Abstract
Many insects possess endosymbiotic bacteria inside their body, wherein intimate interactions occur between the partners. While recent technological advancements have deepened our understanding of metabolic and evolutionary features of the symbiont genomes, molecular mechanisms underpinning the intimate interactions remain difficult to approach because the insect symbionts are generally uncultivable. The bean bug Riptortus pedestris is associated with the betaproteobacterial Burkholderia symbiont in a posterior region of the midgut, which develops numerous crypts harbouring the symbiont extracellularly. Distinct from other insect symbiotic systems, R. pedestris acquires the Burkholderia symbiont not by vertical transmission but from the environment every generation. By making use of the cultivability and the genetic tractability of the symbiont, we constructed a transgenic Burkholderia strain labelled with green fluorescent protein (GFP), which enabled detailed observation of spatiotemporal dynamics and the colonization process of the symbiont in freshly prepared specimens. The symbiont live imaging revealed that, at the second instar, colonization of the symbiotic midgut M4 region started around 6 h after inoculation (hai). By 24 hai, the symbiont cells appeared in the main tract and also in several crypts of the M4. By 48 hai, most of the crypts were colonized by the symbiont cells. By 72 hai, all the crypts were filled up with the symbiont cells and the symbiont localization pattern continued during the subsequent nymphal development. Quantitative PCR of the symbiont confirmed the infection dynamics quantitatively. These results highlight the stinkbug-Burkholderia gut symbiosis as an unprecedented model for comprehensive understanding of molecular mechanisms underpinning insect symbiosis.
Keywords: green fluorescence protein, gut symbiotic bacteria, insect symbiosis, midgut crypt, population dynamics, stinkbug, symbiont colonization
Introduction
Symbiotic associations with microorganisms are omnipresent in nature and have substantially affected organismal evolution (Margulis & Fester 1991; Ruby et al. 2004; Moran 2007). Some microbes are harmful or even lethal to their hosts, being referred to as parasites or pathogens, while others may play pivotal biological roles for their hosts, being regarded as mutualists. Among the amazingly diverse symbiotic associations, the most cohesive forms are found in ‘endosymbiosis’, where the microbes inhabit the host body and the spatial intimacy facilitates intricate interactions between the symbiotic partners. Recent technological advancements in culture-independent molecular methods, including quantitative polymerase chain reaction (PCR), DNA sequencing, molecular phylogenetic analysis, in situ hybridization and secondary ion mass spectrometry (SIMS), as well as genomic and large-scale-omics analyses, have enabled deeper understanding of such endosymbiotic associations (Hentschel et al. 2012; McFall-Ngai et al. 2013). However, cultivation of symbiotic microbes is still crucial for investigating molecular mechanisms underlying host–symbiont interactions (Ruby 2008).
The arthropod class Insecta, consisting of over 1 000 000 described species, is the most diverse animal group. Insects that feed exclusively on nutritionally restricted diets such as plant sap, vertebrate blood and woody material usually possess obligate mutualistic symbionts inside their bodies. These symbiotic bacteria are often essential for hosts' survival and reproduction, and also affect hosts' biology and phenotypes in a variety of ways via, for example, providing essential nutrients (Douglas 1998), digesting food materials (Inoue et al. 2000), influencing host plant usage (Tsuchida et al. 2004), conferring resistance against parasites or pathogens (Oliver et al. 2003; Scarborough et al. 2005), changing host body colour (Tsuchida et al. 2010) and others. For ensuring such host–symbiont associations, many insects have evolved sophisticated mechanisms for vertical symbiont transmission, which may be ovarial transmission, egg surface contamination, coprophagy or other means (Kikuchi 2009; Bright & Bulgheresi 2010). Obligate insect–microbe associations, such as aphid–Buchnera and tsetse–Wigglesworthia symbioses, tend to exhibit host–symbiont phylogenetic congruence, reflecting strict vertical transmission and stable host–symbiont association over evolutionary time (Moran et al. 1993; Clark et al. 2000). Genomes of these symbionts tend to be drastically reduced, which severely hamper their capability of surviving outside their hosts (Shigenobu et al. 2000; Akman et al. 2002; Wernegreen 2002; Baumann 2005; Moran et al. 2008). While genomics of Buchnera, Wigglesworthia and other obligate symbionts has provided invaluable clues to the understanding of their biological roles and evolutionary features (Wernegreen 2002; Baumann 2005; Moran et al. 2008), their fastidious nature has hindered molecular studies on the mechanisms underpinning these symbiotic associations.
On the other hand, elaborate host–symbiont associations without vertical transmission are commonly found in nature, especially among plants and marine invertebrates (Ruby 2008; Bright & Bulgheresi 2010). For example, leguminous plants acquire Rhizobium or allied bacteria from the soil and form root nodules, where the symbiotic bacteria fix atmospheric dinitrogen and provide their hosts with nitrogenous nutrients (Brewin 1991; Schultze & Kondorosi 1998). Bobtail squids harbour a luminous bacterium, Vibrio fischeri, within their symbiotic light organs, which is selectively acquired from the ambient seawater by newborn squids for light production (Nyholm & McFall-Ngai 2004). Reflecting their capability of free life, these symbiotic bacteria are cultivable outside their hosts on standard microbiological media and, therefore, amenable to genetic manipulation (Ruby 2008). Hence, the legume and squid model systems have significantly contributed to our understanding of molecular mechanisms underlying symbiosis.
The bean bug Riptortus pedestris (Hemiptera: Heteroptera: Alydidae; Fig.1a) develops a number of sac-like tissues, called crypts, in a posterior region of the midgut, whose cavity is densely colonized by a betaproteobacterial symbiont of the genus Burkholderia (Kikuchi et al. 2005). The midgut of the bean bug consists of morphologically distinct regions designated as M1, M2, M3 and M4 (Fig.1b). In addition, there is a slightly bulbous region between the M3 and M4 regions, called M4B region (Fig.1b). The M4 region bearing numerous crypts is the principal symbiotic organ harbouring as many as 108 Burkholderia symbiont cells, and the M4B region bearing no crypts also contains some Burkholderia symbiont cells (Goodchild 1963; Futahashi et al. 2013; Kikuchi & Yumoto 2013). Infection with the Burkholderia symbiont improves growth rate and body size of the Riptortus host (Kikuchi et al. 2007), and some Burkholderia strains confer insecticide resistance to the host insect (Kikuchi et al. 2012). Notably, the bean bug does not transmit the Burkholderia symbiont vertically, but acquires the symbiont from the surrounding environment at an early nymphal stage every generation, and the symbiont is easily cultivable on standard microbiological media (Kikuchi et al. 2007, 2011b). Under laboratory conditions, the bean bugs reared in clean containers with soybean seeds and sterilized water cannot acquire the Burkholderia symbiont and become aposymbiotic, whereas the insects reared in the same manner, but with water containing cultured Burkholderia cells, become symbiotic (Kikuchi et al. 2011b; Kikuchi & Yumoto 2013). Hence, the Riptortus–Burkholderia symbiosis represents a novel insect model system wherein the host–symbiont association is tractable experimentally.
Figure 1.
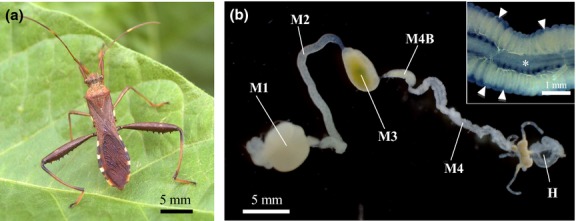
Gut symbiosis in the bean bug R. pedestris. (a) Adult male. (b) Dissected midgut. Inset is an enlarged image of the symbiotic organ. Asterisk and arrowheads indicate midgut main tract and crypts, respectively. Abbreviations: M1, midgut first region; M2, midgut second region; M3, midgut third region; M4B, bulbous midgut region prior to M4 region; M4, midgut fourth region with crypts (symbiotic organ); H, hindgut.
Here, we report that, in the Riptortus–Burkholderia symbiosis, the symbiont is also tractable genetically. We established a genetically engineered strain of the Burkholderia symbiont that expresses a jellyfish-derived green fluorescence protein (GFP; Chalfie et al. 1994; Shimomura 2005), which enabled live imaging of the symbiont localization in the host symbiotic organ. By making use of symbiont visualization by GFP labelling in combination with symbiont quantification by quantitative PCR, we investigated detailed processes of the infection dynamics of the Burkholderia symbiont in vivo during the developmental course of the Riptortus host.
Materials and methods
Insects, bacterial strains and plasmids
The strain of R. pedestris used in this study was originally collected from a soybean (Glycine max) field in Tsukuba, Ibaraki, Japan, and maintained in the laboratory. The insects were reared in petri dishes (90 mm in diameter and 20 mm high) at 25 °C under a long-day regimen (16 h light, 8 h dark) and fed with soybean seeds and distilled water containing 0.05% ascorbic acid (DWA). Burkholderia symbionts used in this study are a rifampicin-resistant (Rfr) spontaneous mutant strain RPE75 and a GFP-expressing mutant strain RPE225 (Table1). These symbiont strains were preserved as frozen stocks at −80 °C and cultured at 25 °C with YG medium (0.5% yeast extract, 0.4% glucose, 0.1% NaCl) as described (Kikuchi et al. 2011b). Escherichia coli strains and plasmids used in this study are listed in Table1.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference or resource |
|---|---|---|
| Bacterial strain | ||
| Burkholderia symbiont | ||
| RPE75 | Wild-type, spontaneous Rfr mutant of Burkholderia symbiont RPE64 | Kikuchi et al. 2011a,b; |
| RPE225 | GFP mutant of Burkholderia symbiont RPE75 | This study |
| Escherichia coli | ||
| WM3064 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4–1360 Δ(araBAD) 567 ΔdapA 1341::[erm pir(wt)] | DK. Newman, California Institute of Technology |
| Plasmid | ||
| pURR25 | Mini Tn7KsGFP, GFP driven by Plac (PA1/04/03) promoter, mobilizable oriTIncPα, suicide oriRR6Kγ; Apr (bla) | Teal et al. 2006; |
| pUX-BF13 | Tn7 transposase genes tnsABCDE, mobilizable oriTIncPα, suicide oriRR6Kγ; Apr (bla) | Bao et al. 1991 |
Administration of Burkholderia symbiont
The symbiont strains were grown to an early log phase in YG medium (containing an adequate antibiotic) on a gyratory shaker (150 rpm) at 30 °C. Colony-forming unit (CFU) values were estimated by plating the culture media on YG agar plates containing adequate antibiotics. Symbiont cells were harvested by centrifuging the culture media, suspended in DWA and adjusted to 104 CFU/μL in DWA. Immediately after first instar nymphs moulted to the second instar, DWA was removed from the rearing containers so that the nymphs were kept without drinking water overnight. Then, DWA containing 104 CFU/μL symbiont cells was supplied to the rearing containers for 24 h, which the second instar nymphs immediately exploited, leading to the acquisition of Burkholderia symbionts. Then, the symbiont-containing DWA was replaced by symbiont-free DWA, and the nymphs were reared to adulthood. To clarify dose dependency of the colonization, symbiont cells were adjusted to 37, 370 and 3,700 CFU/μL in DWA, and inoculated as described above.
Fitness measurement
To reveal fitness effects of the Burkholderia symbiont, 93 and 60 first instar nymphs were prepared for symbiont-infected and uninfected groups, respectively: the former group was inoculated with cultured cells of the symbiont at the second instar stage as described above, while the latter group was left untreated. Nymphs were reared in clean plastic containers (8 cm in diameter, 5.5 cm in depth) to adulthood and subjected to measurements of survival rate (adult emergence rate), nymphal period, body length and thorax width. In each container, 10–11 nymphs were reared and fed with 10 soybean seeds and DWA. For recording time to reproduction and number of eggs, each newly emerged female was mated with two males that emerged from the same experimental group on the same day, and number of eggs the female laid was recorded for 2 weeks from its first oviposition. Each mating pair was reared in a clean plastic container and fed with three soybean seeds and DWA. The fitness parameters were statistically analysed using the software R version 2.12.1 (R development core team 2005). Survival rate was analysed by Fisher's exact test, and the others were analysed by a Mann–Whitney U-test. Colonization of the Burkholderia symbiont was confirmed by diagnostic PCR with specific primer sets (see Table S1, Supporting information), as previously described (Kikuchi et al. 2007).
Construction of GFP-labelled Burkholderia symbiont
The GFP-labelled Burkholderia symbiont strain RPE225, originating from the Burkholderia symbiont strain RPE75, was generated by inserting a mini-Tn7 transposon containing Plac::GFP into the symbiont chromosome using triparental mating as described (Teal et al. 2006). The donor strain E. coli WM3064 (a diaminopimelic acid [DAP] auxotroph) containing either pURR25 (a plasmid containing the miniTn7KSGFP; Teal et al. 2006) or pUX-BF13 (encoding Tn7 transposase; Bao et al. 1991) was grown and mated with RPE75 cells by triparental mating on YG agar containing 300 μg/mL DAP. GFP-labelled recipient cells were selected for resistance to kanamycin (WM3064 requires DAP to grow) and screened for GFP fluorescence using an epifluorescence microscope (Axiophot, Carl Zeiss).
Observation of dissected symbiotic organ
The insects administered with the GFP-labelled symbiont strain RPE225 were dissected in phosphate-buffered saline (PBS: 137 mm NaCl, 8.1 mm Na2HPO4, 2.7 mm KCl, 1.5 mm KH2PO4 [pH7.4]) using a fine forceps under a dissection microscope. The dissected midguts were observed under the epifluorescence microscope freshly, or after a brief fixation with 4% paraformaldehyde for 10 min and a PBS washing.
Insect rearing on soybean pots inoculated with the GFP-labelled Burkholderia symbiont
Each plant pot (diameter 90 mm, height 120 mm) was filled with sterilized vermiculite and planted with a soybean seed. After the seed budded, approximately 108 cells of the GFP-labelled symbiont strain RPE225 suspended in 10 ml of distilled water were evenly spread across the pot. Newly moulted second instar nymphs were transferred from a rearing cage to the soybean pot. In addition to the soybean plant, ten dry soybean seeds in a small petri dish were supplied to the soybean pot. The infection processes of the GFP-labelled symbiont were observed using the epifluorescence microscope as described above.
Quantitative PCR
Real-time quantitative PCR was performed using SYBR green and MX3000P QPCR system (Stratagene, La Jolla, CA) with primers BSdnaA-F and BSdnaA-R (Table S1, Supporting information) targeting a 0.15 kb region of the dnaA gene of the Burkholderia symbiont as described (Kikuchi et al. 2011b). Total DNA was extracted from M4 and M4B parts by using NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany), and the extracted DNA was eluted in 200 μL of TE buffer (10 mm Tris-HCl [pH 8.0], 0.1 mm EDTA). Each of the PCR mixtures (20 μL in total) consisted of 2 μL of 10× TaqGold buffer (Applied Biosystems, Foster City, CA), 1.2 μL of 25 mm MgCl2, 2 μL of dNTPs (2 mm each of dATP, dTTP, dGTP and dCTP), 1 μL of dimethyl sulphoxide, 0.2 μL of SYBR green I (1/1000 diluted solution; Molecular Probes, Eugene, OR), 0.6 μL of primer mixture (5 μm each of forward and reverse primers), 0.1 μL of AmpliTaq Gold DNA polymerase (Applied Biosystems), 8.9 μL of distilled water and 4 μL of DNA sample. The PCR temperature profile was 40 cycles of 95 °C for 10 s, 60 °C for 15 s and 72 °C for 15 s. A standard curve for the dnaA gene was generated with standard samples that contained 10, 102, 103, 104, 105, 106 and 107 copies per reaction of the target PCR fragment amplified with the primers BSdnaA-F and BSdnaA-R.
Results
Experimental administration and fitness effects of the Burkholderia symbiont
Second instar Riptortus nymphs were orally administrated either with DWA containing cultured Burkholderia cells of the strain RPE75 or sterile DWA without bacteria. When some of the insects were subjected to dissection of the midgut and diagnostic PCR detection of the symbiont at the third instar stage, the former insects were all symbiont positive (30/30) while the latter insects were all symbiont negative (0/30). These insects were reared to adulthood, and their fitness parameters were inspected. While survival rates to adulthood were not different between the symbiotic insects and the aposymbiotic insects (Fig.2a), the symbiotic insects exhibited shorter time to adulthood, larger body length and greater thorax width than the aposymbiotic insects (Fig.2b–d) as previously reported (Kikuchi et al. 2007). In addition, the symbiotic female insects started reproduction earlier and produced more eggs than the aposymbiotic female insects (Fig.2e, f).
Figure 2.
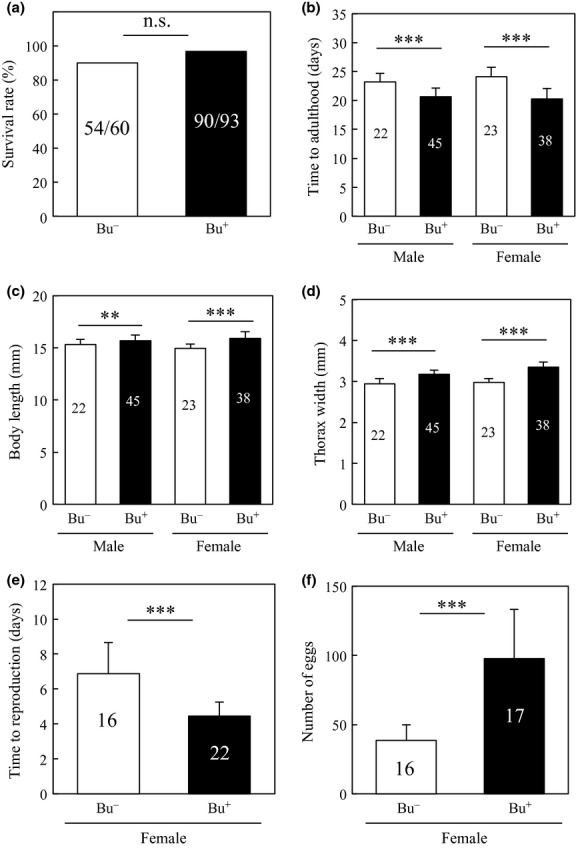
Comparison of fitness parameters between the Burkholderia-infected (Bu+) and uninfected (Bu−) adult insects of R. pedestris. (a) Survival rate (=adult emergence rate) (b) time to adulthood, (c) body length, (d) maximum thorax width, (e) time to reproduction, (f) number of eggs. Numbers inside bars indicate sample sizes. Asterisks indicate statistically significant differences (**, P < 0.001; ***, P < 0.0001).
GFP labelling of the Burkholderia symbiont
We generated a GFP-labelled Burkholderia symbiont strain RPE225 by transforming the strain RPE75 with a Tn7 transposon mutagenesis system. The Tn7:GFP plasmid, pURR25 (Teal et al. 2006), encodes a Tn7 cassette that contains a lac promoter, a GFP gene and a kanamycin resistance (Kmr) gene. Tn7 specifically inserts into the bacterial chromosome at a Tn7 attachment (attTn7) site located at a downstream region of the glmS gene that encodes glucosamine-6-phosphate synthetase (Choi et al. 2006). Because the attTn7 site encodes no functional gene, the Tn7 insertion at the site is expected to be selectively neutral, retaining the intact gene set of the original strain. Reflecting the chromosomal Tn7GFP insertion, the recombinant symbiont strain RPE225 displayed bright and stable fluorescence both in vitro and in vivo, which required neither antibiotic selection nor induction by isopropyl β-D-1-thiogalactopyranoside (IPTG).
Symbiont colonization visualized by GFP labelling
When newly moulted second instar nymphs were administrated with the GFP-labelled Burkholderia symbiont cells, infection of the symbiotic organ was observed around 6 h after inoculation (Fig.3a). At the beginning, the symbiont cells formed an aggregate at the entrance of the M4B region and then gradually migrated into the M4B region. By 24 h after inoculation, dense populations of the symbiont cells appeared in the main tract of the M4 region, and some symbiont cells were detected inside the crypts (Fig.3b). The majority of the crypts were filled with the symbiont cells by 48 h after inoculation (Fig.3c, d). By 72 h after inoculation, all the crypts were filled up with the symbiont cells (Fig.3e), which remained during the subsequent nymphal development (Fig.3f–h).
Figure 3.
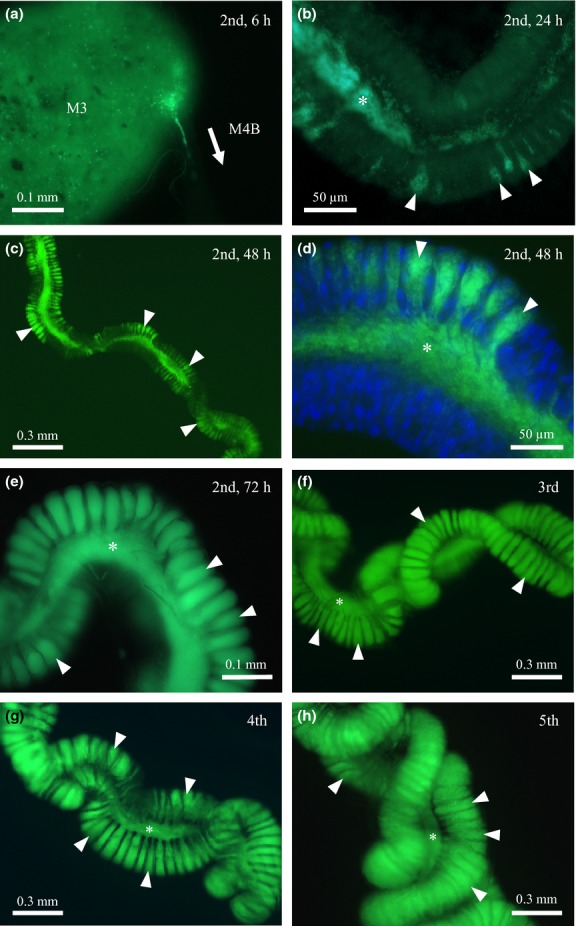
Infection processes of the GFP-labelled Burkholderia symbiont visualized by epifluorescence microscopy. (a) The junction of the M3 and M4B regions of a second instar nymph, 6 h after inoculation. (b) The M4 region of a second instar nymph, 24 h after inoculation. (c, d) The M4 region of a second instar nymph, 48 h after inoculation. (e) The M4 region of a second instar nymph, 72 h after inoculation. (f) The M4 region of a third instar nymph. (g) The M4 region of a fourth instar nymph. (h) The M4 region of a fifth instar nymph. Asterisks and arrowheads indicate the main tract and crypts of the midgut, respectively. In (d), host nuclear DNA was counterstained by 4',6-diamidino-2-phenylindole.
Population dynamics of the Burkholderia symbiont during colonization
Figure4 shows the titres of the Burkholderia symbiont in the dissected symbiotic organ (M4 plus M4B) during colonization. Quantitative PCR targeting the symbiont dnaA gene revealed that the symbiont titres in the infected insects exponentially increased as the host development proceeded and reached a plateau around 107 per individual by the third instar stage.
Figure 4.
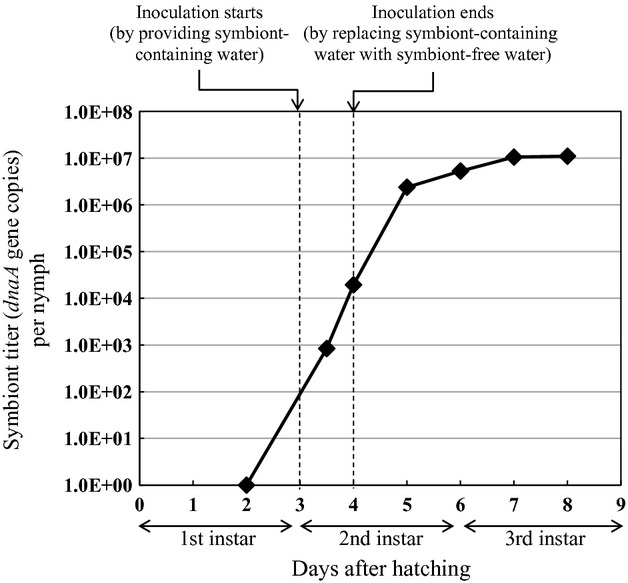
Population dynamics of the Burkholderia symbiont during colonization to and proliferation in the midgut symbiotic organ. Dissected midguts were subjected to quantitative PCR of symbiont dnaA gene copies. Mean of 17 or 18 insects is indicated at each data point. The symbiont inoculation window is indicated by dotted lines.
Dose dependency of the symbiont colonization process
To clarify the dose dependency of the symbiont colonization process, newly moulted second instar nymphs were inoculated with different doses (37, 370 and 3700 CFU/μL) of the GFP-labelled Burkholderia symbiont. At the low inoculum dose (37 CFU/μL), the second instar nymphs exhibited delayed symbiont infection to the midgut crypts in comparison with the insects inoculated at higher doses (370 and 3,700 CFU/μL; Fig.5). At the third instar, all the insects possessed symbiotic organs that were full of symbiont cells irrespective of the inoculum doses (data not shown).
Figure 5.
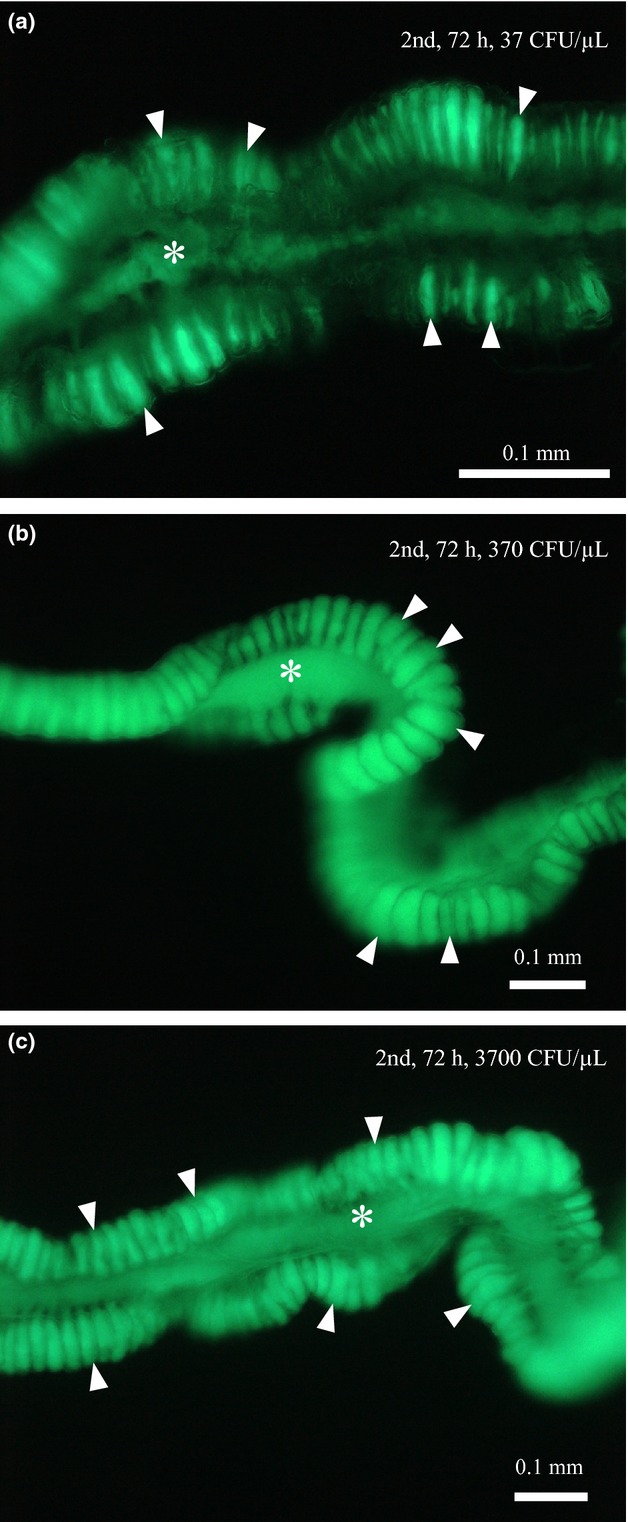
Infection processes of the GFP-labellled Burkholderia symbiont in second instar nymphs of R. pedestris inoculated at different doses (37, 370 and 3700 CFU/μL). The insects were dissected 72 h after inoculation, and their midgut M4 regions were observed by epifluorescence microscopy. (a) 37 CFU/μL, (b) 370 CFU/μL and (c) 3700 CFU/μL. Asterisks and arrowheads indicate the main tract and crypts of the midgut M4 region, respectively.
Symbiont colonization in soybean plant pot
The colonization process of the Burkholderia symbiont to the Riptortus host was investigated in soybean plant pots inoculated with the GFP-labelled Burkholderia symbiont. Under the simulated seminatural condition, the symbiont colonization to the midgut crypts proceeded as in the petri dish experiments of the artificial symbiont administration (see Fig.3). The majority of the crypts were filled with the symbiont cells by 48 h after the second instar moult (Fig.6a), and all crypts were filled up with the symbiont cells by 72 h after the second instar moult (Fig.6b).
Figure 6.
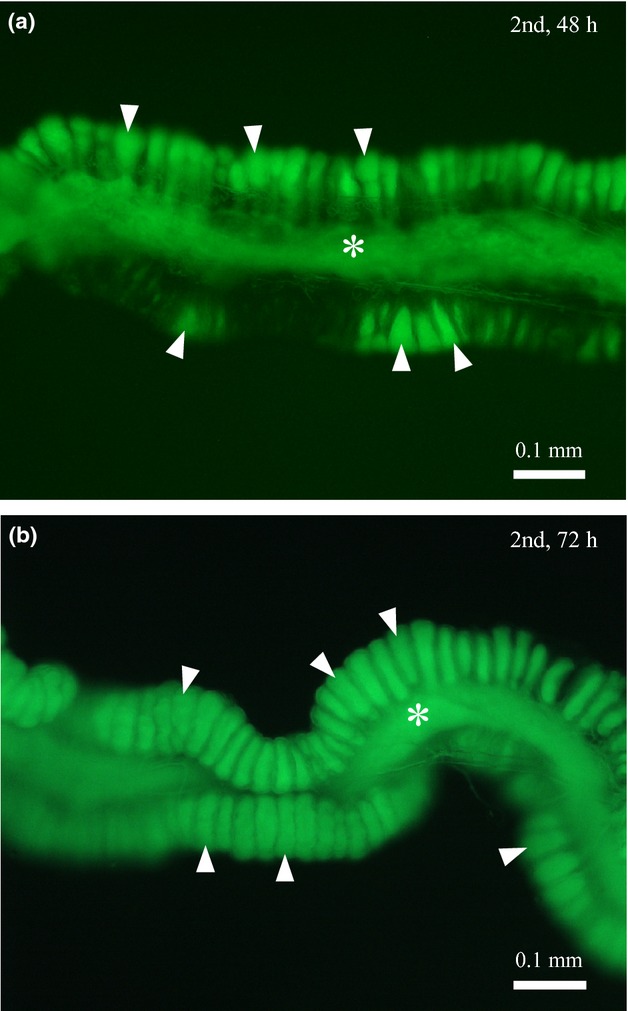
Infection processes of the GFP-labelled Burkholderia symbiont in R. pedestris reared in soybean pots. Cultured symbiont cells were inoculated to the soybean pots, to which newly moulted second instar nymphs were introduced. The insects were dissected, and their midgut M4 regions were observed by epifluorescence microscopy. (a) 48 h after introduction and (b) 72 h after introduction. Asterisks and arrowheads indicate the main tract and crypts of the midgut M4 region, respectively.
Discussion
Like many insect symbionts of mutualistic nature thus far reported, the Burkholderia symbiont of R. pedestris exhibited beneficial effects on the host insect (Fig.2) and specific localization to the symbiotic organ (Figs1b and 3). Distinct from other symbionts, however, the Burkholderia symbiont is easily cultivable outside the host on standard microbiological media and therefore genetically manipulatable. In this study, we successfully established a GFP recombinant strain of the Burkholderia symbiont by making use of standard plasmid and conjugation systems commonly employed for genetic studies of E. coli and other proteobacteria. The GFP-labelled symbiont strain enabled live imaging of individual symbiont cells within dissected midgut symbiotic organs of the host insect, which offered an unprecedented opportunity to closely inspect the symbiont infection dynamics in the insect–microbe symbiosis.
Our observations of the infection processes using the GFP-labelled Burkholderia strain revealed that the symbiont colonization proceeds rapidly in second instar Riptortus nymphs, initiating within a few hours and completing by 72 h after inoculation. The colonization patterns under a laboratory rearing condition in petri dishes where the symbiont was administrated via drinking water (Fig.3) were similar to the colonization patterns under a simulated seminatural condition in soybean plant pots where soil-mediated symbiont acquisition occurred (Fig.6). Previous studies on the Riptortus–Burkholderia symbiosis have demonstrated that the host insect orally acquires the specific symbiont from the complex soil microbiota (Kikuchi et al. 2007), the symbiont acquisition preferentially occurs at the second instar stage (Kikuchi et al. 2011b) and even less than 100 symbiont cells are sufficient for establishing the symbiotic association (Kikuchi & Yumoto 2013). On the basis of these results, it seems likely that the symbiont infection processes are finely programmed and regulated in the host–symbiont interactions.
Our rearing experiments clearly showed beneficial effects of the Burkholderia symbiont on the Riptortus host (Fig.2). However, it is currently unclear what the symbiont actually does for the host. A number of plant-sucking hemipteran insects possess beneficial symbionts inside their bodies, which are primarily located in the cytoplasm of specific cells called bacteriocytes (Buchner 1965; Kikuchi 2009). Recent genomic analyses of these symbiotic bacteria, in parallel with conventional nutritional and physiological studies, have shown that these symbionts provide their hosts with essential amino acids and some vitamins that are scarce in their plant sap diets (Douglas 1998; International aphid genomics consortium 2010; McCutcheon & Moran 2012; Husnik et al. 2013). While R. pedestris is a seed feeder rather than a sap feeder, it seems plausible, although speculative, that the Burkholderia symbiont is provisioning some nutritional factors that are deficient in food sources of the Riptortus host. It is notable that the recently determined genome of the Burkholderia symbiont encodes complete metabolic pathways for essential amino acids and most vitamins (Shibata et al. 2013). Further nutritional and physiological studies are required to clarify biological roles of the Burkholderia symbiont for the Riptortus host.
The quantitative PCR analysis unveiled the initial and subsequent infection dynamics of the Burkholderia symbiont in the development of Riptortus nymphs: after inoculation, the symbiont titres sharply increased during the second instar period and reached a plateau at around 107 dnaA gene copies towards the third instar (Fig.4). It should be noted that the symbiont titres in the symbiotic organ exhibited continuous increase even after the symbiont-containing drinking water was replaced with sterile drinking water (Fig.4), indicating that the symbiont population growth within the second instar nymphs is mainly attributable to within-host proliferation of the symbiont cells. The symbiont population plateau towards the third instar is probably relevant to the filling up of the crypts in the host symbiotic organ (Fig.3e, f).
Vertical symbiont transmission from mother to offspring is among the most pivotal processes for maintaining intimate host–symbiont associations (Bright & Bulgheresi 2010). Hence, researchers have paid considerable attention to the infection processes of symbiotic bacteria within their host insects (Buchner 1965). One of the best-described systems is the aphid–Buchnera symbiosis, wherein the symbiont cells are transmitted from maternal bacteriocytes to early embryos by specific exo- and endocytotic mechanisms (Buchner 1965; Miura et al. 2003; Koga et al. 2012). Such transovarial mechanisms for vertical symbiont transmission have also been described from many other systems including mealybug–Tremblaya, bedbug–Wolbachia, louse–Riesia and other endosymbiotic associations (Buchner 1965; Fukatsu & Nikoh 2000; Sasaki-Fukatsu et al. 2006; Hosokawa et al. 2010). In the tsetse–Wigglesworthia symbiosis, the symbiont cells are not transovarially transmitted but passed to the fly larva via milk gland secretion in utero (Attardo et al. 2008). In these systems, the symbiont transmission processes occur within the insect bodies and tissues, and the symbionts are genetically intractable due to their obligatory dependence on the intrahost environment. Hence, visualization of these symbiont transmission processes has been conducted on fixed specimens using such histological techniques as light microscopy, in situ hybridization, immunohistochemistry, and scanning and transmission electron microscopy.
On the other hand, some insects like stinkbugs have evolved posthatch symbiont transmission mechanisms outside their bodies. The stinkbugs or true bugs, belonging to the insect suborder Heteroptera with over 40 000 described species (Schuh & Slater 1995; Weirauch & Schuh 2011), are often in intimate association with specific symbiotic bacteria in the lumen of the midgut (Buchner 1965; Kikuchi 2009). These symbiotic associations are generally mutualistic, and symbiont-deprived insects often suffer growth defects, higher mortality, reduced fecundity and/or complete sterility (Buchner 1965; Abe et al. 1995; Fukatsu & Hosokawa 2002; Hosokawa et al. 2006, 2012; Kikuchi et al. 2007, 2009; Prado et al. 2009; Tada et al. 2011; Boucias et al. 2012; Salem et al. 2013). In the families Pentatomidae, Acanthosomatidae, Scutelleridae and Pyrrhocoridae, vertical symbiont transmission occurs upon oviposition via egg surface contamination with symbiont-containing excretions (Buchner 1965; Abe et al. 1995; Fukatsu & Hosokawa 2002; Kikuchi et al. 2007, 2009; Kaltenpoth et al. 2009; Prado et al. 2009; Kaiwa et al. 2010; Boucias et al. 2012). In the family Plataspidae, vertical symbiont transmission also occurs upon oviposition, but uniquely, the symbiont is provided within mother-made symbiont-containing capsules (Schneider 1940; Fukatsu & Hosokawa 2002; Hosokawa et al. 2006). In some subsocial species of the families Cydnidae and Parastrachiidae, mother insects stay with and take care of their offspring and provide the newborn nymphs with symbiont-containing fluid from the anus (Schorr 1957; Hosokawa et al. 2012). In these systems, the symbiont transmission processes occur outside the insect body, and, therefore, can be observed directly, often as specialized behaviour, ecology and structure of the host stinkbugs (Hosokawa et al. 2008, 2012). In the stinkbug superfamilies Lygaeoidea and Coreoidea (including the family Alydidae to which R. pedestris belongs), the uniqueness of the symbiosis is highlighted by the exceptional features that the symbiont is not vertically transmitted but acquired from the environment, and is easily cultivable, experimentally tractable and genetically manipulatable (Kikuchi et al. 2007, 2011a,b; Kikuchi & Yumoto 2013).
Genetically modified symbiotic bacteria labelled with GFP or other fluorescent proteins have provided a powerful tool for understanding spatiotemporal infection dynamics of the symbiont as well as intrahost ecological interactions of the symbiotic microbiota in legume–Rhizobium, squid–Vibrio, nematode–Photorhabdus/Xenorhabdus, zebrafish–Pseudomonas and other model symbiotic systems (Gage 2002; Dunn et al. 2006; Rawls et al. 2007; Ciche et al. 2008). The introduction of the GFP-labelled symbiont into the Riptortus–Burkholderia system facilitates the utility of the emerging model for insect symbiosis studies. The Riptortus–Burkholderia system has been regarded as promising in that (i) the symbiont is easily cultivable on standard microbiological media, which is exceptional among insect symbiotic bacteria of a beneficial nature; (ii) the symbiont is orally acquired by young nymphs from the environment every generation, which allows easy preparation of symbiotic and aposymbiotic insects under controlled genetic backgrounds; (iii) both symbiotic and aposymbiotic insects are able to reach sexual maturity and reproduce in the laboratory, which are suitable for comparative physiological, transcriptomic, proteomic and metabolomic analyses on the effects of symbiosis; (iv) RNA interference of the host gene expression works beautifully, which enables straightforward functional analyses of host insect genes involved in symbiosis (Kikuchi et al. 2007, 2011b; Futahashi et al. 2011; Kikuchi & Yumoto 2013). Recently, in addition, both the symbiont genome sequence (Shibata et al. 2013) and the host transcriptome of the midgut symbiotic organ (Futahashi et al. 2013) have been determined, and a homologous recombination procedure to disrupt the symbiont genes has been established (Kim et al. 2013a,b). The accumulated knowledge and tools will enable approaches to investigate molecular mechanisms and associated genetic bases of the insect–microbe symbiosis from both the host side and the symbiont side. A number of important questions still remain unanswered. Why does the Burkholderia symbiont specifically infect and stably colonize the gut symbiotic organ of the Riptortus host? What factors affect the population dynamics of the symbiont in the intrahost ecosystem? How does the symbiont interact (and probably cope) with host immunity? We expect that molecular, genetic, cytological, physiological and ecological work with the Riptortus–Burkholderia model system will lead to deeper and wider understanding of the mechanisms underlying the insect–microbe symbiosis in the near future.
Acknowledgments
This study was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant number 24117525 and by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry. We thank Dianne K. Newman (California Institute of Technology) and Joerg Graf (University of Connecticut) for providing plasmids, and Naruo Nikoh (Open University of Japan) for genomic information of the Burkholderia symbiont.
Data accessibility
Data of the fitness experiments and quantitative PCR: DRYAD doi:10.5061/dryad.q1r19.
Supporting Information
Additional supporting information may be found in the online version of this article.
Primer sets used for diagnostic and quantitative PCR.
References
- Abe Y, Mishiro K, Takanashi M. Symbiont of brown-winged green bug, Plautia stali Scott. Japanese Journal of Applied Entomology and Zoology. 1995;39:109–115. [Google Scholar]
- Akman L, Yamashita A, Watanabe H, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nature Genetics. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Attardo GM, Lohs C, Heddi A, et al. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. Journal of Insect Physiology. 2008;54:1236–1242. doi: 10.1016/j.jinsphys.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annual Review of Microbiology. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Boucias DG, Garcia-Maruniak A, Cherry R, et al. Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis. FEMS Microbiology Ecology. 2012;82:629–641. doi: 10.1111/j.1574-6941.2012.01433.x. [DOI] [PubMed] [Google Scholar]
- Brewin NJ. Development of the legume root nodule. Annual Review of Cell Biology. 1991;7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nature Reviews Microbiology. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York, New York: Interscience; 1965. [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Choi KH, DeShazer D, Schweizer HP. mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nature Protocols. 2006;1:162–169. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- Ciche TA, Kim KS, Kaufmann-Daszczuk B, Nguyen KC, Hall DH. Cell Invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Applied and Environmental Microbiology. 2008;74:2275–2287. doi: 10.1128/AEM.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P, Wernegreen JJ. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution; International Journal of Organic Evolution. 2000;54:517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annual Review of Entomology. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Applied and Environmental Microbiology. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Hosokawa T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Applied and Environmental Microbiology. 2002;68:389–396. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Nikoh N. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera) Applied and Environmental Microbiology. 2000;66:643–650. doi: 10.1128/aem.66.2.643-650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Tanaka K, Matsuura Y, et al. Laccase2 is required for cuticular pigmentation in stinkbugs. Insect Biochemistry and Molecular Biology. 2011;41:191–196. doi: 10.1016/j.ibmb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Tanaka K, Tanahashi M, et al. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS ONE. 2013;8:e64557. doi: 10.1371/journal.pone.0064557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. Journal of Bacteriology. 2002;184:7042–7046. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild AJP. Studies on the functional anatomy of the intestines of Heteroptera. Proceedings of the Zoological Society of London. 1963;141:851–910. [Google Scholar]
- Hentschel U, Weis VM, McFall-Ngai MJ. Biological bulletin virtual symposium: discoveries in animal symbiosis in the “omics” age. The Biological Bulletin. 2012;223:5–6. doi: 10.1086/BBLv223n1p5. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biology. 2006;4:e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Symbiont acquisition alters behaviour of stinkbug nymphs. Biology Letters. 2008;4:45–48. doi: 10.1098/rsbl.2007.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Hironaka M, Mukai H, et al. Mothers never miss the moment: a fine-tuned mechanism for vertical symbiont transmission in a subsocial insect. Animal Behaviour. 2012;83:293–300. [Google Scholar]
- Husnik F, Nikoh N, Koga R, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- Inoue T, Kitade O, Yoshimura T, Yamaoka I. Symbiotic association with protists. In: Abe T, Bignell DE, Higashi M, editors. Termites: Evolution, Sociality, Symbioses, Ecology. Dordrecht: Kluwer Academic Publishers; 2000. pp. 275–288. [Google Scholar]
- International aphid genomics consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biology. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa N, Hosokawa T, Kikuchi Y, et al. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Applied and Environmental Microbiology. 2010;76:3486–3494. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M, Winter SA, Kleinhammer A. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiology Ecology. 2009;69:373–383. doi: 10.1111/j.1574-6941.2009.00722.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes and Environments. 2009;24:195–204. doi: 10.1264/jsme2.me09140s. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Yumoto I. Efficient colonization of the bean bug Riptortus pedestris by an environmentally transmitted Burkholderia symbiont. Applied and Environmental Microbiology. 2013;79:2088–2091. doi: 10.1128/AEM.03299-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Meng XY, Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Applied and Environmental Microbiology. 2005;71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Applied and Environmental Microbiology. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Nikoh N, et al. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biology. 2009;7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. The ISME Journal. 2011a;5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. Specific developmental window for establishment of an insect-microbe gut symbiosis. Applied and Environmental Microbiology. 2011b;77:4075–4081. doi: 10.1128/AEM.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hayatsu M, Hosokawa T, et al. Symbiont-mediated insecticide resistance. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Lee HJ, Kikuchi Y, et al. Bacterial cell wall synthesis gene uppP is required for Burkhoderia colonization of the stinkbug gut. Applied and Environmental Microbiology. 2013a;79:4879–4886. doi: 10.1128/AEM.01269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Won YJ, Nikoh N, et al. Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:E2381–E2389. doi: 10.1073/pnas.1303228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Meng XY, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Fester R. Symbiosis as a Source of Evolutionary Innovation. Cambridge, Massachusetts: MIT Press; 1991. [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nature Reviews Microbiology. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, et al. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) Journal of Experimental Zoology Part B, Molecular and Developmental Evolution. 2003;295:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proceedings of the Royal Society B. 1993;253:167–171. [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annual Review of Genetics. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Annual Review of Microbiology. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado SS, Golden M, Follett PA, Daugherty MP, Almeida RP. Demography of gut symbiotic and aposymbiotic Nezara viridula L. (Hemiptera: Pentatomidae) Environmental Entomology. 2009;38:103–109. doi: 10.1603/022.038.0112. [DOI] [PubMed] [Google Scholar]
- R development core team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nature Reviews Microbiology. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby E, Henderson B, McFall-Ngai M. We get by with a little help from our (little) friends. Science. 2004;303:1305–1307. doi: 10.1126/science.1094662. [DOI] [PubMed] [Google Scholar]
- Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae) Environmental Microbiology. 2013;15:1956–1968. doi: 10.1111/1462-2920.12001. [DOI] [PubMed] [Google Scholar]
- Sasaki-Fukatsu K, Koga R, Nikoh N, et al. Symbiotic bacteria associated with stomach discs of human lice. Applied and Environmental Microbiology. 2006;72:7349–7352. doi: 10.1128/AEM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- Schneider G. Beiträge zur Kenntnis der symbiontischen Einrichtungen der Heteropteren. Zeitschrift für Morphologie und Ökologie der Tiere. 1940;36:565–644. [Google Scholar]
- Schorr H. Zur Verhaltensbiologie und Symbiose von Brachypelta aterrima Först (Cydnidae, Heteroptera) Zeitschrift für Morphologie und Ökologie der Tiere. 1957;45:561–602. [Google Scholar]
- Schuh RT, Slater JA. True Bugs of the World (Hemiptera: Heteroptera) New York, New York: Cornell University Press; 1995. [Google Scholar]
- Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annual Review of Genetics. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- Shibata TF, Maeda T, Nikoh N, et al. Complete genome sequence of Burkholderia sp. strain RPE64, bacterial symbiont of the bean bug R. pedestris. Genome Announcements. 2013;1:e00441–e00413. doi: 10.1128/genomeA.00441-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Shimomura O. The discovery of aequorin and green fluorescent protein. Journal of Microscopy. 2005;217:1–15. doi: 10.1111/j.0022-2720.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- Tada A, Kikuchi Y, Hosokawa T, et al. Obligate association with gut bacterial symbiont in Japanese populations of the southern green stinkbug Nezara viridula (Heteroptera: Pentatomidae) Applied and Entomology and Zoology. 2011;46:483–488. [Google Scholar]
- Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Applied and Environmental Microbiology. 2006;72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Horikawa M, et al. Symbiotic bacterium modifies aphid body color. Science. 2010;330:1102–1104. doi: 10.1126/science.1195463. [DOI] [PubMed] [Google Scholar]
- Weirauch C, Schuh RT. Systematics and evolution of Heteroptera: 25 years of progress. Annual Review of Entomology. 2011;56:487–510. doi: 10.1146/annurev-ento-120709-144833. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nature Reviews Genetics. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sets used for diagnostic and quantitative PCR.
Data Availability Statement
Data of the fitness experiments and quantitative PCR: DRYAD doi:10.5061/dryad.q1r19.


