Abstract
l-Aspartate is a regulatory feedback inhibitor of the biotin-dependent enzyme pyruvate carboxylase in response to increased levels of tricarboxylic acid cycle intermediates. Detailed studies of l-aspartate inhibition of pyruvate carboxylase have been mainly confined to eukaryotic microbial enzymes, and aspects of its mode of action remain unclear. Here we examine its inhibition of the bacterial enzyme Rhizobium etli pyruvate carboxylase. Kinetic studies demonstrated that l-aspartate binds to the enzyme cooperatively and inhibits the enzyme competitively with respect to acetyl-CoA. l-Aspartate also inhibits activation of the enzyme by MgTNP-ATP. The action of l-aspartate was not confined to inhibition of acetyl-CoA binding, because the acetyl-CoA-independent activity of the enzyme was also inhibited by increasing concentrations of l-aspartate. This inhibition of acetyl-CoA-independent activity was demonstrated to be focused in the biotin carboxylation domain of the enzyme, and it had no effect on the oxamate-induced oxaloacetate decarboxylation reaction that occurs in the carboxyl transferase domain. l-Aspartate was shown to competitively inhibit bicarbonate-dependent MgATP cleavage with respect to MgATP but also probably inhibits carboxybiotin formation and/or translocation of the carboxybiotin to the site of pyruvate carboxylation. Unlike acetyl-CoA, l-aspartate has no effect on the coupling between MgATP cleavage and oxaloacetate formation. The results suggest that the three allosteric effector sites (acetyl-CoA, MgTNP-ATP, and l-aspartate) are spatially distinct but connected by a network of allosteric interactions.
Pyruvate carboxylase (PC, EC 6.4.1.1) is a biotin-dependent carboxylase, which catalyzes carboxylation of pyruvate to oxaloacetate. This reaction is considered to be an important anaplerotic reaction because it replenishes tricarboxylic acid cycle intermediates that have been withdrawn for anabolic purposes.1 PC is found in wide variety of organisms, including eubacteria, yeast, fungi, and animals (for reviews, see refs (1) and (2)). In mammals, PC is also involved in gluconeogenesis in liver, de novo fatty acid synthesis in liver and adipose tissue, and neurotransmitter synthesis in astrocytes.2,3 Furthermore, PC is also necessary for glucose-induced insulin secretion in pancreatic β-cells.4 As PC has such diverse metabolic roles, dysregulation of this enzyme is involved in many diseases, including type 2 diabetes, obesity, and cancers.3,5,6
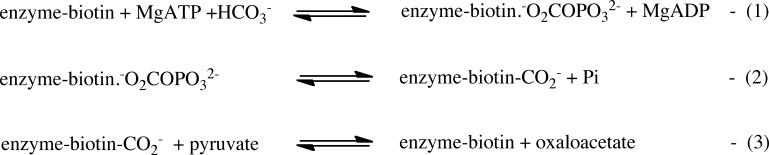
Pyruvate carboxylation catalyzed by PC proceeds through a series of reactions shown in Figure 1. Reactions 1 and 2, in which the biotin cofactor is carboxylated via a carboxyphosphate intermediate (−O2COPO32–), occur in the biotin carboxylase (BC) domain. Reaction 3, in which the carboxyl group is transferred from the carboxybiotin to pyruvate to form oxaloacetate, occurs in the carboxyl transferase (CT) domain. PC is commonly an α4 tetramer, and the overall pyruvate carboxylation reaction has been shown to proceed via intersubunit catalysis where the subunits act in pairs so that the biotin of one subunit is carboxylated in its own BC domain but transfers its carboxyl group to pyruvate in its partner subunit’s CT domain.7
Figure 1.
Partial reactions catalyzed by pyruvate carboxylase. Reactions 1 and 2 occur in the BC domain, and reaction 3 occurs in the CT domain.
In the majority of organisms, the activity of PC is positively regulated by the allosteric activator acetyl-CoA as a result of an increased rate of fatty acid oxidation. This mechanism allows sufficient levels of oxaloacetate to oxidize β-oxidation-derived acetyl-CoA. In microbes, PC is negatively regulated by l-aspartate, which signals an abundance of tricarboxylic acid cycle intermediates. From structural studies of RePC7 and Staphylococcus aureus PC,8 the binding site for acetyl-CoA has been identified as an allosteric domain that is surrounded by the BC, CT, and biotin carboxyl carrier protein (BCCP) domains. The binding site for l-aspartate has yet to be identified.
While much has been learned about the action of acetyl-CoA in a wide variety of organisms (see ref (9) for a review), the action of l-aspartate has been most extensively studied in the eukaryotic microbial PCs from Aspergillus nidulans(10) and Saccharomyces cerevisiae.11,12 In both cases, the inhibition was reported to be competitive with respect to acetyl-CoA; in A. nidulans, PC l-aspartate decreased the cooperativity of activation of the enzyme by acetyl-CoA, while in S. cerevisiae PC, it increased the cooperativity. Much less is known about the inhibitory effects of l-aspartate in bacterial PCs, and in general, the loci and mechanisms of action of l-aspartate are not well understood. In this study, we have performed a detailed steady-state kinetic analysis of the inhibitory effects of l-aspartate on RePC, which has been extremely thoroughly characterized in structural and mechanistic terms,7,13−15 to investigate the loci of action and inhibitory mechanisms of l-aspartate.
Experimental Procedures
Materials
Sodium pyruvate, sodium oxaloacetate, ATP, sodium phosphoenolpyruvate, acetyl-CoA, NADH, malate dehydrogenase, lactate dehydrogenase, and pyruvate kinase were purchased from Sigma. 2′,3′-O-(2,4,6-Trinitrophenyl)adenosine 5′-triphosphate (TNP-ATP) was obtained from Jena Bioscience. l-Aspartic acid was purchased from Fluka.
RePC Expression and Purification
N-Terminally His-tagged RePC was expressed in Escherichia coli BL21(DE3) and purified as described previously.16 The purified RePC was resuspended and stored at −80 °C in storage buffer containing 30% (v/v) glycerol, 100 mM Tris-HCl (pH 7.8), and 1 mM dithioerythreitol.17
Pyruvate Carboxylation Activity Assay
The pyruvate carboxylating activities in the absence or presence of acetyl-CoA were determined by a coupled spectrophotometric assay in which the oxaloacetate produced was converted to malate with concomitant oxidation of NADH in a reaction catalyzed by malate dehydrogenase.13 The assays were performed at 30 °C in 1 mL reaction mixtures containing 0.1 M Tris-HCl (pH 7.8), 20 mM NaHCO3, 6 mM MgCl2, 1 mM MgATP, 0.2 mM NADH, 10 mM sodium pyruvate, and 5 units of malate dehydrogenase. The concentrations of acetyl-CoA and l-aspartate were varied from 0 to 150 μM and from 0 to 8 mM, respectively.
To determine the effect of l-aspartate inhibition on the MgTNP-ATP activation of pyruvate carboxylation, MgTNP-ATP concentrations were varied from 0 to100 μM at different l-aspartate concentrations (140 μg of enzyme was used per assay).
The kapp values were calculated by dividing the initial velocity by the enzymic biotin concentration in each assay.
Oxamate-Induced Oxaloacetate Decarboxylation
The activity of oxamate-induced oxaloacetate decarboxylation in the absence of acetyl-CoA was determined by a coupled spectrophotometric assay in which the pyruvate produced was converted to lactate with concomitant oxidation of NADH in a reaction catalyzed by lactate dehydrogenase.18 All assays were performed at 30 °C in 1 mL of a reaction mixture containing 0.1 M Tris-HCl (pH 7.8), 0.22 mM NADH, 1 mM oxamate, and 4 units of lactate dehydrogenase. The concentrations of oxaloacetate and l-aspartate were varied between 0 and 250 μM and between 0 and 8 mM, respectively. The kcat value was calculated from the initial velocity divided by the enzymic biotin concentration in each assay.
Bicarbonate-Dependent ATP Cleavage Activity Assay
The activity of bicarbonate-dependent ATP cleavage reactions in the absence of acetyl-CoA was determined by a coupled spectrophotometric assay in which the ADP produced was converted to ATP with concomitant dephosphorylation of phosphoenolpyruvate to form pyruvate in a reaction catalyzed by pyruvate kinase. The pyruvate thus formed was converted to lactate with concomitant oxidation of NADH in a reaction catalyzed by lactate dehydrogenase.19 The reactions were performed at 30 °C in 1 mL mixture containing 0.1 M Tris-HCl (pH 7.8), 20 mM NaHCO3, 5 mM MgCl2, 0.22 mM NADH, 10 mM phosphoenolpyruvate, 5 units of pyruvate kinase, and 4 units of lactate dehydrogenase. Activities were determined by varying concentrations of MgATP and l-aspartate from 0 to 1000 μM and from 0 to 8 mM, respectively. The assay was initiated by the addition of the purified enzyme (493 μg/each assay).
Coupling between Oxaloacetate Formation and Pi Release in the Pyruvate Carboxylation Reaction
To determine the effect of l-aspartate on the coupling between oxaloacetate formation and Pi release in the pyruvate carboxylation assay, the initial rates of oxaloacetate formation and Pi release were determined in the absence and presence (8 mM) of l-aspartate. The rate of Pi release was measured using the assay of Black and Jones.20 Briefly, 2% (w/v) ammonium molybdate·4H2O, 14% (w/v) ascorbic acid in 50% (v/v) trichloroacetic acid, and 2% (w/v) trisodium citrate·2H2O with 2% (w/v) sodium arsenite in 2% (v/v) acetic acid were prepared as reagents A–C, respectively; 200 μL of reagent A, 300 μL of reagent B, and 800 μL of water were added to the cuvette followed by the addition of 200 μL samples of the pyruvate carboxylation reaction mixture 4, 8, 12, and 16 min after the start of the reaction. The solution was immediately mixed and stood for 1 min, after which 1 mL of reagent C was added. The blue color developed within 4 min, and the absorbance was measured at 700 nm. A standard curve of a range of Pi concentrations was prepared using a standard solution of NaH2PO4 in 0.1 M Tris (pH 7.8) and the assay described above. The Pi content of the pyruvate carboxylation reaction mixtures was calculated from the standard curve. From plots of Pi released versus time, the initial rates of Pi release were calculated by linear regression analysis for triplicate time courses. The initial rates of oxaloacetate formation were determined from spectrophotometric assays using the malate dehydrogenase coupling reaction, as described above. The reactions were initiated by addition of the purified RePC (35 μg/assay). The coupling between oxaloacetate formation and Pi release was calculated as the ratios of initial rates of these two reactions, and three separate sets of measurements were taken so that ratios are reported as the mean and standard deviations of these measurements.
Data Analysis
The acetyl-CoA dependence or MgTNP-ATP dependence of the pyruvate carboxylation reaction at each concentration of l-aspartate was analyzed by using nonlinear least-squares regression fits of the data to eq 1.
| 1 |
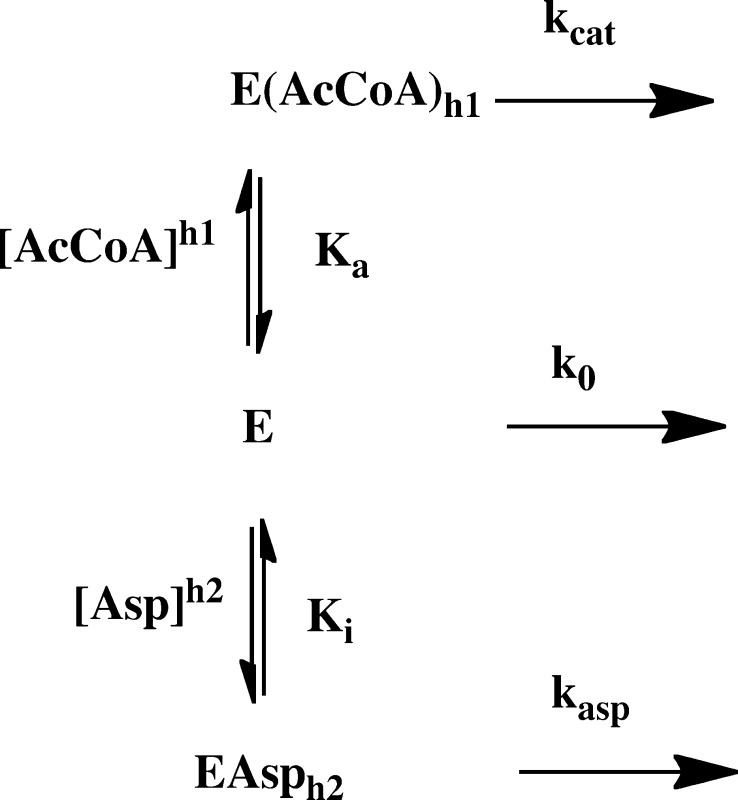
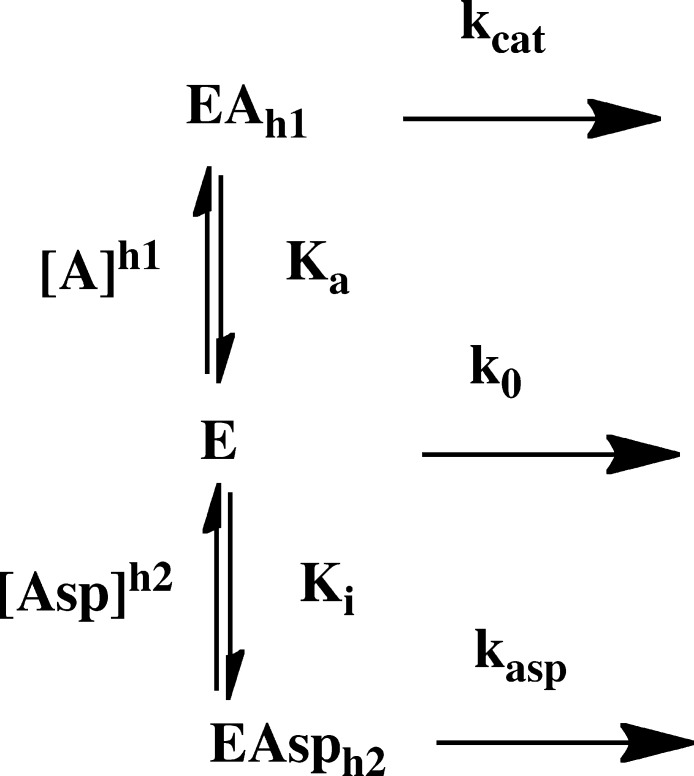
where [A] is either the acetyl-CoA concentration or the MgTNP-ATP concentration, Ka is the activation constant, h1 is the Hill coefficient of cooperativity, kapp is the apparent catalytic rate constant for the reaction at each concentration of l-aspartate, k0 is the catalytic rate constant of the acetyl-CoA-independent reaction (determined experimentally), and kcat is the catalytic rate constant of the acetyl-CoA-dependent reaction when acetyl-CoA is saturating. A global analysis of the acetyl-CoA activation of pyruvate carboxylating activities at the different, fixed concentrations of l-aspartate was performed by a nonlinear least-squares regression fit of the complete set of data initially to eq 2 that was derived for the reaction scheme in Figure 2, which describes competitive inhibition by l-aspartate with respect to acetyl-CoA but where kasp is set to zero.
| 2 |
where k0, kcat, Ka, and h1 are as described for eq 1 except they are the values of these parameters in the absence of l-aspartate, [Asp] is the concentration of l-aspartate, Ki is the dissociation constant of the enzyme–aspartate complex, and h2 is the Hill coefficient of cooperativity for the binding of l-aspartate to the RePC tetramer.
Figure 2.
Reaction scheme showing the proposed inhibitory mechanism of pyruvate carboxylating activity by l-aspartate in the presence of acetyl-CoA, where k0 and kcat are catalytic rate constants for the acetyl-CoA-independent and -dependent reactions catalyzed by the enzyme (E) and enzyme·acetyl-CoA complex (EAh1), respectively. The catalytic rate constant for the reaction catalyzed by the enzyme·l-aspartate complex (EAsph2) is kasp. Ka is the apparent dissociation constant of the EAh1 complex, and h1 is the Hill coefficient for the activation process. Ki is the apparent dissociation constant of the enzyme·l-aspartate complex (EAsph2), and h2 is the Hill coefficient for the inhibition by l-aspartate.
Inactivation of pyruvate carboxylating activity by l-aspartate in the absence of acetyl-CoA was analyzed by using nonlinear least-squares regression fits of the data to eq 3.
| 3 |
where the parameters are as described for eq 2, except for kasp, which is the catalytic rate constant for the pyruvate carboxylation reaction conducted by the enzyme·l-aspartate complex. The value of k0 was determined experimentally.
The data in Figure 3 were subsequently fit to eq 4, where the value of kasp was set at the value determined from the analysis of the effect of l-aspartate on acetyl-CoA-independent pyruvate carboxylation described above.
| 4 |
Figure 3.
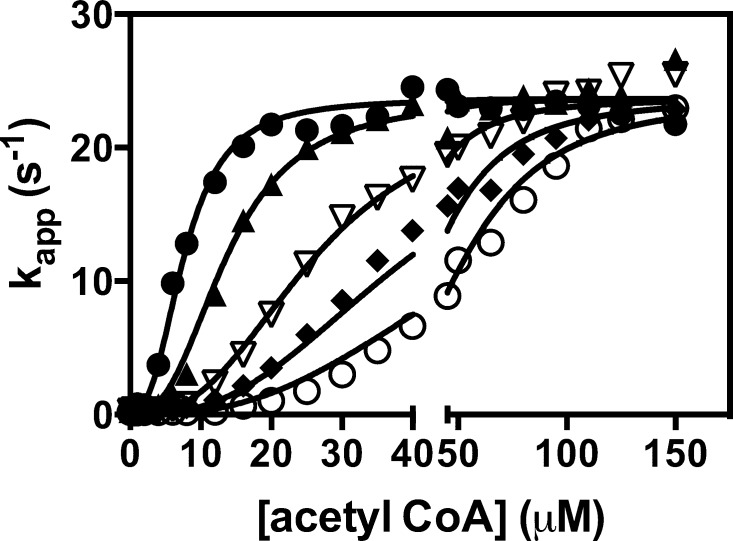
Effect of increasing concentrations of l-aspartate [(●) 0, (▲) 2, (▽) 4, (◆) 6, and (○) = 8 mM] on the activation of the pyruvate carboxylation reaction by acetyl-CoA. The lines represent a global, nonlinear least-squares regression fit of the data to eq 2. Reaction conditions were as described in Experimental Procedures.
The kinetics of oxamate-induced oxaloacetate decarboxylation in the absence of acetyl-CoA at different l-aspartate concentrations were analyzed by nonlinear least-squares regression fits of the data to eq 5.
| 5 |
where KOAA and [OAA] are the Michaelis–Menten constant for and the concentration of oxaloacetate, respectively. KI is the substrate inhibition constant for oxaloacetate.
The data describing inhibition of the bicarbonate-dependent MgATP cleavage reaction by l-aspartate were analyzed by a nonlinear least-squares regression fit initially to eq 6 that describes mixed inhibition and then to eq 7 that describes competitive inhibition.
| 6 |
| 7 |
where Km is the Michaelis–Menten constant for MgATP, Ki is the dissociation constant of the enzyme·l-aspartate complex, and Kis is that for the enzyme·l-aspartate·MgATP complex.
Results
Effect of Increasing l-Aspartate Concentrations on the Activation of Pyruvate Carboxylation by Acetyl-CoA
Table 1 shows the parameters estimated from the fits of eq 1 to each individual set of data at each concentration of l-aspartate shown in Figure 3 and k0. This clearly shows that Ka increases with increasing l-aspartate concentrations, whereas there is no strong trend of change in either kcat or h1. However, k0 does appear to decrease with increasing l-aspartate concentrations. As mentioned earlier, inhibition of fungal PCs by l-aspartate has been found to be competitive with respect to acetyl-CoA. To determine if this could be the case here, we developed a model (Figure 2) that describes this type of inhibition and from which eq 2 was derived. The data in Figure 3 were globally fit to this equation, and the solid lines in Figure 3 represent the fit. As one can see, the fit was good with an R2 of 0.992. The following parameters were estimated from the fit: kcat = 23.7 ± 0.2 s–1, Ka = 7.70 ± 0.27 μM, h1 = 2.65 ± 0.09, Ki = 1.26 ± 0.08 mM, and h2 = 2.78 ± 0.11. The value of h2 suggests that the binding of l-aspartate to RePC is also cooperative with a Hill coefficient similar to that for acetyl-CoA. A fit of eq 2 in which h2 was set to 1 (no allosteric binding of l-aspartate) failed to give a good fit to the data.
Table 1. Kinetic Parameters for Acetyl-CoA Activation of the Pyruvate Carboxylation Reaction Catalyzed by RePC in the Presence of Different Concentrations of l-Aspartatea.
| [l-aspartate] (mM) | Ka for acetyl-CoA (μM) | Hill coefficient (h1) | kcat (s–1) | k0 (s–1) |
|---|---|---|---|---|
| 0 | 7.3 ± 0.9 | 2.47 ± 0.21 | 22.9 ± 0.5 | 0.32 |
| 2 | 14.5 ± 0.5 | 2.87 ± 0.26 | 23.9 ± 0.6 | 0.27 |
| 4 | 23.7 ± 0.7 | 2.48 ± 0.16 | 24.9 ± 0.6 | 0.23 |
| 6 | 35.6 ± 1.1 | 2.74 ± 0.22 | 22.4 ± 0.6 | 0.21 |
| 8 | 60.5 ± 3.0 | 2.54 ± 0.21 | 25.3 ± 1.2 | 0.19 |
Assay conditions: 0.1 M Tris-HCl (pH 7.8), 20 mM NaHCO3, 6 mM MgCl2, 1 mM MgATP, 0.2 mM NADH, 10 mM sodium pyruvate, and 5 units of malate dehydrogenase (MDH). The concentrations of acetyl-CoA and l-aspartate were varied from 0 to 150 μM and from 0 to 8 mM, respectively. The parameters (±standard errors) were estimated from a nonlinear regression fit of the data at each l-aspartate concentration to eq 1 except k0, which was measured directly.
Effect of Increasing l-Aspartate Concentrations on the Pyruvate Carboxylation Reaction in the Absence of Acetyl-CoA
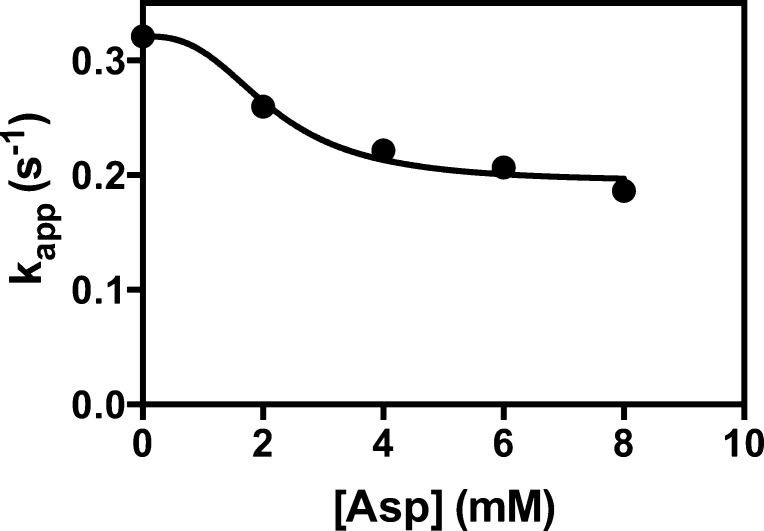
Figure 4 shows a plot of kapp at different concentrations of l-aspartate in the absence of acetyl-CoA. The data were fit to eq 3 that describes inhibition that depends on [l-aspartate]h2, using the value of h2 estimated above. The fit to eq 3 gave values of Ki and kasp of 2.2 ± 0.2 mM and 0.19 ± 0.01 s–1, respectively. The value of Ki is somewhat higher than that estimated for the global fit of the data in Figure 3, but the data set in Figure 4 is much smaller. The positive value of kasp, which is significantly different from zero (t test; p < 0.01), indicated that at a saturating concentration of l-aspartate, the enzyme retains some residual activity. Refitting the data in Figure 3 to eq 4, which accounts for this residual activity in the enzyme·l-aspartate complex, resulted in only very small changes to the estimated parameters: kcat = 23.7 ± 0.2 s–1, Ka = 7.70 ± 0.27 μM, h1 = 2.69 ± 0.09, Ki = 1.25 ± 0.07 mM, and h2 = 2.81 ± 0.11.
Figure 4.
Effect of increasing concentrations of l-aspartate on the acetyl-CoA-independent pyruvate carboxylation reaction. Data points at zero acetyl-CoA concentration were replotted from Figure 3. The line represents a nonlinear least-squares regression fit of the data to eq 3.
Effect of Increasing l-Aspartate Concentrations on the Activation of Pyruvate Carboxylation by MgTNP-ATP
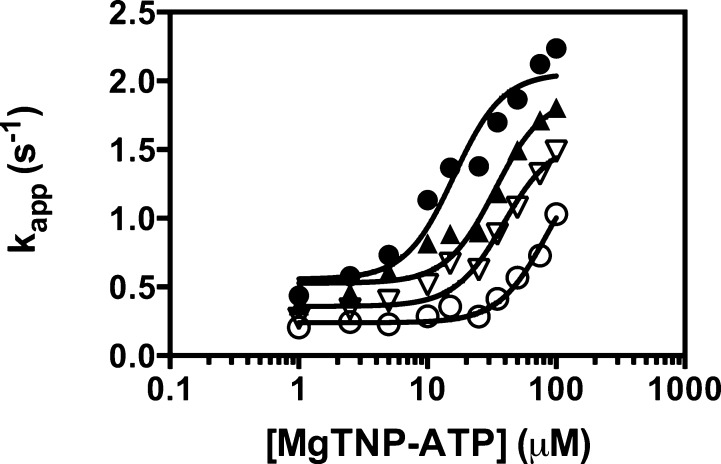
Figure 5 shows the effects of l-aspartate on the activation of pyruvate carboxylation by MgTNP-ATP, with each set of data at individual l-aspartate concentrations fit to eq 1. The estimated parameters from these fits are listed in Table 2. It is clear that with increasing concentrations of l-aspartate the value of Ka increases markedly and, as expected, that of k0 decreases. The value of kcat also appears to decrease with increasing concentrations of l-aspartate; however, the value of kcat at 8 mM l-aspartate is not significantly different from that at 0 mM l-aspartate (t test; p > 0.1).
Figure 5.
Effect of increasing concentrations of l-aspartate [(●) 0, (▲) 2, (▽) 4, and (○) 8 mM] on the activation of the pyruvate carboxylation reaction by MgTNP-ATP. The lines represent a nonlinear least-squares regression fit of each set data at a particular concentration of l-aspartate to eq 1, where h1 was set to 2.3, the value determined previously.13 Reaction conditions were as described in Experimental Procedures.
Table 2. Kinetic Parameters for MgTNP-ATP Activation of the Pyruvate Carboxylation Reaction Catalyzed by RePC in the Presence of Different Concentrations of l-Aspartatea.
| [l-aspartate] (mM) | Ka for MgTNP-ATP (μM) | kcat (s–1) | k0 (s–1) |
|---|---|---|---|
| 0 | 16.1 ± 2.9 | 2.1 ± 0.1 | 0.48 |
| 2 | 33.5 ± 5.6 | 1.9 ± 0.1 | 0.43 |
| 4 | 39.1 ± 6.2 | 1.6 ± 0.1 | 0.23 |
| 8 | 85.1 ± 17.6 | 1.5 ± 0.3 | 0.16 |
Assay conditions: 0.1 M Tris-HCl (pH 7.8), 20 mM NaHCO3, 6 mM MgCl2, 1 mM MgATP, 0.2 mM NADH, 10 mM sodium pyruvate, and 5 units of malate dehydrogenase (MDH). The concentrations of MgTNP-ATP and l-aspartate were varied from 0 to 100 μM and from 0 to 8 mM, respectively. The parameters (±standard errors) except k0, which was measured directly, were estimated from a nonlinear regression fit of the data at each l-aspartate concentration to eq 1, where h1 was set to 2.3, the value determined previously.13
Effect of l-Aspartate on the Coupling between MgATP Cleavage and Pyruvate Carboxylation
The coupling between MgATP cleavage in the BC domain and pyruvate carboxylation in the CT domain was examined by measuring the rates of the formation of oxaloacetate and release of Pi in the absence and presence (8 mM) of l-aspartate under identical reaction conditions. The ratios of oxaloacetate formation to Pi release in the absence and presence of l-aspartate (8 mM) were 0.96 ± 0.06 and 0.89 ± 0.06, respectively. The means ± SD of coupling ratios in the absence and presence of l-aspartate were found to be not significantly different using a t test (p > 0.2). This suggests that binding of l-aspartate to the enzyme has no effect on the coupling between the reactions in the BC and CT domain reactions.
Effect of Increasing l-Aspartate Concentrations on the Partial Reactions of RePC in the Absence of Acetyl-CoA
To examine how l-aspartate has the observed inhibitory effect on pyruvate carboxylation in the absence of acetyl-CoA and to determine the locus of its action, the kinetics of oxamate-dependent oxaloacetate decarboxylation (CT domain)18 and bicarbonate-dependent MgATP cleavage in the absence of pyruvate (BC domain)19 were measured in the presence of increasing concentrations of l-aspartate.
Kinetic parameters for oxamate-dependent oxaloacetate decarboxylation (data not shown) in the absence of l-aspartate and in the presence of 8 mM l-aspartate were obtained by fitting the data to eq 5. Comparison of the estimates of these parameters and their accompanying standard errors using t tests showed that there were no significant differences among the values of kcat, KOAA, and KI in the presence of 0 and 8 mM l-aspartate (p > 0.2 in all cases).
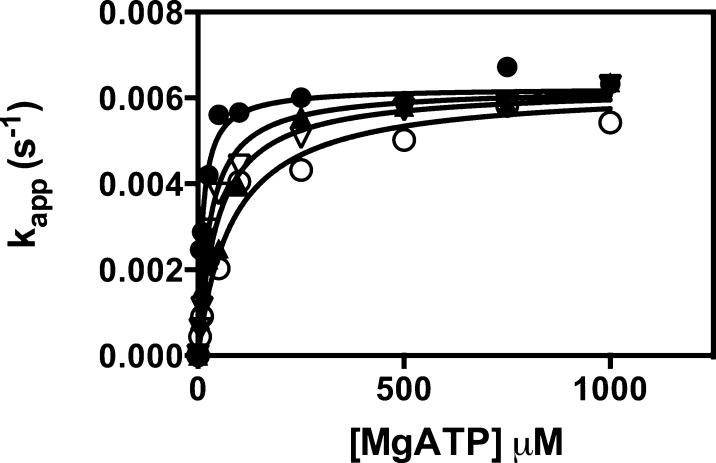
The effects of increasing concentrations of l-aspartate on the kinetics of bicarbonate-dependent MgATP cleavage in the absence of pyruvate are shown in Figure 6. The solid lines represent global fits of the data to eq 7, which describes competitive inhibition and gave the following estimates for parameters: kcat = 0.0062 ± 0.0001 s–1, Km = 11 ± 2 μM, and KI = 1.2 ± 0.3 mM. A fit to eq 6, which describes mixed inhibition, gave a fit of the data that was not significantly better than the fit to eq 7 by the variance ratio test (p > 0.1).
Figure 6.
Effect of increasing concentrations of l-aspartate [(●) 0, (▲) 2, (▽) 4, and (○) 8 mM] on the bicarbonate-dependent MgATP cleavage reaction in the absence of acetyl-CoA and pyruvate. The lines represent global fits of the data to eq 7. Reaction conditions were as described in Experimental Procedures.
Discussion
Previously, IC50 values for the inhibition by l-aspartate for PC have been reported to be in the range of 0.6–4 mM,10,11,21 while Dunn et al.22 reported that 10 mM l-aspartate only marginally inhibited the enzyme from Sinorhizobium meliloti. Thus, the Ki value of 1.3 mM obtained for RePC in this study is at the lower end of the range.
As with PC from S. cerevisiae,11,12 increasing concentrations of l-aspartate resulted in an increase in the apparent Ka for activation of the enzyme by acetyl-CoA. However, unlike in the S. cerevisiae enzyme where increasing l-aspartate concentrations markedly increased the Hill coefficient of cooperativity for the activation by acetyl-CoA,11,12l-aspartate had little effect on the cooperativity of acetyl-CoA activation in RePC. The competitive nature of the inhibition of RePC by l-aspartate with respect to acetyl-CoA activation was also noted in the fungal PCs,10−12 and the antagonistic nature of the inhibition with respect to acetyl-CoA was noted but not analyzed for PC from the bacterial enzyme Rhodobacter capsulatus.23 The cooperative nature of the binding of l-aspartate to RePC has also been observed in the fungal PCs,10,11 where Hill coefficients of 1.53 and 2.1 have been reported for the enzymes from S. cerevisiae and A. nidulans, respectively. In a bacterial PC from Arthrobacter globiformus,24 a Hill coefficient of 2.0 was reported. Thus, the value of the Hill coefficient of 2.8 reported here for RePC inhibition by l-aspartate was somewhat higher than these values, indicating increased cooperativity of binding. In RePC, the value of the Hill coefficient for l-aspartate inhibition is similar to that of acetyl-CoA activation, but in the case of PC from S. cerevisiae,11 the Hill coefficient for l-aspartate inhibition is greater than that of acetyl-CoA binding; in A. globiformus,24 it is lower and acetyl-CoA does not activate A. nidulans PC in the absence of l-aspartate.10
The inhibition of RePC in the absence of acetyl-CoA in this work has also been reported to occur in the fungal PCs,10,11,13 but there has been little investigation of the mechanism of this inhibition. We have shown that l-aspartate affects reactions in the BC domain but not the CT domain of RePC. It is clear that one mode of action is that of competitive inhibition with respect to MgATP binding in the bicarbonate-dependent MgATP cleavage reaction. However, this does not explain the inhibition of the pyruvate carboxylation reaction of RePC that was performed in the presence of close to saturating concentrations of MgATP.13 Because the effect of l-aspartate does not lie in the CT domain, it is likely that this inhibition of pyruvate carboxylation occurs in the BC domain. It has been reported that acetyl-CoA enhances the coupling between the cleavage of MgATP (as measured by Pi release) and the formation of carboxybiotin25 and oxaloacetate in RePC.15 However, l-aspartate does not appear to significantly affect the coupling between MgATP cleavage and oxaloacetate formation. This indicates that the inhibitory effect of l-aspartate at saturating concentrations of MgATP may be an effect on the rate of either carboxybiotin formation or the translocation of carboxybiotin to the CT domain.
As with activation of PCs by acetyl-CoA, there seems to be considerable variation in the parameters that describe the inhibition of PCs by l-aspartate, even between bacterial species. Generally, however, l-aspartate inhibition appears to be competitive with respect to acetyl-CoA and cooperative. The question of the mechanism of this competitive inhibition by l-aspartate then arises. The simplest model would be one in which l-aspartate and acetyl-CoA compete for the same binding site on PC. There is, however, some evidence to suggest that this is not the case. First, Libor et al.21 showed that modification of the thermophilic Bacillus PC with trinitrobenzenesulfonate specifically inhibited acetyl-CoA-dependent enzyme activity but had no effect on the acetyl-CoA-independent activity or its inhibition by l-aspartate. Moreover, the presence of acetyl-CoA protected the enzyme against modification. Second, Adina-Zada et al.26 showed that mutation of key residues in the acetyl-CoA binding site that increased the Ka between 76- and 252-fold had virtually no effect on the inhibition of RePC by l-aspartate in the absence of acetyl-CoA. Thus, it is probable that there is a distinct binding site for l-aspartate that exerts its effects on acetyl-CoA binding through allosteric effects. RePC has been crystallized in the presence of l-aspartate, but because of the high B factors the binding site for the inhibitor could not be located.27 Adina-Zada et al.13 showed that inhibition of RePC by MgTNP-ATP was also competitive with respect to acetyl-CoA but that its binding site was separate from that of the allosteric activator. Like l-aspartate, MgTNP-ATP was also shown to have allosteric effects in the absence of acetyl-CoA, but MgTNP-ATP allosterically activated the enzyme. In this work, we have shown that l-aspartate also inhibits MgTNP-ATP activation of RePC. Thus, it is possible that the binding site for l-aspartate is separate from both that of acetyl-CoA and that of MgTNP-ATP but that there are allosteric interactions among all three binding sites.
In summary, we have shown that the inhibition of RePC by l-aspartate is multifaceted. As with other microbial PCs, the inhibition is competitive with respect to the allosteric activator, acetyl-CoA, but there is a component of the inhibition that is independent of acetyl-CoA. The locus of this part of the inhibition lies in the BC domain and is comprised in part of competitive inhibition of MgATP binding and biotin carboxylation and/or carboxybiotin translocation. Because it is unlikely that the l-aspartate binding site is in the MgATP binding site, the acetyl-CoA binding site, or the MgTNP-ATP binding site, the effects of l-aspartate on acetyl-CoA activation, MgTNP-ATP activation, and MgATP binding and the effects on biotin carboxylation and/or carboxybiotin translocation are allosteric, induced from a remote l-aspartate binding site. Further understanding of the mechanism of action of l-aspartate must await a structural resolution of its binding site.
Glossary
Abbreviations
- acetyl-CoA
acetyl-coenzyme A
- PC
pyruvate carboxylase
- RePC
Rhizobium etli pyruvate carboxylase
- TNP-ATP
2′,3′-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate
- BC
biotin carboxylase
- CT
carboxyl transferase.
This work was supported by National Institutes of Health Grant GM070455 and the Thailand Research Fund (BRG5480002) to S.J. C.S. was supported by the RGJ-PhD scholarship (PHD/0308/2551) from the Thailand Research Fund.
The authors declare no competing financial interest.
References
- Jitrapakdee S.; St Maurice M.; Rayment I.; Cleland W. W.; Wallace J. C.; Attwood P. V. (2008) Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 413, 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S.; Wallace J. C. (1999) Structure, function and regulation of pyruvate carboxylase. Biochem. J. 340(Part 1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S.; Vidal-Puig A.; Wallace J. C. (2006) Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell. Mol. Life Sci. 63, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N. M.; Longacre M. J.; Stoker S. W.; Boonsaen T.; Jitrapakdee S.; Kendrick M. A.; Wallace J. C.; MacDonald M. J. (2008) Impaired anaplerosis and insulin secretion in insulinoma cells caused by small interfering RNA-mediated suppression of pyruvate carboxylase. J. Biol. Chem. 283, 28048–28059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.; Sudderth J.; Yang C.; Mullen A. R.; Jin E. S.; Mates J. M.; DeBerardinis R. J. (2011) Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. U.S.A. 108, 8674–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T. W.; Lane A. N.; Higashi R. M.; Farag M. A.; Gao H.; Bousamra M.; Miller D. M. (2009) Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Maurice M.; Reinhardt L.; Surinya K. H.; Attwood P. V.; Wallace J. C.; Cleland W. W.; Rayment I. (2007) Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science 317, 1076–1079. [DOI] [PubMed] [Google Scholar]
- Xiang S.; Tong L. (2008) Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat. Struct. Mol. Biol. 15, 295–302. [DOI] [PubMed] [Google Scholar]
- Adina-Zada A.; Zeczycki T. N.; Attwood P. V. (2012) Regulation of the structure and activity of pyruvate carboxylase by acetyl CoA. Arch. Biochem. Biophys. 519, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S. A.; Marston F. A.; Selmes I. P.; Chapman A. G.; Scrutton M. C. (1981) Pyruvate carboxylase from Aspergillus nidulans. Regulatory properties. Eur. J. Biochem. 118, 271–278. [DOI] [PubMed] [Google Scholar]
- Cazzulo J. J.; Stoppani A. O. (1968) The regulation of yeast pyruvate carboxylase by acetyl-coenzyme A and l-aspartate. Arch. Biochem. Biophys. 127, 563–567. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S.; Adina-Zada A.; Besant P. G.; Surinya K. H.; Cleland W. W.; Wallace J. C.; Attwood P. V. (2007) Differential regulation of the yeast isozymes of pyruvate carboxylase and the locus of action of acetyl CoA. Int. J. Biochem. Cell Biol. 39, 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adina-Zada A.; Hazra R.; Sereeruk C.; Jitrapakdee S.; Zeczycki T. N.; Maurice M. S.; Cleland W. W.; Wallace J. C.; Attwood P. V. (2011) Probing the allosteric activation of pyruvate carboxylase using 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate as a fluorescent mimic of the allosteric activator acetyl CoA. Arch. Biochem. Biophys. 509, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeczycki T. N.; Menefee A. L.; Adina-Zada A.; Jitrapakdee S.; Surinya K. H.; Wallace J. C.; Attwood P. V.; St Maurice M.; Cleland W. W. (2011) Novel insights into the biotin carboxylase domain reactions of pyruvate carboxylase from Rhizobium etli. Biochemistry 50, 9724–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeczycki T. N.; Menefee A. L.; Jitrapakdee S.; Wallace J. C.; Attwood P. V.; St Maurice M.; Cleland W. W. (2011) Activation and inhibition of pyruvate carboxylase from Rhizobium etli. Biochemistry 50, 9694–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adina-Zada A.; Jitrapakdee S.; Wallace J. C.; Attwood P. V. (2014) Coordinating role of His216 in MgATP binding and cleavage in pyruvate carboxylase. Biochemistry 53, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adina-Zada A.; Zeczycki T. N.; St Maurice M.; Jitrapakdee S.; Cleland W. W.; Attwood P. V. (2012) Allosteric regulation of the biotin-dependent enzyme pyruvate carboxylase by acetyl-CoA. Biochem. Soc. Trans. 40, 567–572. [DOI] [PubMed] [Google Scholar]
- Attwood P. V.; Cleland W. W. (1986) Decarboxylation of oxalacetate by pyruvate carboxylase. Biochemistry 25, 8191–8196. [DOI] [PubMed] [Google Scholar]
- Attwood P. V.; Graneri B. D. (1992) Bicarbonate-dependent ATP cleavage catalysed by pyruvate carboxylase in the absence of pyruvate. Biochem. J. 287(Part 3), 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M. J.; Jones M. E. (1983) Inorganic phosphate determination in the presence of a labile organic phosphate: Assay for carbamyl phosphate phosphatase activity. Anal. Biochem. 135, 233–238. [DOI] [PubMed] [Google Scholar]
- Libor S. M.; Sundaram T. K.; Scrutton M. C. (1978) Pyruvate carboxylase from a thermophilic Bacillus. Studies on the specificity of activation by acyl derivatives of coenzyme A and on the properties of catalysis in the absence of activator. Biochem. J. 169, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. F.; Araiza G.; Finan T. M. (2001) Cloning and characterization of the pyruvate carboxylase from Sinorhizobium meliloti Rm1021. Arch. Microbiol. 176, 355–363. [DOI] [PubMed] [Google Scholar]
- Modak H. V.; Kelly D. J. (1995) Acetyl-CoA-dependent pyruvate carboxylase from the photosynthetic bacterium Rhodobacter capsulatus: Rapid and efficient purification using dye-ligand affinity chromatography. Microbiology 141(Part 10), 2619–2628. [DOI] [PubMed] [Google Scholar]
- Gurr J. A.; Jones K. M. (1977) Purification and characterization of pyruvate carboxylase from Arthrobacter globiformis. Arch. Biochem. Biophys. 179, 444–455. [DOI] [PubMed] [Google Scholar]
- Legge G. B.; Branson J. P.; Attwood P. V. (1996) Effects of acetyl CoA on the pre-steady-state kinetics of the biotin carboxylation reaction of pyruvate carboxylase. Biochemistry 35, 3849–3856. [DOI] [PubMed] [Google Scholar]
- Adina-Zada A.; Sereeruk C.; Jitrapakdee S.; Zeczycki T. N.; St Maurice M.; Cleland W. W.; Wallace J. C.; Attwood P. V. (2012) Roles of Arg427 and Arg472 in the binding and allosteric effects of acetyl CoA in pyruvate carboxylase. Biochemistry 51, 8208–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzan A. D.; Menefee A. L.; Zeczycki T. N.; Kumar S.; Attwood P. V.; Wallace J. C.; Cleland W. W.; St Maurice M. (2011) Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli. Biochemistry 50, 9708–9723. [DOI] [PMC free article] [PubMed] [Google Scholar]