Abstract
In natural habitats, bacteria often occur in multicellular communities characterized by a robust extracellular matrix of proteins, amyloid fibres, exopolysaccharides and extracellular DNA. These biofilms show pronounced stress resistance including a resilience against antibiotics that causes serious medical and technical problems. This review summarizes recent studies that have revealed clear spatial physiological differentiation, complex supracellular architecture and striking morphology in macrocolony biofilms. By responding to gradients of nutrients, oxygen, waste products and signalling compounds that build up in growing biofilms, various stress responses determine whether bacteria grow and proliferate or whether they enter into stationary phase and use their remaining resources for maintenance and survival. As a consequence, biofilms differentiate into at least two distinct layers of vegetatively growing and stationary phase cells that exhibit very different cellular physiology. This includes a stratification of matrix production with a major impact on microscopic architecture, biophysical properties and directly visible morphology of macrocolony biofilms. Using Escherichia coli as a model system, this review also describes our detailed current knowledge about the underlying molecular control networks – prominently featuring sigma factors, transcriptional cascades and second messengers – that drive this spatial differentiation and points out directions for future research.
Introduction
In their natural habitats, bacteria preferably adopt a sedentary lifestyle in communities known as biofilms (Costerton et al., 1987). Interest in biofilms increased considerably when microbiologists began to appreciate bacteria as ‘social’ organisms and, in particular, when it was recognized that such social conduct was also the cause of severe medical and technological problems (Costerton et al., 1999; Beech and Sunner, 2004). Still unsolved problems associated with biofilms, which can be as important and dissimilar as their function as environmental reservoirs of pathogens (Hall-Stoodley and Stoodley, 2005), their resistance against antibiotics (Hoiby et al., 2010) or key roles in chronic infection (Bjarnsholt et al., 2013) and biocorrosion (Beech and Sunner, 2004), but also various beneficial aspects of biofilms have kept microbiologists busy trying to elucidate the fundamental principles driving biofilm formation.
The sessile and multicellular biofilm lifestyle is accompanied by the production of an extracellular matrix composed of exopolysaccharides, proteins, amyloid fibres and even extracellular nucleic acids, which promotes surface and intercellular adhesion and provides protection to the resident bacteria (Flemming and Wingender, 2010). Actual matrix composition is highly diverse among species and subject to intricate environmental regulation and sometimes mutational variation. Some species seem to strongly rely on combinations of exopolysaccharides. Thus, small colony variants of Pseudomonas aeruginosa produce high levels of the ‘aggregative’ exopolysaccharides Pel and Psl with distinct roles in early biofilm formation, while mucoid variants overproduce alginate (Yang et al., 2011; Mann and Wozniak, 2012). Other species combine amyloid protein fibres, which are prominently found in biofilms (Larsen et al., 2007), with exopolysaccharide(s) and/or specific additional proteins. Examples include amyloid curli fibres and cellulose in Salmonella spp., Escherichia coli and other enterobacteriaceae (Römling, 2005), or the TasA amyloid, the EPS exopolysaccharide and BslA hydrophobin in the Gram-positive Bacillus subtilis (Ostrowski et al., 2011; Vlamakis et al., 2013). On the other hand, Vibrio cholera relies on a specific combination of matrix proteins – RbmA, RbmC and Bap1, which have distinct functions during early biofilm formation – and the VPS exopolysaccharide (Yildiz and Schoolnik, 1999; Fong et al., 2006; Fong and Yildiz, 2007). Extracellular DNA in biofilm matrices has only recently gained attention. It derives from cell lysis mediated by prophages and various autolysins (Das et al., 2013), contributes to viscoelasticity of biofilms (Peterson et al., 2013) and facilitates twitching motility-related expansion at the leading edge of colonies and interstitial biofilms (Gloag et al., 2013).
The high diversity of natural biofilm settings has been experimentally mimicked using a series of biofilm models, which differ in the nature of the surface support of the biofilm as well as in the spatial distribution of nutrients, oxygen and water (Fig. 1). Thus, biofilm growth in microtiter plates or flow cells represents submerged biofilms growing on solid inert surfaces typically found in natural aquatic environments or on catheters (Christensen et al., 1999; O'Toole, 2011). Subaerial biofilms, which often contain cyanobacteria together with fungi, grow also on solid support (e.g. rocks in deserts or on buildings), which may provide micronutrients, while oxygen, carbon dioxide and ambiental water is provided by the surrounding air (Gorbushina and Broughton, 2009). Pellicle biofilms – as first stunningly described by Ferdinand Cohn (1876) in the 19th century – grow on liquid–air interfaces with nutrients coming from the liquid phase (Ostrowski et al., 2011; Vlamakis et al., 2013). Finally, macrocolony biofilms on nutrient-providing semi-solid agar plates reflect conditions of biofilms growing on decaying organic material such as soil or human food materials. Unlike classical colonies that arise from single cells, macrocolonies are inoculated by spotting a cell suspension (a few microlitres) on agar-solidified medium and are grown for extended times (Aguilar et al., 2007).
Figure 1.
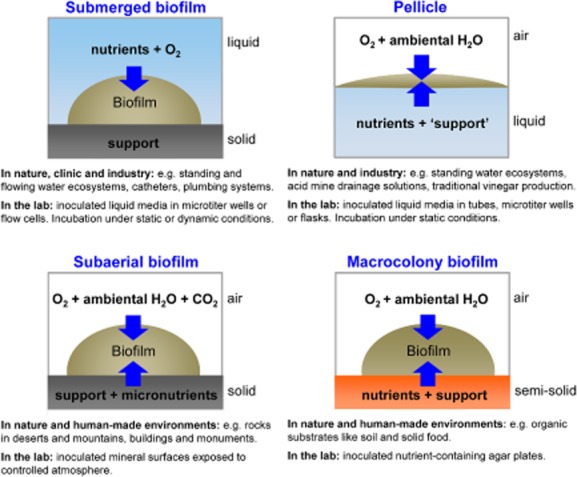
Biofilm models. Schematic representation of four types of biofilms that typically occur in nature and human-made environments. The schematic illustrates differences in surface support and spatial distribution of nutrients, oxygen and water. The figure provides examples of ‘real-world’ settings in which these types of biofilm commonly develop and laboratory settings designed to mimic these different biofilms.
The mechanisms by which gradients of nutrients, oxygen, waste products or signalling compounds that build up within a biofilm as well as additional external stress conditions shape biofilm structure and drive physiological differentiation have only recently emerged as a major topic in biofilm research [summarized by Haussler and Fuqua (2013)]. Two major attributes make macrocolonies a highly valuable model to study physiological differentiation and architectural development in biofilms: (i) at the microscale, macrocolonies exhibit a remarkably high level of organization, where it is even possible to identify distinct cell lineages; and (ii) macrocolony morphogenesis involves the elaboration of macroscopic complex three-dimensional forms – sharing characteristics with tissues of higher organisms – that are strongly modulated by different environmental or stress conditions.
This review will first point out the many links between stress responses and biofilm physiology that have emerged over the past years. It will then proceed to describe how stress conditions such as nutrient and oxygen limitation that occur heterogeneously in virtually every biofilm give rise to specific patterns of physiological stratification within macrocolonies. Special emphasis will be put on the role extracellular matrix components and the link to environmental stresses that so far have been studied in a handful of bacterial model organisms such as E. coli, Salmonella spp., Pseudomonas aeruginosa and B. subtilis. Finally, taking E. coli as an example, this review describes how already existing detailed knowledge of global stress regulatory networks that control bacterial physiology in liquid cultures also provides a conceptual framework to understand the molecular basis of physiological stratification and biofilm microarchitecture.
Links between stress responses and biofilm physiology
Even early biofilm studies already indicated that community life can be stressful for bacteria. Due to cellular crowding in biofilms, bacteria obviously have to cope with nutritional competition, non-optimal oxygen supply and the accumulation of waste products (such as acids produced by fermentation in oxygen-depleted zones of a biofilm). Not surprisingly, biofilm cells in general were thus found to grow relatively slowly (Moller et al., 1996; Anderl et al., 2003). This raised the question why biofilms then seem to be the dominant form of bacterial life in natural environments. However, slow or even no growth is generally associated with pronounced stress resistance induced by general stress responses such as those triggered by σS (RpoS) in E. coli and other gamma-proteobacteria (Hengge, 2011) or by σH and σB in low GC Gram-positives such as B. subtilis (Price, 2011). Bacteria in a biofilm in general also exhibit increased antibiotic resistance, which e.g. in P. aeruginosa has been observed to depend on nutrient starvation signalling by the stringent response alarmone (p)ppGpp (Nguyen et al., 2011). Moreover, matrix-surrounded community life can provide protection against protozoan grazing and other forms of predation in natural environments (Matz et al., 2005).
With the advent of gene expression profiling approaches, a large number of known stress-induced and stationary-phase genes were found to be activated in biofilm cells of different bacterial species (Schembri et al., 2003; Beloin et al., 2004; Ren et al., 2004; Hentzer et al., 2005; Domka et al., 2007). These and several other evidences suggested an overall picture of biofilm cells resembling stationary phase cells (Stoodley et al., 2002; Beloin and Ghigo, 2005). In fact, in E. coli, the expression of many regulatory and structural genes involved in the biosynthesis of curli fibres and cellulose, i.e. key biofilm matrix components, depends on the general stress response master regulator σS (Römling et al., 1998; 2000,), whose own expression requires (p)ppGpp (Hengge, 2011). Also the expression of many enzymes involved in the synthesis and degradation of the biofilm-promoting second messenger c-di-GMP is under σS control (Weber et al., 2006; Sommerfeldt et al., 2009). Production of flagella, which contribute to the initial stages of biofilm formation (Pratt and Kolter, 1998; Wood et al., 2006), requires the second messenger cAMP (Soutourina et al., 1999), which accumulates under conditions of non-optimal carbon source supply (Botsford and Harman, 1992). Furthermore, highly complex stress/starvation-triggered developmental processes such as the formation of endospores in B. subtilis have been found to naturally occur within biofilms with regulatory circuits of sporulation and biofilm formation being tightly interconnected (Branda et al., 2001; Vlamakis et al., 2013). In social bacteria such as Myxococcus, starvation-induced sporulation is obligatorily coupled to the formation of a highly structured biofilm that culminates in the formation of fruiting bodies (Kroos, 2007).
Nevertheless, the simplistic view of biofilms representing aggregates of highly stress resistant and slowly growing or even stationary phase cells has been debated on the basis of the recognized heterogeneity of microenvironments within biofilms (An and Parsek, 2007). Recent more comprehensive experimental designs have actually brought heterogeneity into context by identifying distinct cell subpopulations and starting to establish links to global stress regulatory networks (Vlamakis et al., 2008; Williamson et al., 2012; Serra et al., 2013b).
Physiological differentiation within the three-dimensional space of macrocolony biofilms
Chemical gradients and patterns of physiological stratification
Residing at different locations within structurally complex communities implies for bacteria to be non-uniformly exposed to growth resources and extracellular signalling molecules that diffuse along gradients. Such chemical gradients impact on gene expression, which ultimately determines the formation of physiologically distinct subpopulations (Stewart and Franklin, 2008).
Nutrients and oxygen gradients are particularly crucial in physiological differentiation. These gradients occur in any biofilm and result from the dynamic balance between diffusion and consumption. For P. aeruginosa, oxygen is critical to keep cell metabolism active in the absence of alternative electron acceptors. In regions close to the surface of P. aeruginosa macrocolonies, oxygen is consumed faster than it can diffuse into deeper layers, thereby becoming inaccessible for the rest of the community (Werner et al., 2004). Thus, acting as a growth-limiting resource, oxygen defines a two-layer pattern with a top layer of cells (about 60 μm thick) that exhibit active protein synthesis and a thick middle and bottom layer of cells (about two third of the macrocolony) that are metabolically inactive (Werner et al., 2004; Williamson et al., 2012). Due to this inactivity, cells in the lower layer allow nutrients to diffuse from the agar interface through the colony bottom layer to the upper layer where they are actually consumed.
Interestingly, less oxygen-dependent bacterial species can completely reverse this physiological stratification. As described in further detail below, E. coli macrocolonies feature actively growing cells in the lower layer close to the nutrient-providing agar support, while the upper layer is inhabited by non-growing starving cells (Serra et al., 2013b). For E. coli cells, oxygen limitation (or the absence of alternative respiratory electron acceptors) is not crucial for growth since they can obtain energy through fermentation. Therefore, nutrients are in this case the growth resource that becomes limiting in the upper zone and determines the physiological switch that results in stratification.
An interesting example of macrocolonies combining these stratification patterns was reported for Staphylococcus aureus and Staphylococcus epidermidis. Based on the detection of DNA and protein biosynthesis, staphylococcal macrocolonies can show a three-layer pattern with cells growing aerobically and fermentatively in zones adjacent to the air and agar interfaces, respectively, and non-growing cells found in a relatively thick middle zone (Rani et al., 2007). The interpretation of this pattern is that the cells at the macrocolony bottom consume fermentable substrates such as glucose, fructose or serine. No further growth is possible except along the top zone close to the air interface, where oxygen and non-fermentable substrates diffusing up from the bottom zone are available. The absence of both oxygen and fermentable carbon sources makes the large inner zone a growth-inactive area (Rani et al., 2007).
Importantly, the formation of such a growth-inactive zone is a key determinant of antibiotic resistance of the biofilm. Beta-lactamase-negative Klebsiella pneumoniae macrocolony biofilms exhibit an essentially similar three-layered pattern of physiological differentiation as Staphylococcus biofilms (Anderl et al., 2003). When macrocolonies were subject to ampicillin treatment for 12 h, cells were killed in the bottom and top layers, i.e. where cells were growing fermentatively or aerobically, respectively, but not in the central part of the macrocolony inhabited by cells that grow slowly or not at all (Anderl et al., 2003). It should be noted that while in macrocolonies nutrients and oxygen diffuse in opposite directions, in submerged biofilms grown on inert surfaces, both resources come from the surrounding liquid medium (Fig. 1). This logically explains the finding of growing cells in the outer layers and starving cells inside the ‘mushroom'-like structures of submerged biofilms (An and Parsek, 2007; Stewart and Franklin, 2008).
The question of the existence of distinct physiological states within macrocolony biofilms was also addressed in B. subtilis, where fluorescence reporters under the control of gene promoters specific for motile, matrix-producing and sporulating cells were used to visually localize these three cell types in cross-sections of a macrocolony (Vlamakis et al., 2008). The results demonstrated that cellular differentiation within a biofilm follows a precise spatiotemporal order. In terms of spatial organization, flagellated cells were predominantly found at the bottom and outer edge of the macrocolony. Matrix-producing cells were present in patches throughout the macrocolony, while sporulating cells were primarily found at the top of the macrocolony (Vlamakis et al., 2008). The latter was in agreement with earlier studies showing sporulation to take place preferentially in aerial projections, i.e. at the tip of colony wrinkles (Branda et al., 2001; Veening et al., 2006). In terms of temporal order, flagellated bacteria primarily differentiated into matrix-producing cells, with those located in the most nutrient-deprived zones ultimately initiating sporulation (Vlamakis et al., 2008). Thus, cell fate within B. subtilis macrocolonies also seems dictated by a ‘nutritional state'-dependent logic with the final transition to sporulating cells occurring not only far away from the nutrient source, but also after an energy-expensive process such as matrix production probably intensified nutrient depletion. Furthermore, the dynamics of these local differentiation events seems strongly modulated by self-produced small molecules. The ComX pheromone, which stimulates B. subtilis natural competence, also triggers differentiation to surfactin producers (Lopez et al., 2009a). Surfactin, in turn, acts paracrinally on a different subpopulation stimulating both matrix biosynthesis and cannibal toxin production (Lopez et al., 2009a,b,). Thus, acting like ‘cannibals’, matrix-producing cells thrive at the expense of other cell types, as they grow on the nutrients released by cannibalized cells which delays their differentiation into sporulating cells. Promoting the increase and maintenance of the matrix-producing population seems therefore a strategy to prioritize nutrient utilization for structural consolidation and protection of the community.
Nutrient-dependent physiological differentiation in E. coli macrocolonies
The extensively studied physiology of the E. coli growth cycle in liquid cultures was recently shown to provide also a unique basis for understanding physiological differentiation within a biofilm (Serra et al., 2013b). With abundant carbon/energy sources, as for instance present in a complex medium such as the commonly used Luria–Bertani broth (LB), E. coli initially grows exponentially with short division cycles. When composition and quality of these resources become suboptimal due to ongoing consumption, bacteria start getting ‘hungry’ and enter a phase of post-exponential growth with decreasing growth rates (Amsler et al., 1993; Ferenci, 2001; Liu et al., 2005). To optimize nutrient scavenging, post-exponential-phase E. coli cells, which are still rod-shaped and divide, not only express additional high affinity transport and metabolic systems, but also produce flagella and become highly motile (Adler and Templeton, 1967; Amsler et al., 1993; Liu et al., 2005; Zhao et al., 2007). However, when this ‘foraging’ strategy is no longer successful and available nutrients decrease further and become insufficient to support growth, E. coli cells undergo a global physiological switch to concentrate on maintenance and survival (Lange and Hengge-Aronis, 1991a). The molecular basis of this switch is a sigma factor exchange at RNA polymerase, with σS now taking the lead (see below for details). Cells stop the costly production of flagella and undergo a couple of reductive cell divisions, which generates smaller and ovoid cells (Lange and Hengge-Aronis, 1991b). Finally, they enter into stationary phase with cells arresting growth and becoming multiple stress resistant. If this happens below 30°C, much of the remaining resources are spent on producing extracellular matrix components like amyloid curli fibres and cellulose [summarized in Hengge (2011)].
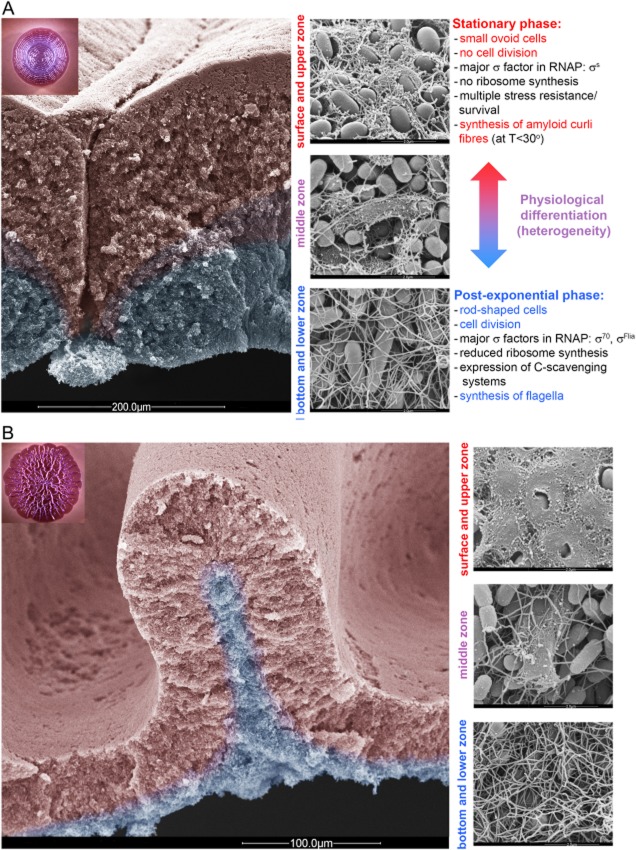
Using flagella, curli fibres, cell size and shape and the abundance of cell division as ‘anatomical’ hallmarks in high-resolution microscopic studies, these physiological changes known to occur in a temporal sequence in liquid cultures were found to translate into a precise spatial order within E. coli K-12 macrocolonies (Serra et al., 2013b) (Fig. 2A). This order is determined by the relative distance of bacteria from the nutrient source. Thus, bacteria at the bottom layers as well as at the thin outer edges of mature macrocolonies, i.e. close to the nutrient-providing agar interface, exhibit morphological features of post-exponential growth physiology: cells are elongated and rod-shaped; they divide and produce flagella, which form a tight mesh at the bottom of the macrocolony (Fig. 2D). By contrast, bacteria in the upper layer of the macrocolony, i.e. far away from nutrients, show morphological features characteristic of stationary phase: small ovoid cells that only very rarely show division are encased in a network of curli fibres that form ‘custom-moulded baskets’ around the producing cells (Fig. 2B). Similarly, curli-producing and H2O2-resistant bacteria (also the latter a phenotype dependent on σS) were found exclusively in the upper layer of macrocolonies of a uropathogenic E. coli strain (DePas et al., 2013). In conclusion, the basic architecture of E. coli macrocolonies has a two-layer structure with a zone of vegetative growth at the bottom and the outer edges and a zone of σS-determined stationary phase physiology in the top layer.
Figure 2.
Two-layer pattern of physiological differentiation in Escherichia coli macrocolony biofilms.A. Side-view scanning electron microscopic (SEM) image showing a cross-section of a macrocolony of E. coli K-12 strain W3110, which produces curli fibres but no cellulose, grown on salt-free LB plates for 5 days. Areas false-colored in red and blue represent zones of cells exhibiting stationary and post-exponential-phase-like physiology, respectively. The area false-colored in purple represents the physiological transition zone in between the lower and upper layers. The inset shows a top-view image of a W3110 macrocolony grown under the same conditions (plates supplemented with Congo red).The three high-resolution SEM images and adjacent descriptions of characteristic features show (from top to bottom) stationary phase-like cells in the surface/upper layer of a W3110 macrocolony, physiological heterogeneity in the middle transition zone (with curli-encased bacteria right next to curli-free cells entangled by their flagella), and post-exponential phase cells at the bottom layer.B. Side-view SEM image showing a cross-section through a ridge of a macrocolony of the curli- and cellulose-producing E. coli K-12 strain AR3110 grown on salt-free LB plates for 5 days. The false-color code is the same as in (A). Note that the physiological transition zone (false colored in purple) is very narrow compared to that in the curli-only macrocolony shown in (A). The inset displays a top-view image of a AR3110 macrocolony grown under the same conditions (plates supplemented with Congo red).The three SEM images at the right side show (from top to bottom) a composite of curli and cellulose covering stationary phase cells at the AR3110 macrocolony surface, bacteria fully encased in curli and cellulose right next to ‘naked’ cells with entangled flagella in the middle transition zone, and details of the mesh of entangled flagella formed by post-exponentially growing cells at the bottom of the AR3110 macrocolony.These images belong to a series, from which several others of similar content have been published before (Serra et al., 2013a). Only the large image shown in (B) is identical to a previously published one and is shown here with permission.
Between these two layers of E. coli K-12 macrocolonies, a transition zone of physiological heterogeneity can be discerned, in which small cells that already have switched to the stationary-phase mode and thus have turned on curli fibre production are found right next to elongated curli-free and flagellated cells (Fig. 2C). Remarkably, the curli-covered bacteria in this middle zone are found arranged in small chains (often consisting of four cells) and patches, suggesting that once cells turn on curli production, then this state is maintained in their progeny (Serra et al., 2013b). Since these adjacent curliated and curli-free cells are exposed to the same environmental conditions, this heterogeneity points to a stochastic element and possibly bistability in the outcome of sigma factor competition and/or the control of curli fibre production (see below).
Unlike many commensal or pathogenic E. coli ‘wild-type’ strains (Bokranz et al., 2005), E. coli K-12 laboratory strains do not produce cellulose as a matrix component (Da Re and Ghigo, 2006). This defect could be repaired by changing an E. coli K-12-specific chromosomal single nucleotide polymorphism that generates a stop codon in the bcsQ gene, which is polar on the expression of the downstream genes encoding the subunits of cellulose synthase (Serra et al., 2013a). The production of curli fibres and cellulose in this ‘dedomesticated’ strain resulted in pronounced phenotypic changes (Fig. 2E). At the high-resolution microscopic level, curli fibres and cellulose form a composite material that not only covers but also firmly connects cells at the surface and in the upper part of the top layer of the macrocolony. In addition, cellulose forms long filaments and extended sheets at the interface between the top and bottom layers (Fig. 2F). Macroscopically, colony morphology is drastically different from a curli-only producing strain: whereas the latter generates a colony pattern of concentric rings (Fig. 2A) that form by breakage of the brittle non-elastic curli fibre network of the top cell layer, the ‘dedomesticated’ strain produces very large and flat macrocolonies with complex patterns of long ridges and wrinkles (Fig. 2E). It seems that the cell-connective properties of the cellulose-curli composite interfere with vertical expansion, thus forcing the growing colony into lateral expansion. These macrocolonies in fact show tissue-like, i.e. cohesive and elastic properties, which results in their buckling up into macroscopic structures when cellular crowding generates lateral tension. However, even though matrix structure and physical properties as well as macrocolony morphology differ rather strikingly from the ‘classical’ curli-only producing E. coli K-12, the basic stratification into two layers with growing flagellated cells at the bottom and the outer edges and matrix-producing stationary phase cells in the top zone is essentially the same (Serra et al., 2013a).
This two-layer architecture was also found in mutant macrocolonies that produce cellulose only or that even did not produce curli and cellulose at all (Serra et al., 2013a). Thus, production of the biofilm matrix itself does not cause physiological and spatial differentiation of E. coli macrocolonies, but rather is a consequence of the differentiation into layers of vegetatively growing and starving stationary phase cells. However, once stationary phase cells start to produce matrix components, then this process can indeed modulate differentiation further by accelerating the use of remaining carbon/energy sources and by affecting macrocolony morphology, which involves relocation of entire regions by tissue-like folding and buckling and thereby impacts on nutrient and oxygen gradients.
Stress response networks controlling cellular differentiation in E. coli macrocolony biofilms
Overall, these recent studies also suggest that bacterial biofilm formation should not be considered as a programme of specific physiological differentiation. Rather, cells generating a biofilm respond to their self-conditioned local microenvironment by adapting their gene expression to suit their specific needs similarly as planktonic cells do in liquid cultures. The major difference is that bacteria residing in biofilm do so in a stable spatially structured environment. At the molecular level, the regulatory mechanisms are provided by already well-studied stress responses (Storz and Hengge, 2011), which essentially guide bacteria in making the decision between two fundamentally different life strategies – either to opt for growth and proliferation or for maintenance and survival. Using E. coli as a model system, we show here how the regulatory networks that control the transition from post-exponential to stationary phase provide a conceptual framework to understand spatial physiological differentiation in structured macrocolonies. The principle components of these networks are different sigma subunits of RNAP, several master regulators controlling transcriptional cascades and the nucleotide second messengers cAMP, (p)ppGpp and c-di-GMP.
Sigma subunits competing for RNAP core provide the basis for physiological differentiation into post-exponentially growing and stationary phase cells
With available resources always being limited, bacteria cannot maximize proliferation and general stress resistance at the same time (Ferenci, 2001; 2005; Nystrom, 2004; Hengge, 2011). At the molecular level, this is reflected by a limited cellular amount of RNAP core enzyme for which the various sigma factors, which activate different spectra of target genes, have to compete (Ishihama, 2000; Grigorova et al., 2006). While in growing cells the master sigma subunit is the vegetative σ70 (RpoD) which essentially drives expression of ‘housekeeping’ genes involved in growth and metabolism, such a global role is taken over by σS (RpoS) in stationary phase cells, which redirects transcription to hundreds of genes involved in multiple stress resistance, maintenance metabolism and other stationary phase functions (Weber et al., 2005). These global activities of σ70 and σS are complemented and – sometimes transiently replaced if need be – by several alternative sigma factors that control relatively smaller sets of stress genes. In E. coli, these are σFliA (σ28 or RpoF), which – together with σ70 – drives the expression of genes involved in assembling and operating flagella during the post-exponential growth phase, as well as σ32 (RpoH), σE (σ24 or RpoE) and σ54 (RpoN), which control sets of genes involved in coping with heat shock, extracytoplasmic stress and nitrogen limitation respectively (Storz and Hengge, 2011).
The point at which σS ‘takes over’ RNAP and then drives cells into stationary phase physiology is determined by the integration of a plethora of environmental cues and cellular signals that impact on rpoS expression, rpoS mRNA translation, proteolysis of σS protein and its binding to RNAP core. These cues include reduced growth rate (reflected in increased cellular (p)ppGpp), low concentration of energy and carbon sources (reflected in increased cAMP, oxidized components of the respiratory chain and reduced ATP levels) and low concentrations of phosphorus or nitrogen sources. Despite its complexity – involving multiple signal integration, complex homeostatic feedback as well as positive feedbacks – the switch from a state of low production rates of unstable σS in growing cells to high synthesis and stabilization of σS during entry into stationary phase is now well understood. In addition, a series of different acute stress signals – such as sudden exposure to high osmolarity, acid, radiation, etc. – can rapidly throw this switch even in fast growing cells in case of emergency, which makes σS also a general stress sigma factor [for reviews focusing on different aspects of this control network, see Hengge (2009a; 2011), and Klauck and Hengge (2011)].
Notably, the threshold for switching to σS-driven physiology can be shifted by mutation [reviewed in Hengge (2011)]. Thus, when E. coli is kept under constant long-term carbon limitation in a chemostat (in the absence of additional stress) or during long-term stationary phase in LB medium, where balanced cryptic growth is established (i.e. still viable cells grow slowly on the debris of lysed cells), mutants with attenuated σS levels or activity emerge. These mutants have a competitive advantage by remaining in the growth state under the very limiting conditions, where the parental strains already switch to stationary phase physiology (Finkel, 2006; Ferenci, 2008).
The second messengers cAMP and (p)ppGpp and the transcriptional master regulators CRP and FlhDC determine the physiology of post-exponentially growing cells
Competition for RNAP core of the vegetative σ70 and the stationary phase σS, with each one – when in the dominant role – operating at the expense of the other, constitutes the fundamental framework, in which mutually exclusive transcriptional cascades can drive biofilm-relevant properties such as flagella production in growing cells and curli and cellulose production in stationary phase cells (Pesavento et al., 2008). These regulatory cascades involve several key transcription factors as master regulators, which in turn respond to the crucial second messengers cAMP, (p)ppGpp and c-di-GMP (Fig. 3).
Figure 3.
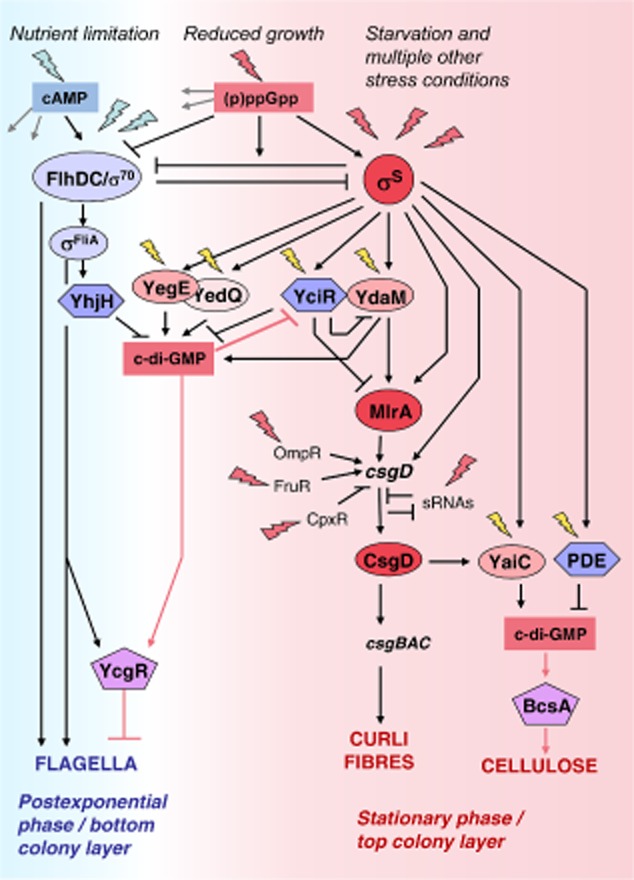
Master regulators and second messenger signalling in the control of physiological differentiation within E. coli macrocolony biofilms. In E. coli, the physiological stratification into two layers (bottom and top, symbolized by blue and light red colour) within macrocolonies follows essentially the same nutrient-dependent logic that drives the physiological switch from post-exponential to stationary phase. At the molecular level, this switch involves sigma factor competition for RNA polymerase core enzyme (mainly σ70/σFliA and σS) and the inverse coordination of two feedforward transcriptional cascades that control flagella (cAMP-CRP/FlhDC/σ70/σFliA; left side of the figure, with transcription factors represented by light blue ovoids) and curli/cellulose synthesis (σS/MlrA/CsgD; right side of the figure, with transcription factors represented by red ovoids) that operate in post-exponential phase cells and stationary phase cells respectively (see text for details and references). c-di-GMP producing DGCs are shown as light red ovoids, c-di-GMP-degrading PDEs as blue hexagons and c-di-GMP-binding PilZ-like effector proteins as purple pentagons. Blue and red bolts indicate stress signal input typically associated with post-exponential and stationary phase physiology respectively. Yellow bolts indicate additional unknown signal input via the N-terminal sensory domains of most DGCs and PDEs.
The cellular levels of cAMP and (p)ppGpp are enhanced in cells entering the post-exponential state, with cAMP accumulating in response to quantitatively and qualitatively low carbon/energy sources and increasing (p)ppGpp being associated with decreasing growth rates. While (p)ppGpp redirects vegetative RNAP away from ribosomal gene expression towards the expression of other genes, many of which have biosynthetic functions (a process called the ‘stringent response’), cAMP bound to the master regulator CRP activates the expression of numerous carbon scavenging systems (Grainger et al., 2005; Shimada et al., 2011). In addition, the production of flagella is under control of cAMP-CRP (Soutourina et al., 1999). This global regulator activates flhDC, which in the hierarchical flagellar transcriptional cascade represents the single class 1 operon and encodes the master regulator (Liu and Matsumura, 1994; Wang et al., 2006). FlhDC and the σ70-containing RNAP then activate the class 2 flagellar operons that encode proteins for the components and assembly of the hook-basal body, the flagellar sigma factor σFliA, its antisigma factor FlgM and many other proteins. Once the hook-basal body is assembled, FlgM is secreted which relieves inhibition of σFliA to permit transcription of class 3 genes, including those encoding flagellin (fliC), i.e. the major subunit of the flagellar filament, and the flagellar motor proteins (Chilcott and Hughes, 2000; Chevance and Hughes, 2008).
Already in parallel to these processes and under the positive influence of (p)ppGpp, rpoS transcription increases and – in response to dwindling carbon/energy sources – σS protein, which in growing cells is degraded by the ClpXP protease with the help of the targeting factor RssB, becomes stabilized. Consequently, σS accumulates, but it still remains inefficient in outcompeting σ70/σFliA for core RNAP binding (Hengge, 2011). Moreover, the FlhDC-controlled cascade prioritizes its action over σs-controlled gene expression via FliZ, an abundant nucleoid-associated protein expressed from the same operon as σFliA that shows a DNA-binding specificity with a strong sequence overlap with the σS consensus promoter, thus acting as a global repressor at many σS-dependent promoters (Pesavento et al., 2008; Pesavento and Hengge, 2012).
Overall, σ70, cAMP-CRP, FlhDC, σFliA and FliZ thus establish a transcriptional pattern that includes flagella production and allows cells to optimize growth under the no longer optimal conditions in the post-exponential phase. In an E. coli macrocolony biofilm, this physiological state is found in the bottom layer, which is a highly crowded environment but nevertheless close enough to the nutrient-providing agar support to still support growth and proliferation.
Activation of σS and the σS/MlrA/CsgD transcriptional cascade in the switch to stationary phase physiology
As nutrient conditions worsen – i.e. later in post-exponential phase in liquid culture or further away from the agar support in a macrocolony – the flhDC operon is no longer transcribed and, due to proteolytic degradation of FlhDC and σFliA, expression of the entire flagellar cascade is shut down (Barembruch and Hengge, 2007; Pesavento et al., 2008). In the macrocolony, this is apparent as a ‘dilution’ of flagella per cell with increasing distance from the agar support (Serra et al., 2013b). This is an effect of increasing levels of (p)ppGpp (Traxler et al., 2011), which interferes with flagellar expression (Lemke et al., 2009), further drives σS expression (Lange et al., 1995) and shifts sigma factor competition for RNAP core in favour of alternative sigma factors such as σS (Jishage et al., 2002). Binding to RNAP core and thus activation of σS is further promoted by the σS-binding Crl protein and the anti-σ70 factor Rsd, a process that includes a positive feedback due to expression of Rsd being further enhanced by σS itself (Jishage and Ishihama, 1998; 1999; Bougdour et al., 2004; Typas et al., 2007; Banta et al., 2013). In addition, the negative homeostatic feedback in the control of σS levels provided by the σS-dependent expression of its own proteolytic targeting factor RssB (a response regulator that is rate limiting for σS degradation in growing cells) no longer operates for two reasons: (i) σS-containing RNAP levels get higher than required to maximally activate the rssAB promoter (Pruteanu and Hengge-Aronis, 2002), and (ii) energy limitation (in the presence of oxygen) leaves the quinones of the respiratory chain in an oxidized state, which inactivates the histidine sensor kinase ArcB that is required to phosphorylate and thus activate RssB (to drive σS proteolysis) and ArcA (a globally acting transcription factor, which also represses rpoS transcription) (Malpica et al., 2004; Mika and Hengge, 2005). All these highly integrated regulatory processes allow σS to strongly accumulate and basically ‘take over’ genome-wide gene expression which results in multiple stress resistance, switching to a maintenance metabolism and cells becoming smaller and ovoid due to ‘reductive’ cell division [summarized in Hengge (2011)].
Another major consequence of this switch is that σS now deploys essentially the entire regulatory machinery for the synthesis of biofilm matrix components such as curli fibres and cellulose (Fig. 3). This includes the expression of a σS-controlled feedforward cascade consisting of the transcriptional regulators MlrA and CsgD (Brown et al., 2001; Weber et al., 2006) as well as several diguanylate cyclases (DGC) and phosphodiesterases (PDE) that produce and degrade the second messenger c-di-GMP, respectively, which is essential for the expression of CsgD (Weber et al., 2006; Pesavento et al., 2008; Sommerfeldt et al., 2009; Lindenberg et al., 2013) (see also below). CsgD expression and therefore curli and cellulose production occurs below 30°C in most E. coli strains (Bokranz et al., 2005). In some strains, however, this still uncharacterized temperature regulation does not operate and for some pathogenic E. coli curli fibres contribute to adhesion and even to invasion into host cells (Gophna et al., 2001).
The csgD gene (also called agfD in Salmonella) is in fact a key node or regulatory hub in this complex biofilm control network (Fig. 3). Its transcriptional regulation is remarkably complex and receives additional modulatory inputs from an entire series of transcription factors (including OmpR, CpxR and FruR) in response to diverse conditions such as changing osmolarity, envelope stress or nutrient limitation (Prigent-Combaret et al., 2001; Brombacher et al., 2003; 2006; Gerstel and Römling, 2003; Jubelin et al., 2005; Ogasawara et al., 2010b; Lindenberg et al., 2013). In addition, csgD mRNA levels and translation are negatively modulated by several stress-regulated small regulatory RNAs (sRNA), including RprA, McaS and OmrAB [recently summarized by Mika and Hengge (2013)]. In general, these sRNAs have multiple targets. Thus, McaS inversely regulates FlhDC and CsgD expression and thereby contributes to the mutual exclusion of flagella and curli/cellulose production (Jørgensen et al., 2012; Thomason et al., 2012). In a feedforward loop, RprA directly targets not only csgD mRNA, but also ydaM mRNA, which encodes a DGC that is essential for transcriptional initiation at the csgD promoter (Mika et al., 2012). Another intriguing and not fully characterized effect of this mRNA/sRNA interaction network is that high levels of csgD mRNA can also sequester these sRNAs and thereby affect the expression of their other targets (Mika et al., 2012; Mika and Hengge, 2013).
CsgD protein then drives the formation of curli fibres by directly activating transcription of the csgBAC operon (Römling et al., 2000; Weber et al., 2006), which encodes the major curli subunit CsgA, the nucleator protein CsgB and the small periplasmic protein CsgC that is proposed to play a role in subunit secretion (Evans and Chapman, 2013). As described below, CsgD control of cellulose production is indirect involving c-di-GMP signalling. Attempts to define a csgD regulon have yielded mixed results. Chromatin immunoprecipitation (ChIP) analysis of a CsgD-overproducing strain has detected around a dozen genes (besides csgBAC and yaiC) that may be positively or negatively regulated by CsgD protein (Ogasawara et al., 2011). However, using a csgD mutant in a microarray-based transcriptome analysis, a different and rather large csgD regulon (again including csgBAC and yaiC) has been observed, but currently, it is unclear which of these target genes are controlled by CsgD protein at the level of transcription initiation and which are indirectly regulated by csgD mRNA sequestering sRNAs which target the mRNAs of these genes (Mika et al., 2012).
In conclusion, σS-dependent and, in particular, CsgD-dependent gene expression literally shapes the stationary phase upper layer of macrocolony biofilms. While this is most apparent for the matrix network surrounding the cells as well as for the size, shape and stress resistance of cells residing in this region of a macrocolony (Fig. 2), it may well extend to not yet characterized cellular and community properties regulated by σS, CsgD and the sRNA network described above.
Role of the second messenger c-di-GMP in the inverse coordination of flagellar activity and biofilm matrix synthesis
c-di-GMP is a ubiquitous second messenger that in general promotes biofilm formation by driving the expression of adhesins and matrix components while usually downregulating expression and/or activity of flagella [for reviews focusing on different aspects, see Jenal and Malone (2006), Hengge (2009b; 2010), Krasteva et al. (2012) and Römling et al. (2013)]. c-di-GMP is synthesized by DGCs characterized by the GGDEF domain, with this amino acid motif representing the active site (A-site) of these enzymes. The activities of most DGCs are also allosterically feedback-inhibited by their own product, with c-di-GMP binding to a secondary site (I-site). Dedicated PDEs containing EAL or HD-GYP domains degrade c-di-GMP (Schirmer and Jenal, 2009). These antagonistic enzymes regulate the intracellular levels of c-di-GMP, which is bound by effector components, which belong to diverse families of proteins or riboswitches in the 5'-untranslated regions of mRNAs. Acting for instance as transcription factors, as components of exopolysaccharide synthesis and excretion machineries or – in the case of riboswitches – in translational control, these effectors directly control the activity of highly diverse target components (Boyd and O'Toole, 2012; Krasteva et al., 2012).
The complexity of c-di-GMP signalling networks also relies on the often large numbers of different DGCs and PDEs that exist in most bacterial species. The E. coli K-12 genome encodes 12 proteins with a GGDEF domain, 10 proteins with an EAL domain and 7 proteins that contain both domains. In terms of enzymatic activity, these proteins represent a set of 12 DGCs, 13 PDEs and 4 ‘degenerate’ proteins, with the latter not displaying enzymatic activity and acting by direct protein–protein interactions (Hengge, 2010). Expression of these proteins is highly differentially controlled, but many – DGCs as well as PDEs – are induced during transition of E. coli from post-exponential to stationary growth phase (Sommerfeldt et al., 2009). In addition, some of these DGCs and PDEs seem to have highly specific functions that cannot be complemented by other DGCs and PDEs. This specificity has led to assumptions of ‘local’ action of some of these proteins in larger protein–protein complexes, with such interactions actually having been observed for many of these proteins [summarized in Hengge (2009b)].
A specific subset of these DGCs and PDEs becomes crucially involved in the operational switch between the flagellar and curli/cellulose control cascades (Fig. 3) (Weber et al., 2006; Pesavento et al., 2008). The first key player in this switch is YhjH, the most strongly expressed PDE in E. coli (Sommerfeldt et al., 2009), whose expression from a class 3 flagellar gene is driven by the cAMP-CRP/FlhDC/σFliA cascade (Ko and Park, 2000; Frye et al., 2006; Girgis et al., 2007) and which keeps c-di-GMP levels low in post-exponentially growing cells (Spangler et al., 2010). Yet together with that of the other flagellar genes, also yhjH expression is shut down during the later post-exponential phase, whereas the activation of σS then promotes the expression of the DGCs YegE and YedQ, which drive up c-di-GMP levels to the point that the c-di-GMP-binding effector protein YcgR (itself previously expressed from a class 3 flagellar gene) can now slow down flagellar rotation by acting as a direct brake at the flagellar basal body (Pesavento et al., 2008; Boehm et al., 2010; Fang and Gomelsky, 2010; Paul et al., 2010). This YcgR-dependent slowing down of flagellar rotation, which may save remaining energy, was actually directly observed under the microscope as a reduced swimming speed of late post-exponential phase cells from a liquid culture (Pesavento et al., 2008). In the bottom layer of still growing cells in a crowded macrocolony biofilm, it may prevent flagella that get entangled by rotation from becoming overstretched (Serra et al., 2013b).
The next key player in the switch is the PDE YciR, a ‘trigger enzyme’ that – in response to the now increasing c-di-GMP level – allows the equally σS-controlled DGC YdaM and the transcription factor MlrA to activate csgD transcription by an intriguing mechanism: YciR initially inhibits both YdaM and MlrA by direct specific interactions, which are relieved when c-di-GMP levels get high enough for YciR to efficiently bind and degrade c-di-GMP. Thus, YciR combines several activities: it is (i) a direct antagonist for YdaM and MlrA, (ii) an effector for c-di-GMP (controlled by the YegE/YhjH module) and (iii) a PDE (which reduces the level of its own inhibitor of its second activity, i.e. its antagonism to YdaM and MlrA) (Lindenberg et al., 2013). This makes YciR a perfect switch protein – initially it can contribute to keeping c-di-GMP levels low, but then becomes titrated when c-di-GMP production is ramped up by increasing levels of YegE and a kicking-in positive feedback due to the DGC activity of YdaM, whose inhibition by YciR is relieved when the latter is exposed to sufficient c-di-GMP to efficiently operate as a PDE (Fig. 3). Moreover, YdaM – besides its DGC activity that provides for the positive feedback – also activates MlrA by directly interacting with the C-terminal ligand-binding domain of this MerR-like transcription factor (Lindenberg et al., 2013). The result is an efficient initiation of csgD transcription by MlrA, which binds in the csgD promoter region (Ogasawara et al., 2010a), and acts together with σS-containing RNAP (Weber et al., 2006). The direct interactions of YciR and YdaM with MlrA as well as the role of additional transcription factors (see above) actually imply the formation of a large nucleoprotein complex at the csgD promoter, which, however, remains to be demonstrated experimentally.
Downstream of CsgD expression, c-di-GMP is again necessary to allosterically activate cellulose synthase by directly binding to the regulatory PilZ-like domain of its larger subunit BcsA (Ross et al., 1987; Römling et al., 2005). For this process, the key player is the DGC YaiC (AdrA in Salmonella), which is expressed under the control of CsgD and σs-containing RNAP (Römling et al., 2000; Zogaj et al., 2001; Weber et al., 2006; Sommerfeldt et al., 2009). yaiC/adrA mutants do express CsgD and curli normally, but are specifically defective in producing cellulose (Römling et al., 2000; Simm et al., 2004). The reason for this high specificity of YaiC for activating cellulose synthase is unclear. Perhaps only YaiC – due to a high Ki for product inhibition at its I-site – is able to drive c-di-GMP levels high enough to efficiently activate cellulose synthase, whose PilZ domain has a relatively high Kd for c-di-GMP binding (Pultz et al., 2012).
In summary, during transition from the post-exponential to the stationary growth phases cells essentially undergo a fine-tuned switch from a low to a high cellular c-di-GMP state. This switch, however, does not depend exclusively on isolated c-di-GMP signalling proteins, but rather on the more global regulatory networks directed by a few master regulators that precisely determine when and where the c-di-GMP signalling components are expressed and operate. Based on the stratification into post-exponentially growing and stationary phase bacteria within macrocolonies (Serra et al., 2013a, b), the known regulatory patterns of DGCs and PDEs (Sommerfeldt et al., 2009), and increasing cellular c-di-GMP level along the growth cycle in liquid cultures (Spangler et al., 2010), one can logically predict a corresponding heterogeneity of cellular c-di-GMP levels within these biofilms. An exciting challenge for the future will be to determine the precise levels of c-di-GMP and associated signalling components in single cells within structurally complex macrocolonies.
Short-range heterogeneity within macrocolonies and bistability of gene expression
The response to gradients of nutrients and oxygen, which extend over long distances within a macrocolony biofilm, establishes long-range heterogeneity of gene expression and global physiology as described above. In addition, short-range heterogeneity has been observed, with cells residing right next to each other displaying different morphology and physiology. In the physiological ‘transition’ zones between the bottom and upper strata as well as at the surface behind a certain distance from the outer growing edge of E. coli macrocolonies, shorter and highly curliated cells are found directly adjacent to ‘naked’, elongated and fully flagellated cells. In addition, expression of the CsgD regulator shows the same pattern of heterogeneity in these zones (Serra et al., 2013b). These directly adjacent cells must be exposed to identical environmental conditions, which points to an endogenous source of this micro-heterogeneity. Most likely, this is due to regulatory patterns that can generate bistability of gene expression. These patterns include the activity of antagonistic enzymes generating ultrasensitivity, long transcriptional cascades, mutual inhibition of regulatory components and positive feedbacks (Hooshangi et al., 2005; Dubnau and Losick, 2006; Lipshtat et al., 2006). All of these patterns are actually prominently present in the network that controls CsgD (Fig. 3). In particular, mutual inhibition is found at three levels, i.e. in (i) sigma factor competition, (ii) YciR degrading its own inhibitor and (iii) mutual inhibition of csgD mRNA and sRNAs. It will be a challenge for future work to sort out the contributions of these network motifs to bistable expression of CsgD and the matrix components curli and cellulose.
The combination of long-range heterogeneity of gene expression due to gradients of resources and short-range heterogeneity due to bistability of expression of a key regulator may be a general principle in biofilm architecture. Also in macrocolonies of B. subtilis, a stratification of expression of flagellar genes in the bottom layer and of matrix and sporulation genes in the upper layer was shown (Branda et al., 2001; Vlamakis et al., 2008) (and see above). In addition, biofilm matrix-producing cells are found side-by-side with non-producer cells, with this micro-heterogeneity based on bistability in the control of the regulator Spo0A activating expression of SinI, which in the producer cells antagonizes the repressor SinR that is expressed in all cells in the relevant regions of the macrocolonies (Chai et al., 2008).
Conclusions and perspectives
Based on a number of recent studies summarized here, it can be concluded that the heterogeneous cellular physiology and supracellular architecture of macrocolony biofilms is essentially established by stress responses that react to gradients of nutrients and oxygen that build up during growth of these biofilms. These stress responses require cells to make the decision between growth and proliferation or maintenance and survival, which – when happening along nutrient and oxygen gradients – results in a physiological stratification of the biofilm. An important consequence is a heterogeneous production of matrix components, which include various exoproteins, amyloid fibres and exopolysaccharides that provide protection, cohesion and even tissue-like elasticity to the biofilm. Overall, this also means that biofilm formation is not the result of a specific genetic programme of developmental differentiation, but rather provides a spatially differentiated multicellular environment, in which specific developmental processes such as sporulation can then be triggered.
As only shortly mentioned in this review, macrocolony biofilms also exhibit striking macroscopic morphological patterns of ridges, rings and wrinkles, a phenotype that has been termed ‘wrinkled’, ‘rugose’ or ‘rdar’ (‘rough, dry and red’, with ‘redness’ depending on the use of the dye Congo Red), although these simple designations do not adequately reflect the complexity and diversity of these structures, which are drastically modulated by oxygen content, the presence of reactive oxygen species or salt or the humidity at the agar surface (Römling, 2005; Aguilar et al., 2007; Beyhan et al., 2007; DePas et al., 2013; Dietrich et al., 2013; Kolodkin-Gal et al., 2013; Serra et al., 2013a). A common signature of these structures is their strict dependence on extracellular matrix components (Friedman and Kolter, 2004; Römling, 2005; Romero et al., 2010; Colvin et al., 2012; Serra et al., 2013a, b), which confer the connectivity and elasticity that allow growing bacterial biofilms to essentially behave as tissues that buckle up under the spatial constraints and therefore tension generated by cellular crowding (Serra et al., 2013a; Trejo et al., 2013). It will be a major challenge for future research to characterize the underlying morphogenetic molecular mechanisms, i.e. how distinct matrix components, the network of multiple c-di-GMP-controlling DGCs and PDEs, the asymmetric expression of these components in different biofilm strata established by long-range nutrient and oxygen gradients as well as the role of additional stress signal input contribute to the microarchitecture and macroscopic morphology of biofilms.
Acknowledgments
Funding by the European Research Council under the European Union's Seventh Framework Programme (ERC-AdG 249780 to RH) is gratefully acknowledged. DOS is the recipient of a post-doctoral fellowship from the Alexander von Humboldt Foundation.
References
- Adler J. Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- Aguilar C, Vlamakis H, Losick R. Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsler CD, Cho M. Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D. Parsek MR. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol. 2007;10:292–296. doi: 10.1016/j.mib.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Anderl JN, Zahller J, Roe F. Stewart PS. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47:1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta AB, Chumanov RS, Yuan AH, Lin H, Campbell EA, Burgess RR. Gourse RL. Key features of sigmaS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2013;110:15955–15960. doi: 10.1073/pnas.1311642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembruch C. Hengge R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol Microbiol. 2007;65:76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- Beech IB. Sunner J. Biocorrosion: towards understanding interactions between biofilms and metals. Curr Opin Biotechnol. 2004;15:181–186. doi: 10.1016/j.copbio.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Beloin C. Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol. 2004;51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- Beyhan S, Bilecen K, Salama SR, Casper-Lindley C. Yildiz FH. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J Bacteriol. 2007;189:388–402. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sorensen SR, Moser C, Kuhl M, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Bokranz W, Wang X, Tschape H. Römling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Botsford JL. Harman JG. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Lelong C. Geiselmann J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J Biol Chem. 2004;279:19540–19550. doi: 10.1074/jbc.M314145200. [DOI] [PubMed] [Google Scholar]
- Boyd CD. O'Toole GA. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 2012;28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R. Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E, Dorel C, Zehnder AJ. Landini P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Baratto A, Dorel C. Landini P. Gene expression regulation by the Curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol. 2006;188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J. Curtiss R., 3rd MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R. Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF. Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS. Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BB, Sternberg C, Andersen JB, Palmer RJ, Jr, Nielsen AT, Givskov M. Molin S. Molecular tools for study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- Cohn F. Untersuchungen über Bakterien IV: Beiträge zur Biologie der Bacillen. Beitr Biol Pflanzen. 1876;7:249–276. [Google Scholar]
- Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M. Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS. Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Da Re S. Ghigo JM. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol. 2006;188:3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Sehar S. Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep. 2013;5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, et al. Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci USA. 2013;110:2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC. Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T. Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Evans ML. Chapman MR. Curli biogenesis: order out of disorder. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.09.010. Available from URL: http://dx.doi.org/10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X. Gomelsky M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol. 2010;76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Hungry bacteria – definition and properties of a nutritional state. Environ Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- Ferenci T. The spread of a beneficial mutation in experimental bacterial populations: the influence of the environment and genotype on the fixation of rpoS mutations. Heredity (Edinb) 2008;100:446–452. doi: 10.1038/sj.hdy.6801077. [DOI] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Flemming HC. Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fong JC. Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JC, Karplus K, Schoolnik GK. Yildiz FH. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J Bacteriol. 2006;188:1049–1059. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L. Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M. Hughes KT. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U. Römling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Girgis HS, Liu Y, Ryu WS. Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA. 2013;110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J. Ron EZ. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbushina AA. Broughton WJ. Microbiology of the atmosphere-rock interface: how biological interactions and physical stresses modulate a sophisticated microbial ecosystem. Annu Rev Microbiol. 2009;63:431–450. doi: 10.1146/annurev.micro.091208.073349. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J. Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci USA. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK. Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L. Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Haussler S. Fuqua C. Biofilms 2012: new discoveries and significant wrinkles in a dynamic field. J Bacteriol. 2013;195:2947–2958. doi: 10.1128/JB.00239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009a;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009b;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hengge R. Role of c-di-GMP in the regulatory networks of Escherichia coli. In: Visick KL, editor. The Second Messenger Cyclic-di-GMP. Washington, DC: ASM Press; 2010. pp. 230–252. Wolfe AJ and Visick KL(eds). [Google Scholar]
- Hengge R. The general stress response in gram-negative bacteria. In: Hengge R, editor. Bacterial Stress Responses. Washington, DC: ASM press; 2011. pp. 251–289. Storz G, and Hengge R(eds). [Google Scholar]
- Hentzer M, Eberl L. Givskov M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms. 2005;2:37–61. [Google Scholar]
- Hoiby N, Bjarnsholt T, Givskov M, Molin S. Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S. Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci USA. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- Jenal U. Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jishage M. Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc Natl Acad Sci USA. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M. Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J Bacteriol. 1999;181:3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler V. Nystrom T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MG, Nielsen JS, Boysen A, Franch T, Moller-Jensen J. Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P. Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck E. Hengge R. σS-controlling networks in Escherichia coli. In: Filloux AAM, editor; Bacterial Regulatory Networks. Norfolk, UK: Caister Academic Press; 2011. pp. 1–26. [Google Scholar]
- Ko M. Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Elsholz AK, Muth C, Girguis PR, Kolter R. Losick R. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 2013;27:887–899. doi: 10.1101/gad.215244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Giglio KM. Sondermann H. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 2012;21:929–948. doi: 10.1002/pro.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- Lange R. Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991a;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Lange R. Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991b;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Fischer D. Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D. Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T. Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E. Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshtat A, Loinger A, Balaban NQ. Biham O. Genetic toggle switch without cooperative binding. Phys Rev Lett. 2006;96:188101. doi: 10.1103/PhysRevLett.96.188101. [DOI] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ. Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Liu X. Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R. Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009a;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R. Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009b;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O. Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE. Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH. Kjelleberg S. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci USA. 2005;102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F. Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F. Hengge R. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int J Mol Sci. 2013;14:4560–4579. doi: 10.3390/ijms14034560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, et al. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S, Pedersen AR, Poulsen LK, Arvin E. Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K. Ishihama A. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol Lett. 2010a;312:160–168. doi: 10.1111/j.1574-6968.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamada K, Kori A, Yamamoto K. Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010b;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K. Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski A, Mehert A, Prescott A, Kiley TB. Stanley-Wall NR. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;47 doi: 10.3791/2437. doi:103791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WC, Blair DF. Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a ‘backstop brake’ mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C. Hengge R. The global repressor FliZ antagonizes gene expression by sigmaS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res. 2012;40:4783–4793. doi: 10.1093/nar/gks055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A. Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BW, van der Mei HC, Sjollema J, Busscher HJ. Sharma PK. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. MBio. 2013;4:e00497–13. doi: 10.1128/mBio.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA. Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Price CW. The general stress response in Bacillus subtilis and related Gram-positive bacteria. In: Hengge R, editor. Bacterial Stress Responses. Washington, DC: ASM press; 2011. pp. 301–318. Storz G and Hengge R(eds). [Google Scholar]
- Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P. Dorel C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M. Hengge-Aronis R. The cellular level of the recognition factor RssB is rate-limiting for sigmaS proteolysis: implications for RssB regulation and signal transduction in sigmaS turnover in Escherichia coli. Mol Microbiol. 2002;45:1701–1713. doi: 10.1046/j.1365-2958.2002.03123.x. [DOI] [PubMed] [Google Scholar]
- Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B. Miller SI. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol. 2012;86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Bedzyk LA, Setlow P, Thomas SM, Ye RW. Wood TK. Gene expression in Bacillus subtilis surface biofilms with and without sporulation and the importance of yveR for biofilm maintenance. Biotechnol Bioeng. 2004;86:344–364. doi: 10.1002/bit.20053. [DOI] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R. Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Römling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Sierralta WD, Eriksson K. Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Rohde M, Olsen A, Normark S. Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Gomelsky M. Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY. Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri MA, Kjaergaard K. Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- Schirmer T. Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Serra DO, Richter AM. Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013a;195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck G, Mika F. Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013b;4:e103–e113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fujita N, Yamamoto K. Ishihama A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS ONE. 2011;6:e20081. doi: 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M. Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N. Hengge R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology. 2009;155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A. Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler C, Bohm A, Jenal U, Seifert R. Kaever V. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods. 2010;81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Stewart PS. Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG. Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Storz G, editor; Hengge R, editor. Bacterial Stress Responses. Washington, DC: ASM Press; 2011. [Google Scholar]
- Thomason MK, Fontaine F, De Lay N. Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE. Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo M, Douarche C, Bailleux V, Poulard C, Mariot S, Regeard C. Raspaud E. Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc Natl Acad Sci USA. 2013;110:2011–2016. doi: 10.1073/pnas.1217178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Barembruch C, Possling A. Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Kuipers OP, Brul S, Hellingwerf KJ. Kort R. Effects of phosphorelay perturbations on architecture, sporulation, and spore resistance in biofilms of Bacillus subtilis. J Bacteriol. 2006;188:3099–3109. doi: 10.1128/JB.188.8.3099-3109.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R. Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Chai Y, Beauregard P, Losick R. Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P. McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF. Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Pesavento C, Possling A, Tischendorf G. Hengge R. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- Werner E, Roe F, Bugnicourt A, Franklin MJ, Heydorn A, Molin S, et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2004;70:6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS. Franklin MJ. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, Gonzalez Barrios AF, Herzberg M. Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J. Molin S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol. 2011;13:1705–1717. doi: 10.1111/j.1462-2920.2011.02503.x. [DOI] [PubMed] [Google Scholar]
- Yildiz FH. Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Liu M. Burgess RR. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging'-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W. Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]