicated with or without OSAHS. New Med. 2003;34:674–6. [Google Scholar]
- 46.Zhai YL, Fu Y, Wang XD, Liu FQ, Qian J, Fan Z, et al. Relation between plasminogen activator inhibitor-1 and the PAI-1 gene locus 4G/5G polymorphism and their effect on the incidence of coronary heart disease. 2003;5:246–8. [Google Scholar]
- 47.Tobin MD, Braund PS, Burton PR, Thompson JR, Steeds R, Channer K, Cheng S, Lindpaintner K, Samani NJ. Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J. 2004;25:459–67. doi: 10.1016/j.ehj.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Pegoraro RJ, Ranjith N. Plasminogen activator inhibitor type 1 (PAI-1) and platelet glycoprotein IIIa (PGIIIa) polymorphisms in young Asian Indians with acute myocardial infarction. Cardiovasc J S Afr. 2005;16:266–70. [PubMed] [Google Scholar]
- 49.Whiting BM, Anderson JL, Muhlestein JB, Hor- ne BD, Bair TL, Pearson RR, Carlquist JF Intermountain Heart Collaborative Study Group. Candidate gene susceptibility variants predict intermediate end points but not angiographic coronary artery disease. Am Heart J. 2005;150:243–50. doi: 10.1016/j.ahj.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 50.Zak I, Balcerzyk A, Sarecka B, Niemiec P, Ciemniewski Z, Dylag S. Contemporaneous carrier-state of two or three proatherosclerotic variants of APOE, ICAM1, PPARA and PAI-1 genes differentiate CAD patients from healthy individuals. Clin Chim Acta. 2005;362:110–8. doi: 10.1016/j.cccn.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Agirbasli D, Agirbasli M, Williams SM, Phillips JA 3rd. Interaction among 5, 10 methylenetetrahydrofolate reductase, plasminogen activator inhibitor and endothelial nitric oxide synthase gene polymorphisms predicts the severity of coronary artery disease in Turkish patients. Coron Artery Dis. 2006;17:413–7. doi: 10.1097/00019501-200608000-00003. [DOI] [PubMed] [Google Scholar]
- 52.Su S, Chen S, Zhao J, Huang J, Wang X, Chen R, Gu D. Plasminogen activator inhibitor-1 gene: selection of tagging single nucleotide polymorphisms and association with coronary heart disease. Arterioscler Thromb Vasc Biol. 2006;26:948–54. doi: 10.1161/01.ATV.0000204731.17646.f2. [DOI] [PubMed] [Google Scholar]
- 53.Xia DS, Cao J, Song YQ, Hu SY, Guo QY, et al. Association between serotonin transporter and plasminogen activator inhibitor-1 gene polymorphisms and depressive disorder in patients with coronary heart disease. Chin J of Behavioral Med Sci. 2006;15:481–3. [Google Scholar]
- 54.Morange PE, Saut N, Alessi MC, Yudkin JS, Margaglione M, Di Minno G, Hamsten A, Humphries SE, Tregouet DA, Juhan-Vague I. Association of plasminogen activator inhibitor (PAI)-1 (SERPINE1) SNPs with myocardial infarction, plasma PAI-1, and metabolic parameters: the HIFMECH study. Arterioscler Thromb Vasc Biol. 2007;27:2250–7. doi: 10.1161/ATVBAHA.107.149468. [DOI] [PubMed] [Google Scholar]
- 55.Sampaio MF, Hirata MH, Hirata RD, Santos FC, Picciotti R, Luchessi AD, de Quateli Doi S, Armaganijan D, Batlouni M. AMI is associated with polymorphisms in the NOS3 and FGB but not in PAI-1 genes in young adults. Clin Chim Acta. 2007;377:154–62. doi: 10.1016/j.cca.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 56.Taymaz H, Erarslan S, Oner ET, Alkan T, Ağirbaşli M, Kirdar B. Sequence variations within the genes related to hemostatic imbalance and their impact on coronary artery disease in Turkish population. Thromb Res. 2007;119:55–62. doi: 10.1016/j.thromres.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 57.Onalan O, Balta G, Oto A, Kabakci G, Tokgozoglu L, Aytemir K, Altay C, Gurgey A, Nazli N. Plasminogen activator inhibitor-1 4G4G genotype is associated with myocardial infarction but not with stable coronary artery disease. J Thromb Thrombolysis. 2008;26:211–7. doi: 10.1007/s11239-007-0083-z. [DOI] [PubMed] [Google Scholar]
- 58.Saely CH, Muendlein A, Vonbank A, Sonderegger G, Aczel S, Rein P, Risch L, Drexel H. Type 2 diabetes significantly modulates the cardiovascular risk conferred by the PAI-1 -675 4G/5G polymorphism in angiographied coronary patients. Clin Chim Acta. 2008;396:18–22. doi: 10.1016/j.cca.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Sarecka B, Zak I, Krauze J. Synergistic effects of the polymorphisms in the PAI-1 and IL-6 genes with smoking in determining their associated risk with coronary artery disease. Clin Biochem. 2008;41:467–73. doi: 10.1016/j.clinbiochem.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 60.Zhang AY, Ji XW, Guan LX, Zhang AJ, Wang JX. Association of PAI-1 gene polymorphism with prognosis of coronary artery disease. Chin J Med Genet. 2008;2:233–5. [PubMed] [Google Scholar]
- 61.Isordia-Salas I, Leanos-Miranda A, Sainz IM, Reyes-Maldonado E, Borrayo-Sanchez G. Association of the plasminogen activator inhibitor-1 gene 4G/5G polymorphism with ST elevation acute myocardial infarction in young patients. Rev Esp Cardiol. 2009;62:365–72. doi: 10.1016/s1885-5857(09)71663-9. [DOI] [PubMed] [Google Scholar]
- 62.Tassies D, Roque M, Monteagudo J, Martorell T, Sionis A, Arzamendi D, Heras M, Reverter JC. Thrombin-activatable fibrinolysis inhibitor genetic polymorphisms as markers of the type of acute coronary syndrome. Thromb Res. 2009;124:614–8. doi: 10.1016/j.thromres.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Var A, Utük O, Akçali S, Sanlidağ T, Uyanik BS, Dinç G. Impact of hemostatic gene single point mutations in patients with non-diabetic coronary artery disease. Mol Biol Rep. 2009;36:2235–43. doi: 10.1007/s11033-008-9439-5. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Zhao LS, Zheng H, Yang F, Kong RN, Zhou S, et al. The association between PAI-1 and PAI-1 -675 4G/5G polymorphism and coronary artery disease. Shangdong Med J. 2009;5:61–3. [Google Scholar]
- 65.Abboud N, Ghazouani L, Saidi S, Ben-Hadj-Khalifa S, Addad F, Almawi WY, Mahjoub T. Association of PAI-1 4G/5G and -844G/A gene polymorphisms and changes in PAI-1/tissue plasminogen activator levels in myocardial infarction: a case-control study. Genet Test Mol Biomarkers. 2010;14:23–7. doi: 10.1089/gtmb.2009.0039. [DOI] [PubMed] [Google Scholar]
- 66.Cao XL, ZCY , Yin L, Wang SC, Jia XL, et al. Reactive protein, plasminogen activator inhibitor type-1 (PAI-1) levels, PAI-1 promoter 4G/5G polymorphism and acute myocardial infarction. J Geriatr Cardiol. 2010;7:147–51. [Google Scholar]
- 67.Koch W, Schrempf M, Erl A, Mueller JC, Hoppmann P, Schömig A, Kastrati A. 4G/5G polymorphism and haplotypes of SERPINE1 in atherosclerotic diseases of coronary arteries. Thromb Haemost. 2010;103:1170–80. doi: 10.1160/TH09-10-0702. [DOI] [PubMed] [Google Scholar]
- 68.Agirbasli M, Guney AI, Ozturhan HS, Agirbasli D, Ulucan K, Sevinc D, Kirac D, Ryckman KK, Williams SM. Multifactor dimensionality reduction analysis of MTHFR, PAI-1, ACE, PON1, and eNOS gene polymorphisms in patients with early onset coronary artery disease. Eur J Cardiovasc Prev Rehabil. 2011;18:803–9. doi: 10.1177/1741826711398806. [DOI] [PubMed] [Google Scholar]
- 69.Ahmed W, Malik M, Saeed I, Khan AA, Sadeque A, Kaleem U, Ahmed N, Ajmal M, Azam M, Qamar R. Role of tissue plasminogen activator and plasminogen activator inhibitor polymorphism in myocardial infarction. Mol Biol Rep. 2011;38:2541–8. doi: 10.1007/s11033-010-0392-8. [DOI] [PubMed] [Google Scholar]
- 70.Ashavaid TF, Todur SP, Kondkar AA, Nair KG, Shalia KK, Dalal JJ, Rajani R, Ponde CK. Platelet polymorphisms: frequency distribution and association with coronary artery disease in an Indian population. Platelets. 2011;22:85–91. doi: 10.3109/09537104.2010.522275. [DOI] [PubMed] [Google Scholar]
- 71.Lima LM, Carvalho Md, Fonseca Neto CP, Garcia JC, Sousa MO. PAI-1 4G/5G polymorphism and plasma levels association in patients with coronary artery disease. Arq Bras Cardiol. 2011;97:462–389. doi: 10.1590/s0066-782x2011005000110. [DOI] [PubMed] [Google Scholar]
- 72.Zhao XH, Liu P, Han ZQ. The association of plasminogen activator inhibitor type-1 gene 4G/5G polymorphism with coronary heart disease. Chin Morden Doctor. 2012;50:1–5. [Google Scholar]
- 73.Lin ZX, Yan HF, Lin JD, Zou XB. The association between plasminogen activator inhibitor type-1 gene polymorphism and coronary heart disease. Guangdong Med J. 2013;34:3588–90. [Google Scholar]
- 74.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li YY. Plasminogen activator inhibitor-1 4G/5G gene polymorphism and coronary artery disease in the Chinese Han population: a meta-analysis. PLoS One. 2012;7:e33511. doi: 10.1371/journal.pone.0033511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikolopoulos GK, Bagos PG, Tsangaris I, Tsiara CG, Kopterides P, Vaiopoulos A, Kapsimali V, Bonovas S, Tsantes AE. The association between plasminogen activator inhibitor type 1 (PAI-1) levels, PAI-1 4G/5G polymorphism, and myocardial infarction: a Mendelian randomization meta-analysis. Clin Chem Lab Med. 2014;52:937–50. doi: 10.1515/cclm-2013-1124. [DOI] [PubMed] [Google Scholar]
- 77.Carmeliet P, Stassen JM, Schoonjans L, Ream B, van den Oord JJ, De Mol M, Mulligan RC, Collen D. Plasminogen activator inhibitor-1 gene-deficient mice. II. Effects on hemostasis, thrombosis, and thrombolysis. J Clin Invest. 1993;92:2756–60. doi: 10.1172/JCI116893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hamsten A, de Faire U, Walldius G, Dahlén G, Szamosi A, Landou C, Landou C, Blombäck M, Wiman B. Plasminogen activator inhibitor in plasma: risk factor for recurrent myocardial infarction. Lancet. 1987;2:3–9. doi: 10.1016/s0140-6736(87)93050-9. [DOI] [PubMed] [Google Scholar]


