Abstract
Background: The association of glutathione s-transferase P1 (GSTP1) Ile105Val polymorphism with risk of esophageal cancer (EC) has been evaluated in many studies; however, the results from these studies are controversial. Thus, further analysis on association between GSTP1 Ile105Val polymorphism and risk of EC is needed among a larger study population. Method: We searched the relevant electronic databases and performed a meta-analysis based on 21 published case-control studies. The Chi-square based I2-statistic test was performed to evaluate possible heterogeneity across the studies. Additionally, random-effects models were used to calculate crude pooled odds ratios (ORs) with 95% confidence intervals (CIs). Results: Overall, this meta-analysis did support a significant association between GSTP1 Ile105Val polymorphism and risk of EC (pooled OR 1.25, 95% CI, 1.05-1.49). Furthermore, the stratified analysis showed that, in comparison to GSTP1 Ile105Val Ile/Ile genotype, the Val/Val genotype was significantly associated with risk of esophageal squamous cell carcinoma (ESCC) (pooled OR 1.45, 95% CI, 1.07-1.96), particularly in the Caucasian population (pooled OR 1.41, 95% CI, 1.01-1.95). Such a significant association was not observed for esophageal adenocarcinoma (EAC) patients or subjects of an Asian ethnicity. Moreover, substantial evidence of heterogeneity among the studies was not observed. Conclusion: The results from this meta-analysis support a significant association between the GSTP1 Ile105Val polymorphism and risk of EC, particularly in a subgroup with ESCC and in the Caucasian population. Further studies with larger sample sizes are needed to validate our findings.
Keywords: Esophageal cancer, GSTP1, polymorphism, meta-analysis, cancer risk
Introduction
Esophageal cancer (EC) is one of the most common malignancies in the world, with obvious geographical characteristics of its pathogenesis [1]. The development of EC is a multifactorial process. For example, tobacco smoking and alcohol consumption are well-recognized etiological factors for EC [2,3]. However, not every smoker and/or alcohol consumer develops EC, suggesting that individual susceptibility factors might also be involved in the development of this malignancy.
Evidence indicates that genetic polymorphisms in certain carcinogen-metabolizing genes play an important role in modifying the risk for EC [4]. Sequence variations in these genes can alter the expression, function and activity of the encoded enzymes and may consequently increase or decrease carcinogen activation or detoxification. Among them, the glutathiones-transferases (GSTs) represent a superfamily of phase II enzymes which catalyze the conjugation reactions between reduced glutathione and reactive intermediates of a variety of endogenous and exogenous electrophilic compounds. Some of these compounds have carcinogenic potential, thereby making them more water-soluble for easy elimination from the body. GSTP1 has a high level of esophageal expression and plays a central role in the inactivation of toxic and carcinogenic compounds [5,6]. The tendency of GST polymorphisms to alter carcinogen metabolization is well established, with GSTP1 polymorphisms having been extensively studied in human population.
The GSTP1 gene is located on chromosome 11q13 and consists of seven exons. There are two known GSTP1 polymorphic sites, which are characterized by an A-to-G transition at nucleotide 313 (codon 105, exon 5), causing an isoleucine-to-valine change (Ile105Val), and a C-to-T transition at nucleotide 341 (codon 113, exon 6), causing an alanine-to-valine substitution (Ala114Val) [7]. Four allele genotypes are composed of these two polymorphic sites: the wild type GSTP1*A (Ile105, Ala114), GSTP1*B (Val105, Ala114), GSTP1*C (Val105, Val114), and GSTP1*D (Ile105, Val114) [7,8]. The GSTP1 enzyme having Val105 shows a catalytic efficiency for the diol epoxides of polycyclic aromatic hydrocarbons that is seven-fold higher than the isoenzymes having Ile105. In contrast, the catalytic efficiency for 1-chloro-2,4-dinitrobenzene was reduced approximately three-fold in GSTP1/Val105 compared to GSTP1/Ile105 [7,9,10]. The GSTP1/Val105 variant was found to be 2-3 times less stable than the Ile105 variant [11] and was associated with a higher hydrophilic DNA adduct level [12].
The association between GSTP1 polymorphism and risk of EC is unclear, with some studies reporting increased EC risk in subjects carrying GST1/Val105 variant [13-17]; some reporting decreased risk [18]; and others reporting no association [19-28]. A previous meta-analysis investigated the association between EC risk and GSTP1 polymorphism [29]; however, this study also included a limited number of published studies, and new results have since been published. In the current meta-analysis, we gave a more comprehensive overview of risk effect of GSTP1 Ile05Val polymorphism on EC occurrence, including journal articles published up to September 1, 2013.
Materials and methods
Identification of eligible studies
We conducted a literature search through September 1, 2013 using the key words search in the PubMed, Web of Knowledge, MEDLINE, Embase, and Google Scholar electronic databases and search engines. The language of publication was restricted to English. The following search terms were used: glutathione s-transferase p or GST or GSTP1 or glutathione S-transferase P1, and polymorphism or single nucleotide polymorphism or SNP, and esophageal cancer or esophagus or esophageal squamous cell carcinoma or ESCC or esophageal adenocarcinoma or EAC.
Inclusion and exclusion criteria
The following inclusion criteria were used to select studies: (1) case-control study methodology; (2) association of EC with GSTP1 polymorphisms; (3) reported sample size, odds ratios (ORs) and 95% confidence intervals (CIs); and (4) EC cases confirmed using histopathology. The exclusion criteria were as follows: (1) rationale and study design obviously different from our research objectives; (2) not case-control study; (3) malignant tumor cases included in controls; and (4) duplicated studies, reviews, case reports. If duplicate data were presented in more than one study, only the most informative and recent one was included.
Data extraction
Three investigators (Song, Zhou, and Du) reviewed and extracted information independently from selected publications in accordance with the above mentioned inclusion and exclusion criteria. Any conflicts over study/data inclusion were settled by a discussion between the investigators. Following the criteria above, 21 articles [4,13-28,30-33] were included in the present analyses. The steps taken towards article selection are shown below (Figure 1). The following data were extracted from included studies: authors of study, study area and period, the number of cases and controls, OR and 95% CIs. If crude and adjusted ORs and 95% CIs were both offered, we extracted the results that were adjusted for the most potential confounding variables. When the ORs were not presented, we calculated unadjusted ORs from the exposure data given in the articles. The details of each study are shown in Table 1.
Figure 1.
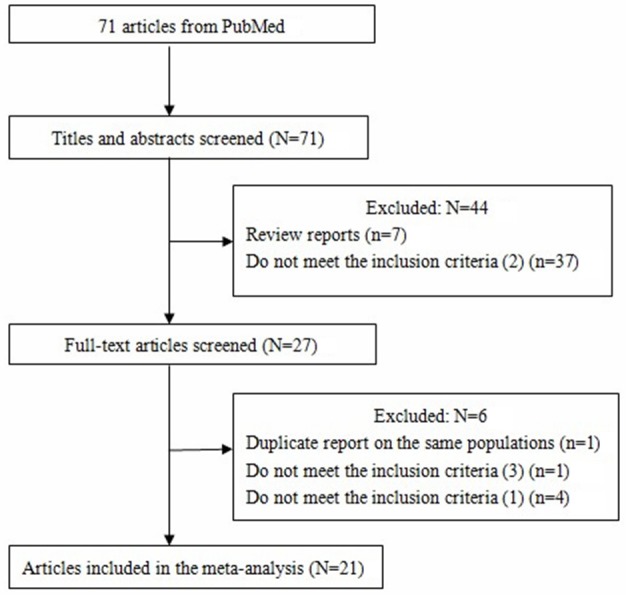
Flow diagram of study identification.
Table 1.
Characteristics of studies on the association of GSTP1 Ile105Val and the risk of esophageal cancer
| Study | Study area | Ethnicity | Cases/controls | OR# (95% CI) | OR& (95% CI) | OR% (95% CI) |
|---|---|---|---|---|---|---|
| Morita 1998 [18] | Japan | Asian | 66/164 | 0.19 (0.07-0.51) | - | 0.18 (0.07-0.48) |
| Lin 1998 [19] | China | Asian | 42/36 | 0.83 (0.31-2.22) | 0.25 (0.03-2.60) | 0.7 (0.3-1.8)a |
| Van Lieshout 1999 [6,13] | Caucasian | Caucasian | 34/247 | 3.45 (1.55-7.65) | 3.65 (0.88-15.07) | 3.47 (1.60-7.57) |
| Tan 2000 [20] | China | Asian | 150/150 | 0.89 (0.55-1.44) | 1.47 (0.50-4.29) | 1.0 (0.8-1.3)a |
| Lee 2000 [21] | China | Asian | 90/270 | - | - | 0.66 (0.39-1.11) |
| Casson 2003 [14] | Canada | Caucasian | 45/45 | 2.5 (1.0-6.3) | 0.8 (0.2-3.1) | 1.8 (0.8-4.3)a |
| Wang 2003 [15] | China | Asian | 62/38 | 1.91 (0.82-4.45) | 2.48 (0.24-25.44) | 5.37 (2.50-11.50) |
| Ribeiro Pinto 2003 [16] | Brazilian | Mixed | 34/68 | - | - | 4.09 (1.29-13.00) |
| Abbas 2004 [22] | France | Caucasian | 70/124 | 1.12 (0.61-2.07) | 1.27 (0.41-3.90) | 1.02 (0.55-1.89)a |
| Jain 2006 [23] | India | Indian | 100/137 | 0.80 (0.47-1.39) | 1.29 (0.48-3.45) | 0.87 (0.52-1.46) |
| Cai 2006 [24] | China | Asian | 218/415 | 0.93 (0.64-1.35) | 0.46 (0.13-1.67) | 0.88 (0.61-1.27) |
| Casson 2006 [25] | Canada | Caucasian | 56/95 | 1.36 (0.65-2.84) | 2.22 (0.81-6.06) | 1.54 (0.77-3.07) |
| Murphy 2007 [26] | Ireland | Caucasian | 207/223 | 0.93 (0.62-1.39) | 1.00 (0.53-1.88) | 0.94 (0.62-1.41) |
| Wideroff 2007 [27] | U.S | Caucasian | 67/206 | 0.71 (0.38-1.32)a | 1.73 (0.75-4.02)a | 0.87 (0.50-1.50) |
| Rossini 2007 [17] | Brazil | Mixed | 162/252 | 1.66 (1.04-2.66) | 1.78 (0.89-3.54) | 2.12 (1.37-3.29)a |
| Zendehdel 2009 [28] | Sweden | Caucasian | 175/471 | 1.21 (0.83-1.75) | 1.41 (0.76-2.62) | 1.24 (0.87-1.77) |
| Liu 2010 [30] | China | Asian | 97/97 | 0.896 (0.478-1.678)a | - | 0.825 (0.447-1.52)a |
| Moaven 2010 [31] | Iran | Mixed | 148/137 | 0.83 (0.50-1.36) | 1.672 (0.678-4.119)a | 1.100 (0.688-1.757)a |
| Li 2010 [32] | South African | Mixed | 245/288 | 1.01 (0.68-1.48)a | 1.21 (0.71-2.07)a | 1.00 (0.70-1.43) |
| Malik 2010 [4] | Kashmir Valley | Indian | 135/195 | 1.00 (0.62-1.6)a | 2.48 (1.03-6.02)a | 1.16 (0.74-1.80) |
| Matejcic 2011 [33] | South African | Mixed | 554/902 | 1.02 (0.79-1.32) | 1.03 (0.77-1.38) | 1.02 (0.81-1.30) |
OR of esophageal cancer associated with GSTP1 Ile105Val: Ile/Val vs. Ile/Ile.
OR of esophageal cancer associated with GSTP1 Ile105Val: Val/Val vs. Ile/Ile.
OR of esophageal cancer associated with GSTP1 Ile105Val: Ile/Val + Val/Val vs. Ile/Ile.
adjusted for potential confounding variables.
OR, odds ratio and CI, confidence interval.
Statistical analysis
Deviations from Hardy--Weinberg equilibrium (HWE) were tested using Fisher’s exact test to evaluate the genetic equilibrium of each study [34]. For all studies, we evaluated the risk of the hetero- and homozygous carriers of the variant Val allele, both together and separately, compared with the wild type Ile allele. Then, we calculated the overall ORs of the polymorphisms.
Tests for heterogeneity were made among studies using the Cochran’s Q and I2 test statistic [35]. For the Cochran’s Q test statistic, a P value < 0.10 was accepted as statistically significant heterogeneity. Random-effects models were used to estimate summary ORs and 95% CIs [36]. To examine potential sources of heterogeneity, we also conducted subgroup analyses by histological types (EAC and ESCC) and by ethnicity (Asian and Caucasian population). Meta regression analysis was also performed to identify sources of heterogeneity according to several variables, such as number of cases, source of controls, covariates adjusted, and publication time.
Sensitivity analyses were conducted to assess the strength of our findings by excluding one study at a time. Begg’s funnel plot and Egger’s regression test [37] were used to evaluate publication bias. In Egger’s test, when P value < 0.10, it was considered statistically significant publication bias. All analyses were conducted using Stata v.12 (StataCorp LP, TX) statistical software.
Results
Literature search and studies’ characteristics
Our keyword search identified 71 papers, from which 44 papers [7 reviews and 37 did not meet criteria (2)] were excluded after review of the abstracts. After reading the full texts of the remaining 27 papers, we eliminated an additional 6 papers, including 1 duplicated report, 4 failure to meet inclusion criterion (1) and 1 failure to meet inclusion criterion (3) (Figure 1). In summary, a total of 21 case-control studies evaluating the association between GSTP1 Ile105Val polymorphism and risk of EC were identified, with 2,757 cases and 4,560 controls included [4,13-28,30-33]. Among these 21 studies, 9 were performed in Asian populations, 7 in Caucasians, and 5 in mixed ethnicity populations. Controls in 7 studies were population-based and controls in the other 14 studies were hospital-based. The study characteristics are shown in Table 1.
Meta-analysis results
The summary OR for Ile/Val + Val/Val vs. Ile/Ile, Ile/Val vs. Ile/Ile and Val/Val vs. Ile/Ile was 1.14 (95% CI 0.94-1.38), 1.05 (95% CI 0.88-1.24), 1.25 (95% CI 1.05-1.49), respectively. (Figures 2, 3 and 4).
Figure 2.
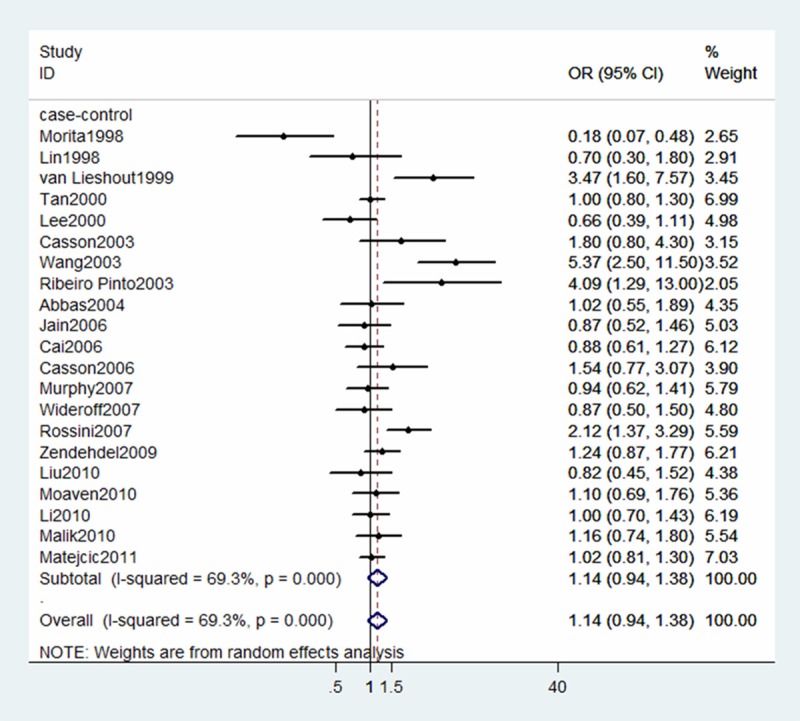
Forest plot for association between GSTP1 Ile105Val and risk of ESCC (Ile/Val + Val/Val vs Ile/Ile). A random-effects model was used. The squares and horizontal lines represent the study-specific OR and 95% CI. The diamond corresponds to the summary OR and 95% CI.
Figure 3.
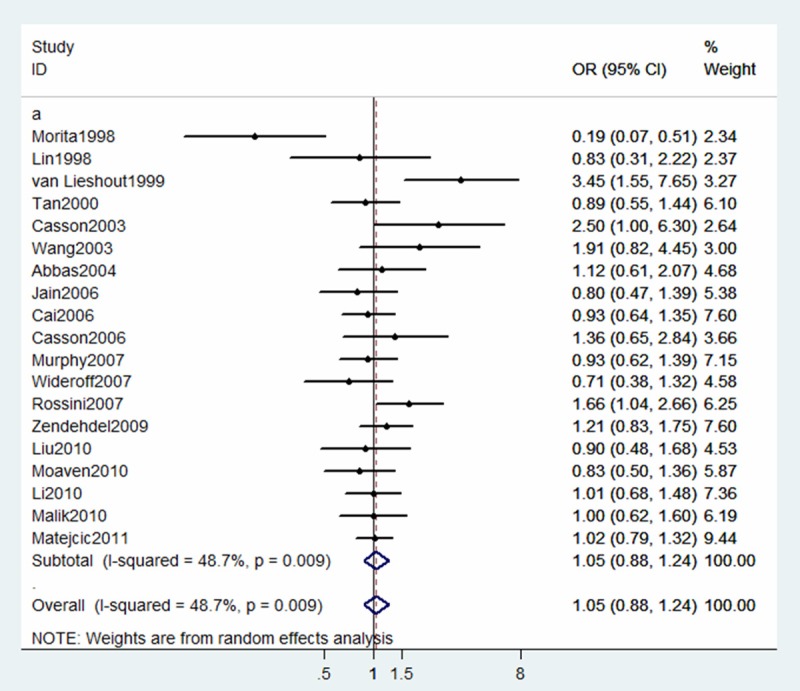
Forest plot for association between GSTP1 Ile105Val and risk of ESCC (Ile/Val vs Ile/Ile). A random-effects model was used. The squares and horizontal lines represent the study-specific OR and 95% CI. The diamond corresponds to the summary OR and 95% CI.
Figure 4.
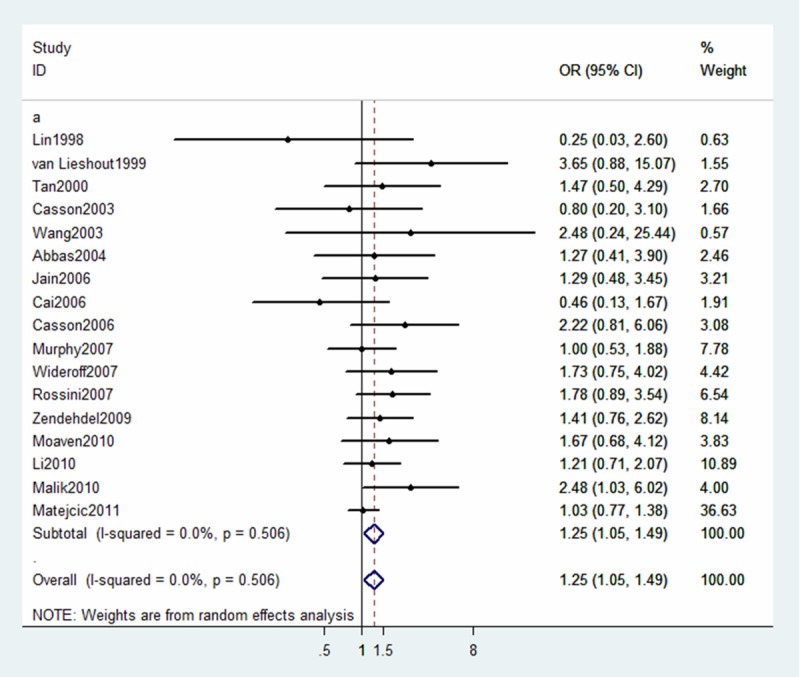
Forest plot for association between GSTP1 Ile105Val and risk of ESCC (Val/Val vs Ile/Ile). A random-effects model was used. The squares and horizontal lines represent the study-specific OR and 95% CI. The diamond corresponds to the summary OR and 95% CI.
Subgroup analyses
Subgroup analyses according to histological types are shown in Table 2. Compared with wild-type (Ile/Ile), variant homozygote (Val/Val) of GSTP1 Ile105Val was associated with a significantly increased risk of ESCC (Val/Val vs. Ile/Ile: OR = 1.45, 95% CI = 1.07-1.96; P = 0.134 for heterogeneity test). However, we failed to find any significant association for GSTP1 Ile105Val with risk of EAC in different genetic models. We also performed stratified analysis by ethnicity (Asian and Caucasian group). As shown in Table 2, a significant association between GSTP1 Ile105Val polymorphism and risk of EC among the Caucasian population was found (OR = 1.41, 95% CI = 1.01-1.95; P = 0.602 for heterogeneity test). Such a significant association was not observed in the Asian study population.
Table 2.
Subgroup analyses for the association of GSTP1 Ile105Val polymorphism and esophageal cancer
| Subgroups | No of studies | OR (95% CI) | I2 statistics (%) | Test for heterogeneity* (p value) |
|---|---|---|---|---|
| Histological subtype | ||||
| ESCC | ||||
| Ile/Val + Val/Val vs. Ile/Ile | 13 | 1.19 (0.93-1.51) | 68.6 | < 0.001 |
| Ile/Val vs. Ile/Ile | 12 | 1.06 (0.91-1.24) | 14.4 | 0.303 |
| Val/Val vs. Ile/Ile | 11 | 1.45 (1.07-1.96) | 33.1 | 0.134 |
| EAC | ||||
| Ile/Val + Val/Val vs. Ile/Ile | 9 | 1.18 (0.90-1.55) | 29.9 | 0.179 |
| Ile/Val vs. Ile/Ile | 9 | 1.21 (0.88-1.67) | 43.9 | 0.075 |
| Val/Val vs. Ile/Ile | 9 | 1.29 (0.90-1.84) | 0 | 0.84 |
| Ethnicity | ||||
| Asian | ||||
| Ile/Val + Val/Val vs. Ile/Ile | 7 | 0.88 (0.55-1.42) | 82.4 | <0.001 |
| Ile/Val vs. Ile/Ile | 6 | 0.83 (0.55-1.27) | 59.9 | 0.029 |
| Val/Val vs. Ile/Ile | 4 | 0.75 (0.33-1.68) | 75.6 | 0.006 |
| Caucasian | ||||
| Ile/Val + Val/Val vs. Ile/Ile | 8 | 1.36 (0.99-1.88) | 53.8 | 0.034 |
| Ile/Val vs. Ile/Ile | 7 | 1.28 (0.91-1.79) | 55.8 | 0.035 |
| Val/Val vs. Ile/Ile | 7 | 1.41 (1.01-1.95) | 0 | 0.602 |
Test for heterogeneity: random effect modeling was used.
Meta-regression analyses
We performed meta-regression analyses regarding the number of cases, ethnicity, adjusted covariates, and the publication time. We found that all of these variables did not appear to be main causes of heterogeneity, with P values equal to 0.542, 0.453, 0.992, and 0.873, respectively.
Sensitivity analyses
We performed sensitivity analyses by removing one study at a time and then estimating summary OR of the remaining studies. We found the results to be stable (data not shown).
Assessment for publication bias
A Begg’s funnel plot was generated, showing nearly symmetrical pattern (Figure 5), indicating low possibility of publication bias. Egger’s test was also used to quantitatively evaluate publication bias, which confirmed no evidence of bias (p = 0.570).
Figure 5.
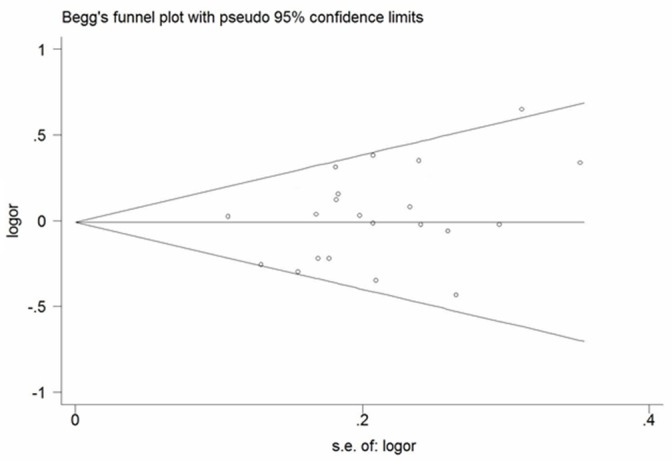
Begg’s funnel plot for publication bias assessment.
Discussion
GSTP1 Ile105Val polymorphism has been evaluated as a potential susceptibility factor contributing to the risk of developing various cancers including cancer of the breast, prostate and lung, with the variant Val105 genotype more likely to be associated with increased risk of cancer [38-40]. Using a meta-analytic approach, we consistently found a statistically significant association between variant homozygote (Val/Val) of GSTP1 Ile105Val and increased risk of EC. A previous meta-analysis of 13 published case-control studies found no significant general main effects for GSTP1 Ile105Val polymorphism on EC risk [29]. In the present study, we analyzed data from 2,757 cases and 4,560 controls in 21 studies to have the statistical power to detect differences and provide more precise risk estimates than the previous meta-analysis and individual studies, the majority of which suffered from limited sample size [4,13-28,30-33].
Results from our meta-analysis stratified by histological types of EC indicate that individuals carrying variant homozygous Val/Val genotype had significantly higher risk of ESCC than individuals carrying wild-type Ile/Ile genotype. However, such a significant association was not observed with EAC. GSTP1 is known to metabolize tobacco-related carcinogens and eliminate the oxidative products of thymidine or uracil propenal [41]. GSTP1 has an effect upon benzo(α)pyrene and its major metabolites [9], which are the major components of cigarette smoke [42]. Moreover, epidemiological studies have shown that GSTP1 polymorphisms seem to be more associated with tobacco-related cancers [19]. ESCC and EAC have significant divergence in etiology while sharing a few etiological factors. Smoking is generally accepted as a risk factor for both ESCC and EAC, but the effect is much stronger in ESCC than in EAC [43]. Thus, the difference in main effects of GSTP1 variant genotype in these two histological types of EC is biologically plausible.
Our stratified analyses by ethnicity revealed a significant association between GSTP1 Ile105Val polymorphism and EC in Caucasians, while the association was not significant in the Asian subgroup. This appears to contradict the above stratification analysis by histological type because ESCC is the major type of EC in the Asian population. However, the result in the Asian subgroup was based on 4 studies in Chinese populations showing a high level of between-study heterogeneity, suggesting that the studies do not estimate the same effect due to different degree of bias. In addition, there are significant differences regarding etiological profiles between high and low incidence areas within China [44]. This difference may also be responsible for variation in EC risk in these studies and the overall non-significance. More large studies are warrant to determine the effect of GSTP1 Ile105Val polymorphism on EC risk in Asians.
Although no significant association between GSTP1 Ile105Val polymorphism and EC risk was observed in dominant model (Ile/Val + Val/Val vs Ile/Ile) or in pair comparison between Ile/Val and Ile/Ile genotype, we cannot exclude that we may fail to detect the effect because of between-study heterogeneity (p < 0.001 for heterogeneity test for both meta-analyses). Although we conducted subgroup analyses by ethnicity and histological type to explore potential sources of heterogeneity, other possible sources such as publication year, case-control matching, and sample size were unable to be examined in our analyses. Uncontrolled confounding factors could be a major source of between-study heterogeneity because most studies used crude ORs and 95% CI. Additionally, the design of some studies was not optimal, some having small sample sizes [14-16,19], and some including populations with highly heterogeneous ethnic backgrounds or of unclear ethnicity [16,17,31-33]. Case-control study design is prone to selection bias, which could be a source of heterogeneity. Furthermore, because GSTP1 polymorphisms might contribute to susceptibility to non-cancer disease, using hospital-based controls may introduce heterogeneity to the meta-analysis. Therefore, more optimal and well-designed studies are required to evaluate the genetic association between GSTP1 Ile105Val polymorphism and EC risk in the future.
Considerable effort was made to test for possible associations between GSTP1 Ile105Val polymorphism and risk of EC, and a significant effect of GSTP1 variant homozygous (Val/Val) genotype on EC risk was found with no significant between-study heterogeneity. However, there are still some limitations to the meta-analysis. First, we cannot control for confounding factors that were not adjusted for in individual studies, such as age, sex, family history of cancer, smoking, alcohol consumption, and other potential risk factors. These factors might modify or even change the direction of observed effect. Second, GSTP1 may interact with environmental factors or interact with other genes in creating susceptibility for EC. However, due to lack of these data in individual data, we were unable to explore potential gene-gene or gene-environment interactions. Thirdly, although we did not find evidence of publication bias, it is possible as we do not have information on unpublished studies.
In conclusion, this meta-analysis demonstrates that GSTP1 Ile105Val significantly modified the risk of EC, especially ESCC, and that the effect of modification was particularly pronounced in the Caucasian population. To confirm our findings, more, well-designed large-scale studies in diverse ethnic populations are warranted.
Abbreviations
- GSTP1
glutathione s-transferase P1
- EC
esophageal cancer
- ORs
odds ratios
- CIs
confidence intervals
- SNP
single-nucleotide polymorphism
- ESCC
esophageal squamous cell carcinoma
- EAC
esophageal adenocarcinoma
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Castellsague X, Munoz N, De Stefani E, Victora CG, Castelletto R. Independent and joint effects of tobacco smoking and alcohol drinking on the risk of esophageal cancer in men and women. Int J Cancer. 1999;82:657–664. doi: 10.1002/(sici)1097-0215(19990827)82:5<657::aid-ijc7>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 4.Malik MA, Upadhyay R, Mittal RD, Zargar SA, Mittal B. Association of xenobiotic metabolizing enzymes genetic polymorphisms with esophageal cancer in Kashmir Valley and influence of environmental factors. Nutr Cancer. 2010;62:734–742. doi: 10.1080/01635581003605904. [DOI] [PubMed] [Google Scholar]
- 5.Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]
- 6.van Lieshout EM, Tiemessen DM, Witteman BJ, Jansen JB, Peters WH. Low glutathione and glutathione S-transferase levels in Barrett’s esophagus as compared to normal esophageal epithelium. Jpn J Cancer Res. 1999;90:81–85. doi: 10.1111/j.1349-7006.1999.tb00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 8.Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikula J, Singhal SS. Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994;224:893–899. doi: 10.1111/j.1432-1033.1994.00893.x. [DOI] [PubMed] [Google Scholar]
- 9.Hu X, Xia H, Srivastava SK, Herzog C, Awasthi YC. Activity of four allelic forms of glutathione S-transferase hGSTP1-1 for diol epoxides of polycyclic aromatic hydrocarbons. Biochem Biophys Res Commun. 1997;238:397–402. doi: 10.1006/bbrc.1997.7311. [DOI] [PubMed] [Google Scholar]
- 10.Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis. 1998;19:433–436. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- 11.Johansson AS, Stenberg G, Widersten M, Mannervik B. Structure-activity relationships and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. J Mol Biol. 1998;278:687–698. doi: 10.1006/jmbi.1998.1708. [DOI] [PubMed] [Google Scholar]
- 12.Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis. 1997;18:1285–1289. doi: 10.1093/carcin/18.7.1285. [DOI] [PubMed] [Google Scholar]
- 13.van Lieshout EM, Roelofs HM, Dekker S, Mulder CJ, Wobbes T. Polymorphic expression of the glutathione S-transferase P1 gene and its susceptibility to Barrett’s esophagus and esophageal carcinoma. Cancer Res. 1999;59:586–589. [PubMed] [Google Scholar]
- 14.Casson AG, Zheng Z, Chiasson D, MacDonald K, Riddell DC. Associations between genetic polymorphisms of Phase I and II metabolizing enzymes, p53 and susceptibility to esophageal adenocarcinoma. Cancer Detect Prev. 2003;27:139–146. doi: 10.1016/s0361-090x(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 15.Wang LD, Zheng S, Liu B, Zhou JX, Li YJ. CYP1A1, GSTs and mEH polymorphisms and susceptibility to esophageal carcinoma: study of population from a high-incidence area in north China. World J Gastroenterol. 2003;9:1394–1397. doi: 10.3748/wjg.v9.i7.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro Pinto LF, Teixeira Rossini AM, Albano RM, Felzenszwalb I, de Moura Gallo CV. Mechanisms of esophageal cancer development in Brazilians. Mutat Res. 2003;544:365–373. doi: 10.1016/j.mrrev.2003.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Rossini A, Rapozo DC, Soares Lima SC, Guimaraes DP, Ferreira MA. Polymorphisms of GSTP1 and GSTT1, but not of CYP2A6, CYP2E1 or GSTM1, modify the risk for esophageal cancer in a western population. Carcinogenesis. 2007;28:2537–2542. doi: 10.1093/carcin/bgm222. [DOI] [PubMed] [Google Scholar]
- 18.Morita S, Yano M, Tsujinaka T, Ogawa A, Taniguchi M. Association between genetic polymorphisms of glutathione S-transferase P1 and N-acetyltransferase 2 and susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 1998;79:517–520. doi: 10.1002/(sici)1097-0215(19981023)79:5<517::aid-ijc12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Lin DX, Tang YM, Peng Q, Lu SX, Ambrosone CB. Susceptibility to esophageal cancer and genetic polymorphisms in glutathione S-transferases T1, P1, and M1 and cytochrome P450 2E1. Cancer Epidemiol Biomarkers Prev. 1998;7:1013–1018. [PubMed] [Google Scholar]
- 20.Tan W, Song N, Wang GQ, Liu Q, Tang HJ. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev. 2000;9:551–556. [PubMed] [Google Scholar]
- 21.Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP, Chen CJ, Hsieh CY, Wu MT. Genetic polymorphisms of p53 and GSTP1, but not NAT2, are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458–464. doi: 10.1002/1097-0215(20000920)89:5<458::aid-ijc10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 22.Abbas A, Delvinquiere K, Lechevrel M, Lebailly P, Gauduchon P. GSTM1, GSTT1, GSTP1 and CYP1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinoma. World J Gastroenterol. 2004;10:3389–3393. doi: 10.3748/wjg.v10.i23.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain M, Kumar S, Rastogi N, Lal P, Ghoshal UC. GSTT1, GSTM1 and GSTP1 genetic polymorphisms and interaction with tobacco, alcohol and occupational exposure in esophageal cancer patients from North India. Cancer Lett. 2006;242:60–67. doi: 10.1016/j.canlet.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 24.Cai L, Mu LN, Lu H, Lu QY, You NC. Dietary selenium intake and genetic polymorphisms of the GSTP1 and p53 genes on the risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:294–300. doi: 10.1158/1055-9965.EPI-05-0680. [DOI] [PubMed] [Google Scholar]
- 25.Casson AG, Zheng Z, Porter GA, Guernsey DL. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423–431. doi: 10.1016/j.cdp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Murphy SJ, Hughes AE, Patterson CC, Anderson LA, Watson RG. A population-based association study of SNPs of GSTP1, MnSOD, GPX2 and Barrett’s esophagus and esophageal adenocarcinoma. Carcinogenesis. 2007;28:1323–1328. doi: 10.1093/carcin/bgm007. [DOI] [PubMed] [Google Scholar]
- 27.Wideroff L, Vaughan TL, Farin FM, Gammon MD, Risch H. GST, NAT1, CYP1A1 polymorphisms and risk of esophageal and gastric adenocarcinomas. Cancer Detect Prev. 2007;31:233–236. doi: 10.1016/j.cdp.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zendehdel K, Bahmanyar S, McCarthy S, Nyren O, Andersson B. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer Causes Control. 2009;20:2031–2038. doi: 10.1007/s10552-009-9399-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Wang F, Shan S, Zhao Y, Qiu X. Genetic polymorphism of p53, but not GSTP1, is association with susceptibility to esophageal cancer risk - a meta-analysis. Int J Med Sci. 2010;7:300–308. doi: 10.7150/ijms.7.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Yin L, Pu Y, Li Y, Liang G. Functional alterations in the glutathione S-transferase family associated with enhanced occurrence of esophageal carcinoma in China. J Toxicol Environ Health. 2010;73:471–482. doi: 10.1080/15287390903523394. [DOI] [PubMed] [Google Scholar]
- 31.Moaven O, Raziee HR, Sima HR, Ganji A, Malekzadeh R. Interactions between Glutathione-S-Transferase M1, T1 and P1 polymorphisms and smoking, and increased susceptibility to esophageal squamous cell carcinoma. Cancer Epidemiol. 2010;34:285–290. doi: 10.1016/j.canep.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Li D, Dandara C, Parker MI. The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet. 2010;11:47. doi: 10.1186/1471-2156-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matejcic M, Li D, Prescott NJ, Lewis CM, Mathew CG. Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case-control study. PLoS One. 2011;6:e29366. doi: 10.1371/journal.pone.0029366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenland S. Quantitative methods in the review of epidemiologic literature. Epidemiol Rev. 1987;9:1–30. doi: 10.1093/oxfordjournals.epirev.a036298. [DOI] [PubMed] [Google Scholar]
- 37.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybicki BA, Neslund-Dudas C, Nock NL, Schultz LR, Eklund L. Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev. 2006;30:412–422. doi: 10.1016/j.cdp.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SA, Fowke JH, Lu W, Ye C, Zheng Y. Cruciferous vegetables, the GSTP1 Ile105Val genetic polymorphism, and breast cancer risk. Am J Clin Nutr. 2008;87:753–760. doi: 10.1093/ajcn/87.3.753. [DOI] [PubMed] [Google Scholar]
- 40.Miller DP, De Vivo I, Neuberg D, Wain JC, Lynch TJ. Association between self-reported environmental tobacco smoke exposure and lung cancer: modification by GSTP1 polymorphism. Int J Cancer. 2003;104:758–763. doi: 10.1002/ijc.10989. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima T, Wang RS, Nimura Y, Pin YM, He M. Expression of cytochrome P450s and glutathione S-transferases in human esophagus with squamous-cell carcinomas. Carcinogenesis. 1996;17:1477–1481. doi: 10.1093/carcin/17.7.1477. [DOI] [PubMed] [Google Scholar]
- 42.Lofroth G. Environmental tobacco smoke: overview of chemical composition and genotoxic components. Mutat Res. 1989;222:73–80. doi: 10.1016/0165-1218(89)90021-9. [DOI] [PubMed] [Google Scholar]
- 43.Pera M, Pera M. Recent changes in the epidemiology of esophageal cancer. Surg Oncol. 2001;10:81–90. doi: 10.1016/s0960-7404(01)00025-1. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–242. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]


