Abstract
An open reading frame with homology to known endolysin genes was identified in the genome of Streptomyces sp. strain 212, which is a newly isolated soil bacterium. The heterologously expressed gene product of this endolysin-like gene, called Mitrecin A, demonstrated bacteriolytic activity against several Gram-negative bacteria. The genome of the bacterial strain was sequenced to draft quality using pyrosequencing followed by genome assembly and gene annotation. Within the sequence, a chromosomally located endolysin-like open reading frame was predicted. The gene product, designated Mitrecin A, was heterologously expressed and isolated from contaminating proteins as a fusion protein to a 6-histidine tag. Mitrecin A consists of 127 amino acids arranged in modular domains of activity. It has an estimated molecular weight of 14·3 kDa and retains sequence homology to the M15C peptidase subfamily of zinc metallocarboxypeptidases. The heat-labile purified recombinant protein has an overall positive charge, has optimal catalytic activities at 26°C in solution of pH 9 with 1% saline and has bacteriolytic activity against Gram-negative bacteria of the medically important genera Aeromonas, Escherichia, Salmonella, Shigella, Vibrio and Yersinia.
Significance and Impact of the Study
The gene of a new protein antimicrobial, Mitrecin A, was discovered in the genome of a soil bacterium. The purified recombinant enzyme, resulting from heterologous over expression of the gene, was found to be tolerant of increased pH conditions and to have bacteriolytic activity against Gram-negative bacteria of the medically important genera Aeromonas, Escherichia, Salmonella, Shigella, Vibrio and Yersinia. Characterization of enzymes such as Mitrecin A from previously uncharacterized bacteria provides potential options for new biocontrol agents in medically and economically important applications like therapeutics, disinfectants, food preservatives, agricultural livestock antimicrobials, and inhibitors of biofilm production.
Keywords: antimicrobials, gene expression, genomics, soil, streptomycetes
Introduction
The biochemical complexity found within soil environments results from their genetic diversity, which is predicted to exceed 4000 bacterial genomes for every gram of soil (Torsvik et al. 1990). Recent studies have utilized metagenomics (Gillespie et al. 2002) and expanded cultivation techniques (Guo et al. 2012) to access the antimicrobial potential from soil microbial communities. The varied antimicrobial agents found within the soil environment include enzymes with bacteriolytic activities (Guo et al. 2012; Singh et al. 2012).
In addition to their importance to basic science, protein antimicrobial enzymes have potential clinical value. Indeed, new antimicrobial compounds are needed to counter medically important strains of pathogenic micro-organisms. In addressing the demand for new therapeutic and disinfectant options against emerging pathogens, previously uncultivated micro-organisms are useful for novel compound discovery (Tiwari and Gupta 2012).
Endolysin-like antimicrobial proteins have advantages as effective biocontrol agents for use in food products (Garcia et al. 2010) and as potential therapeutic compounds (Gupta and Prasad 2011). Expanding databases of predicted proteins from the increasing number of bacterial genomes sequenced and annotated include a growing number of potential endolysins and autolysins.
In this study, we report the selection, identification and primary characterization of a new endolysin-like enzyme, Mitrecin A, from the genome of a soil streptomycete. The heat-labile, cationic enzyme has bacteriolytic activity against Gram-negative bacteria, including the medically important genera of Aeromonas, Escherichia, Salmonella, Shigella, Vibrio and Yersinia. The amino acid sequence of the catalytic domain of the Mitrecin A protein has sequence homology to the M15C peptidase subfamily of zinc metallocarboxypeptidases. The open reading frame encoding Mitrecin A was identified within the draft genome sequence of the producing bacterium, tentatively named Streptomyces sp. strain 212. This organism is a previously uncharacterized actinomycete, isolated from soil samples of a woodland bluff environment in Lynn, Alabama.
Results and discussion
A new endolysin-like protein, Mitrecin A, was isolated and characterized from a soil streptomycete. The producing strain was a Streptomyces species based on 16S rRNA gene analysis (GenBank accession number KC488796) and chemotaxonomic characteristics (data not shown) and considered of interest because of its production of bacteriolytic enzymes against indicator bacteria (Table 1).
Table 1.
Antibacterial spectrum of Mitrecin A. Susceptibility of indicator bacteria to Mitrecin A bacteriolytic activity was measured using heat-killed cultures incorporated into microslide agarose diffusion assays. The substrates were tested against 1 μg of Mitrecin A for 24 h at 37°C. Assays were performed in triplicate and repeated using viable bacteria
| Source | Susceptibility* | |
|---|---|---|
| Gram-negative bacteria | ||
| Aeromonas hydrophila | ATCC† 7965 | + |
| Escherichia coli DH5α | Life Technologies | + |
| Francisella philomiragia O#319L | ATCC 25015 | – |
| Salmonella enterica subsp. enterica ETS34 | ATCC 10708 | + |
| Shigella sonnei | ATCC 9290 | + |
| Vibrio cholerae NCTC 8457 | ATCC 14033 | + |
| Yersinia pseudotuberculosis | ATCC 11960 | + |
| Gram-Positive Bacteria | ||
| Bacillus cereus FDA5 | ATCC 10702 | – |
| Bacillus subtilis 168 | ATCC 23857 | – |
| Bacillus thuringiensis | ATCC 10792 | – |
| Listeria monocytogenes | ATCC 35152 | – |
| Staphylococcus aureus subsp. aureus FDA209P | ATCC 6538P | – |
Susceptibility of the test organisms was determined using the microslide agarose diffusion assay.
American Type Culture Collection (ATCC).
Pyrosequencing of the bacterial genome to draft quality followed by de novo assembly and annotation allowed the identification of the Mitrecin A open reading frame (ORF). The genome sequences of the bacterium were assembled into 1807 contiguous sequences and 152 scaffolds with a mean contiguous sequence length of 5859 bases at an average depth of coverage of 11·6×. The genome of the streptomycete, estimated by analysis in Newbler Assembler, is c. 10 Mbp with a G + C content of 68·4%. The ORF for Mitrecin A (384 bp; GenBank accession number KC488797) was annotated using the Glimmer 3 algorithm in the Institute for Genome Sciences Annotation Engine and was located on a genome fragment of 2864 bp in length. The gene has conserved sequence homology to known and predicted endolysin-like gene sequences, displaying the highest similarity (75% bp identity) to a hypothetical phage endolysin gene from the complete genome of Brevundimonas subvibrioides ATCC 15264 when searched against the NCBI GenBank database using BLASTn. Due to a lack of continuity of the genome fragment containing the Mitrecin A ORF with the remainder of the draft-quality bacterial genome sequence, comprehensive analysis of the genes surrounding Mitrecin A was not possible.
Nucleotide sequence similarities between phage endolysins and bacterial lytic enzymes have been previously reported (Garcia et al. 1988). As shown in this study, Mitrecin A, while located within the genome of strain 212, has significant sequence similarities to phage endolysins. This conservation of sequence with other known phage endolysins suggests the possibility of gene transfer between a bacteriophage and the host bacterial cell. Horizontal gene transfers mediated by integrative elements such as prophages have been reported for other genes (Hambly and Suttle 2005; Cortez et al. 2009).
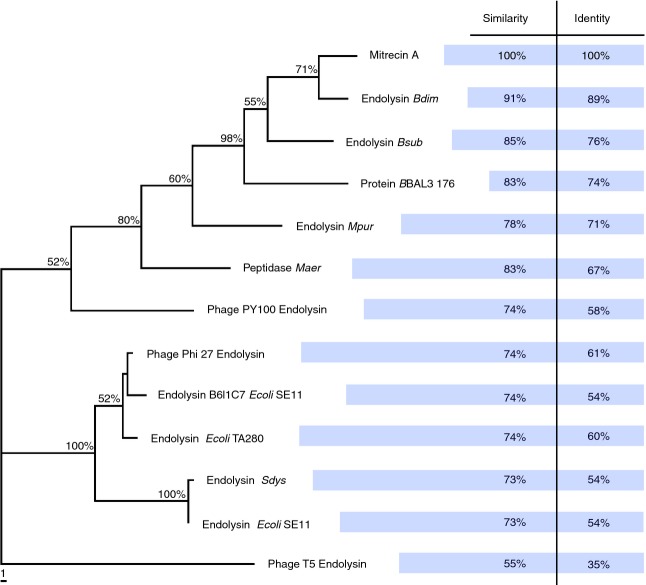
The heterologous expression of the Mitrecin A ORF yielded a 14·3 kDa (127 amino acids) cationic protein with sequence homology to zinc-binding metallocarboxypeptidases. The protein was predicted to have an isoelectric point of 9·4 and a charge of +2·6 at pH 7. Many proteins with sequence similarity to endolysins are predicted from computationally annotated bacterial genome sequences and can be found within protein databases such as UniProtKB, Pfam and MEROPS. For example, protein BBAL3 176 and peptidase Maer (Fig. 1), proteins identified as similar to Mitrecin A as well as other phage endolysins, are also located within the genomes of their hosts Brevundimonas sp. strain BAL3 and Micavibrio aeruginosavorus ARL-13, respectively. A number of these predicted and uncharacterized endolytic proteins and peptidases from these databases have sequence similarity to Mitrecin A (Fig. 1); however, the closest related sequence with characterization of functional activity at the protein level is the endolysin protein of Enterobacteria T5 phage, identified using the UniProtKB database. In the neighbour-joining phylogram (Fig. 1), Mitrecin A shows sequence divergence from all closely related, predicted proteins and marked divergence from the T5 phage endolysin protein.
Figure 1.
Phylogenetic position of Mitrecin A protein as compared to other closely related endolysins. Mitrecin A protein sequence and protein sequences with similarity to and aligned with Mitrecin A were evaluated in PAUP* to construct a neighbour-joining phylogram showing the relationship between Mitrecin A and the similar sequences. Percentages of bootstrap support (>50%) based on 1000 replications are shown at the nodes. The tree was rooted using Enterobacteria phage T5 endolysin as a closely related peptidase endolysin with functional evidence at the protein level. Other endolysins with closely related sequences to Mitrecin A shown in the phylogram have only predicted function. The scale bar indicates the number of substitutes per 10 sites for a unit branch length.
Comparison of the protein sequence within the Pfam protein database identified a conserved d-alanyl-d-alanine carboxypeptidase domain at the C-terminus of Mitrecin A resembling the M15C peptidase family of zinc metallohydrolases (Arthur et al. 1992; Bochtler et al. 2004). Clustalw multiple alignment comparison of Mitrecin A to closely related carboxypeptidase sequences showed conserved protein domain architecture within the C-terminus (Fig. 2), the Mitrecin A catalytic domain defined by three conserved regions containing two histidine residues (H63 and H118) and one aspartic acid residue (D70) predicted in MEROPS to interact with the zinc ion within the binding pocket of the catalytic site (Fig. 2). The active site residue is inferred to be an aspartic acid residue (D115). Within the three conserved regions of the Mitrecin A catalytic domain, the residues H63, D70, D115 and H118 are part of the conserved aspartic acid-histidine (Asp-His) motifs H-x6-d and d-x2-H, which are characteristic of members of the C subfamily of the M15 family of zinc-binding metallopeptidases (Rawlings et al. 2002).
Figure 2.
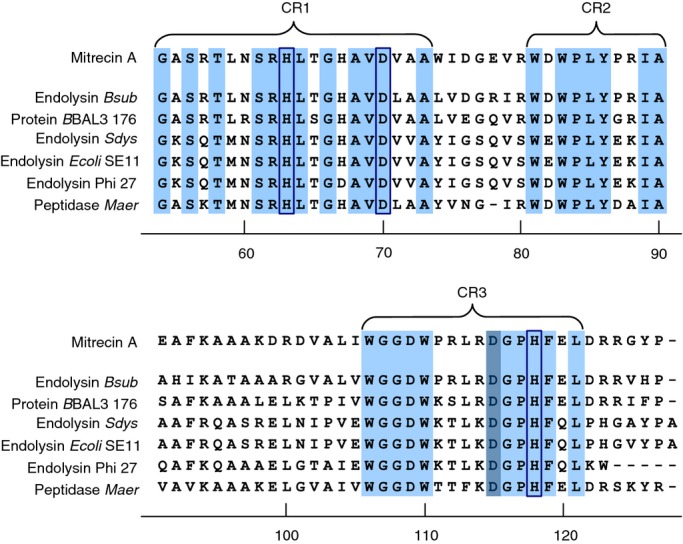
Conserved architecture of the C-terminal catalytic region of Mitrecin A as compared to other similar carboxypeptidases. C-termini of sequences with similarity to Mitrecin A (Fig. 1), identified in MEROPS Blast searches, were aligned against the Mitrecin A C-terminus using the clustalw multiple alignment tool. Conserved amino acid residues and motifs within the C-terminal M15 peptidase subfamily domains as aligned against Mitrecin A are shaded in light blue. Conserved region 1 (CR1), conserved region 2 (CR2), and conserved region 3 (CR3) are indicated. Histidine (H) and aspartic acid (D) residues, inferred by analysis in MEROPS to be necessary for interaction with the metal ion of the catalytic site binding pocket, are enclosed in boxes. The active site residue is shaded in dark blue.
Optimal catalytic activities for Mitrecin A were achieved at 26°C in solution of pH 9 with 1% saline (Fig. 3). The enzyme is sensitive to acidic conditions below pH 6 and shows complete inactivation at temperatures above 55°C. In considering potential downstream applications of the enzyme, pH 7·4 was held constant under varying saline (Fig. 3b) and temperature (Fig. 3c) treatments to illustrate the activity of Mitrecin A at human physiological pH. The bacteriolytic effects of Mitrecin A indicate a resistant population within the bacterial target population, resulting in turbid zones of lysis in microslide assays and a lack of broth clearing during viability bacteriolytic assays. For example, in the kill curve viability assay (Fig. 4a), increased concentrations of Mitrecin A above 0·15 mg ml−1 returned diminished kill results. Similar subpopulation susceptibility to endolysins of actinomycete bacteriophage has been previously described (Higgins and Lechevalier 1969).
Figure 3.
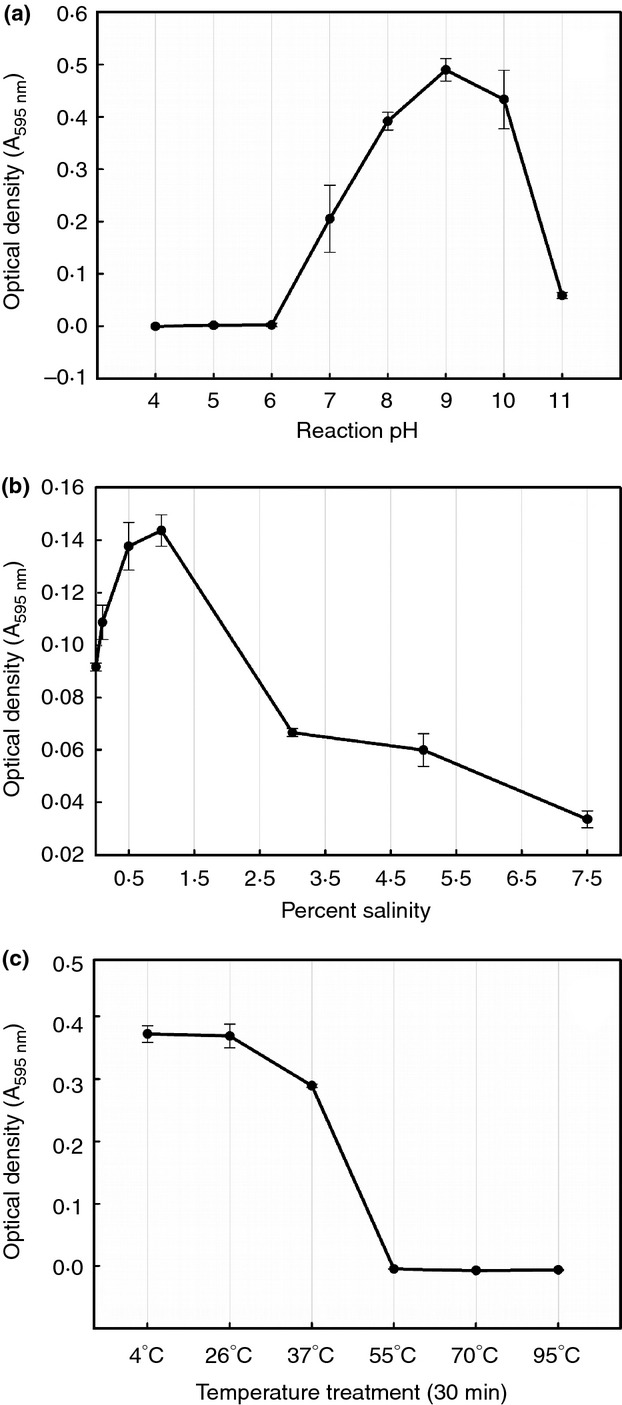
Factors affecting Mitrecin A activity. Residual activity of Mitrecin A was measured against Remazol brilliant blue (RBB)-labelled Y. pseudotuberculosis in quantitative dye-release assays at (a) various pH conditions (pH 4–11), (b) various saline concentrations (0–7·5%) and (c) after various 30-min temperature treatments (4–95°C). Residual activities represent the mean of three independent measurements.
Figure 4.
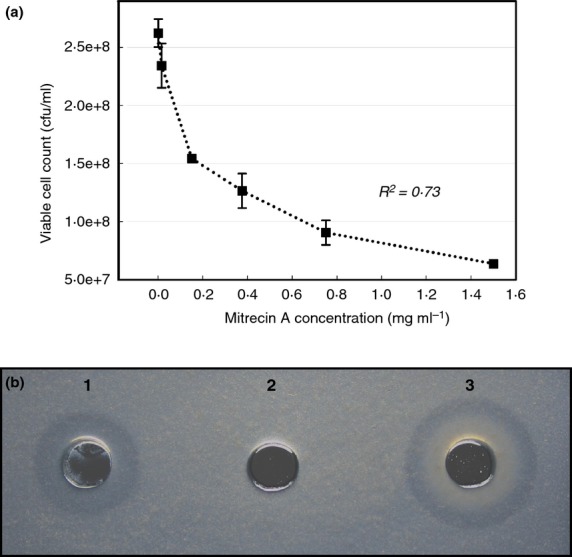
Bacteriolytic activity of Mitrecin A. Residual viability of the Yersinia pseudotuberculosis indicator culture (a) was measured by determining the colony-forming units per millilitre after 16 h exposures of the culture to various concentrations of Mitrecin A. The regression coefficient (R2) of the plot is for the best-fit third-order polynomial. Microslide assays were performed to rapidly visualize the enzymatic activity of Mitrecin A against susceptible bacteria. In an illustration of the microslide assay (b), enzymatic activity for Mitrecin A (well 1, 1 μg), bovine serum albumin (well 2, 1 μg) and lysozyme (well 3, 1 μg) is shown against Aeromonas hydrophila.
Susceptibility of indicator bacteria to the bacteriolytic activity of Mitrecin A was tested using viable and whole-cell heat-killed bacteria in microslide agarose diffusion assays (Fig. 4b). Apart from Francisella philomiragia, Gram-negative bacteria tested against Mitrecin A were susceptible whereas none of the Gram-positive bacteria tested were susceptible (Table 1). The susceptible genera contain pathogenic bacteria of medical and economic importance. Recent studies characterizing new endolysins (Son et al. 2010a; Rodriguez-Rubio et al. 2012; Walmagh et al. 2012) and endolysins with activity against pathogens of medical and economic concern (Son et al. 2010a, b; Eugster et al. 2011) point to the diversity that is displayed by these enzymes. Proposed applications for endolysin-like protein antimicrobials include use as edible films and coatings with antimicrobial properties for fruits and vegetables (Valencia-Chamorro et al. 2011), enzybiotics for treatment of microbial infections (Hermoso et al. 2007; Fischetti 2010; Gupta and Prasad 2011), food preservatives (Settanni and Corsetti 2008; Khan et al. 2010), agricultural livestock antimicrobials (Joerger 2003), biosensors for detection of bacterial cells (Kretzer et al. 2007; Tolba et al. 2012), and inhibitors of biofilm (Son et al. 2010b).
Mitrecin A is organized in a modular arrangement of domains with an N-terminal cell-binding domain and a C-terminal catalytic domain, a common feature in endolysins of Gram-positive bacteria (Garcia et al. 1988) and can be exploited for the development of designed functions. For example, the modular construction of the lytic and binding domains of endolysins has allowed development of chimeric lytic enzymes by combining properties of the two target-independent endolysins. In a study by Fernandes et al. (2012), the lytic domains of enterococcal phage endolysins Lys168 and Lys170 were fused to the cell-binding domain of staphylococcal phage endolysin Lys87, resulting in broadened activity against Gram-positive pathogens. In similar combinatorial studies, LysH5 endolysin was combined with the secretion signal sequence of bacteriocin Lcn972 to increase extracellular excretion of active endolysin (Rodriguez-Rubio et al. 2012), while the streptococcal phage lysin endopeptidase domain was combined with the cell wall-binding domains of the staphylococcal phage lysin LysK and lysostaphin, resulting in killing activity against Staphylococcus aureus isolates from livestock (Schmelcher et al. 2012). Other combinatory studies have fused the peptidoglycan-binding domain with green fluorescent protein, resulting in chimeric proteins with utility for tracking the presence of target bacteria with the fluorescent reporter signal (Briers et al. 2009; Schmelcher et al. 2010). As new endolysin-like proteins such as Mitrecin A are characterized, the development of combinatorial enzymes has the potential to greatly expand the utility of endolysins against pathogens resistant to conventional antibiotics.
Materials and methods
Bacterial strains and culture conditions
Streptomyces sp. strain 212 was isolated from Rainbow Bluff, a woodland rock outcropping in Lynn, Alabama, using the methods described by Farris et al. (2011). The strain was maintained on nutrient agar plates and cultivated in nutrient broth with 5% dextrose for genomic DNA isolations used in de novo genome sequencing assays. Spore suspensions were prepared and stored as described by Kieser et al. (2000). The bacteria listed in Table 1 were used as indicators of bacteriolytic activity of Mitrecin A.
The near complete 16S rRNA gene sequence (GenBank accession number KC488796) of strain 212 was amplified by a polymerase chain reaction using universal bacterial primers 27f (Weisburg et al. 1991) and 1492r (Lane 1991). The partial 16S rRNA gene sequence was searched against the National Center for Biotechnology Information (NCBI) GenBank database using BLASTn.
Enzyme substrate preparation
Enzyme substrates were prepared by growing the respective indicator bacterial cultures to exponential growth phase, followed by autoclaving for 10 min. The heat-killed cells were pelleted by centrifugation, washed three times with ultrapure distilled water (Life Technologies, Grand Island, NY) and stored at −20°C.
Genome sequencing, draft assembly and annotation
Approximately 100 μg of total genomic DNA was extracted and purified from Streptomyces sp. strain 212 using a Qiagen Genomic Tip 20/G kit (Qiagen, Inc., Valencia, CA) according to manufacturer recommendations. Quality and concentration of the genomic DNA was assessed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and ethidium bromide-stained gel electrophoresis (1% agarose) with Low DNA Mass Ladder (Life Technologies; 10068-013). De novo genome sequencing and draft sequence assembly were performed by the Institute for Genome Sciences (IGS) Genomics Resource Center at the University of Maryland School of Medicine using the 454 GS FLX pyrosequencing platform with Titanium chemistry and GS de novo sequence assembly software, Newbler Assembler, ver. 2.3 (454 Life Sciences, Branford, CT). Genome annotation was performed by the IGS Annotation Engine. Open reading frames (ORFs) were identified by the Glimmer 3 algorithm (Delcher et al. 1999). tRNA and rRNA genes were detected by tRNAscan-SE (Lowe and Eddy 1997) and RNAmmer (Lagesen et al. 2007), respectively.
Gene and protein sequence analysis
The gene sequence encoding Mitrecin A (GenBank accession number KC488797) was searched against the NCBI GenBank database using BLASTn. The translated protein sequence of Mitrecin A was searched within the MEROPS peptidase database using wu-blast, the UniProtKB database using wu-blast, the Pfam protein families database using HMMER (Finn et al. 2011) and the NCBI nonredundant(nr) protein database using BLASTp. Protein sequences with high similarity to Mitrecin A identified within the database searches were aligned against the Mitrecin A protein sequence using the clustalw multiple alignment tool (Thompson et al. 1994) within BioEdit biological sequence alignment editor, ver. 7.1.3 (Hall 2011). Phylogenetic relatedness of Mitrecin A to similar protein sequences was determined by performing bootstrap analysis based on 1000 replications and neighbour-joining analysis in paup*, ver. 4.0b10 (Swofford 2003). The generated neighbour-joining phylogram was rooted using Enterobacteria phage T5 endolysin (GenBank accession number AAS19387.1) as a peptidase endolysin with a primary sequence most closely related to Mitrecin A with functional evidence at the protein level. Other predicted peptidases closely related to Mitrecin A had only predicted function. Conserved amino acids within the C-terminal region of Mitrecin A were identified by clustalw multiple alignment against peptidase sequences identified as containing high similarity to Mitrecin A in MEROPS (Fig. 2).
Gene synthesis, gene expression and product purification
The gene encoding Mitrecin A (GenBank accession number KC488797) was fused with a 6-histidine tag sequence at the 3′-end by removing the TGA stop codon and replacing it with the sequence CACCACCACCACCACCACTGA, performed by GenScript (Piscataway, NJ, USA). The fusion gene was synthesized and cloned in pUC57 with an EcoRI restriction site added to the 5′-end and a HindIII restriction site added to the 3′-end. The fusion gene was subsequently subcloned into expression vector pET21a(+), using the EcoRI and HindIII restriction sites. The fusion protein was expressed by IPTG induction (2 mmol l−1) (Sambrook and Russell 2001) in E. coli BL21 Rosetta 2(DE3)pLysS (Life Technologies), and the resultant protein was extracted from inclusion bodies, resolubilized, and partially purified by GenScript using a Ni2+-charged HiTrap IMAC HP column (GE Healthcare Life Sciences, Pittsburgh, PA). The partially purified protein was suspended at a concentration of 0·566 mg ml−1 in freezing buffer (50 mmol l−1 Tris, 0·5 mol l−1 l-arginine, 10% glycerol, pH 9·0) and was stored at −80°C.
Histidine-tagged Mitrecin A was purified using a BioLogic DuoFlow system (Bio-Rad Laboratories, Inc., Hercules, CA) fitted with a Superdex 75 size-exclusion column (GE Healthcare Life Sciences) at a flow rate of 0·8 ml min−1 in phosphate-buffered saline (PBS, 137 mmol l−1 NaCl, 2·7 mmol l−1 KCl, 4·3 mmol l−1 Na2HPO4, 1·4 mmol l−1 KH2PO4, pH 7·4; Teknova, Inc., Hollister, CA). Collected fractions containing histidine-tagged Mitrecin A were concentrated using Amicon Ultra centrifugal filters (10 kDa MWCO; EMD Millipore, Billerica, MA) and analysed for contaminants using a Protein 230 chip on a 2100 Bioanalyzer (Agilent) and in SDS-PAGE. The concentrated enzyme solutions were supplemented with 0·1% sodium azide and stored at 4°C. Sodium azide was removed by buffer exchange using centrifugal filters prior to functional assays. At each point of the purification process and for determining active fractions, histidine-tagged Mitrecin A was located within fractions using the dot-blot assay. Functional activity was monitored using the dye-release and microslide agarose diffusion assays. Fraction purity was assessed using Coomassie blue-stained sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970) and Bioanalyzer analysis.
Mitrecin A protein analysis
The dot-blot assay used in the purification process consisted of spotting 5 μl of the solution onto 0·2-μm nitrocellulose paper (Bio-Rad). Histidine-tagged protein standard (positive control) (Bio-Rad; 161-0375) and PBS (negative control) were also spotted. The spots were allowed to air dry at room temperature. The paper was incubated for 1 h at room temperature in PBS with 5% skim milk (Oxoid Limited, Hampshire, UK) to block residual binding sites. Following the block, the paper was rinsed 3 × in PBS and incubated for 1 h at room temperature with 0·1 μg ml−1 anti-histidine monoclonal antibody conjugated to horse radish peroxidase (GenScript; A00612) in 5% skim milk. After incubation, the paper was rinsed 3 × in PBS and developed using the Immun-Star Western C chemiluminescence kit (Bio-Rad). Development of a black spot, imaged using a VersaDoc Model 4000 imaging system (Bio-Rad), indicated the presence of antibody bound to the 6 × histidine amino acid sequence.
Purity of Mitrecin A during preparation was monitored using a 2100 Bioanalyzer with an Agilent Protein 230 kit and denaturing polyacrylamide gel electrophoresis (PAGE). Denaturing PAGE was performed as described by Laemmli (1970) in a Mini-PROTEAN electrophoresis cell (Bio-Rad) using Mini-PROTEAN TGX Any kD polyacrylamide gels (Bio-Rad) in 0·1% sodium dodecyl sulphate (SDS) Tris-glycine electrophoresis buffer. Molecular weight was estimated using Precision Plus Protein Kaleidoscope standards (10–250 kDa; Bio-Rad). Protein concentrations were determined using the BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL) with concentrations of bovine serum albumin as a standard curve.
Western blot assay was performed by separating protein using SDS-PAGE, followed by transfer to a nitrocellulose membrane (Life Technologies; IB3010-02) using an iBlot gel transfer system (Life Technologies). The membrane was processed under similar conditions to the dot-blot assay papers using 5% skim milk in PBS for membrane blocking, 0·1 μg ml−1 anti-histidine monoclonal antibody conjugated to horse radish peroxidase in 5% skim milk for 6 × histidine detection, and the Immun-Star Western C Chemiluminescence Kit for development. The membrane was imaged using a VersaDoc Model 4000 imaging system (Bio-Rad).
Protein sequencing
Purified Mitrecin A was separated by SDS-PAGE using Mini-PROTEAN TGX Any kD polyacrylamide gels (Bio-Rad) and was transferred to a polyvinylidene difluoride (PVDF) membrane (Life Technologies; IB4010-02) using an iBlot gel transfer system. Protein transferred to the membrane was stained with BioSafe Coomassie blue (Bio-Rad), and the bands corresponding to Mitrecin A protein were excised. The membrane-bound protein was washed six times with Type-I water, and ten residues of the N-terminus were sequenced at the Iowa State University Protein Facility by Edman degradation using a 494 Procise protein sequencer and a 140C Analyzer (Life Technologies).
Functional enzyme detection assays
The presence of the Mitrecin A activity was rapidly measured using modified versions of the quantitative dye-release assay described by Zhou et al. (1988), and the spectrum of Mitrecin A antibacterial activity against specific bacteria was visualized using the microslide agarose diffusion assay described by Lachica et al. (1971). In the dye-release assay, Y. pseudotuberculosis culture, grown to mid-exponential phase in broth, was heat-killed and then washed 3 × with UltraPure distilled water. The cell substrate was covalently labelled with Remazol brilliant blue R dye (RBB; Sigma-Aldrich, St. Louis, MO) in a reaction solution containing 20 mmol l−1 RBB and 250 mmol l−1 NaOH. The substrate labelling reaction solution was incubated with gentle mixing for 6 h at 37°C and then for 12 h at 4°C. Dye-labelled cell substrates were harvested by centrifugation at 5000 g for 20 min, and the unbound dye was decanted from the pellet. The substrate was repeatedly washed with UltraPure distilled water by suspension followed by centrifugation until all free dye was washed from the substrate.
Prior to use in the quantitative dye-release assay, the substrate was washed twice with assay buffer according to the respective assay. The dye-labelled cell suspension was standardized to an optical density (OD595) of 2·0. In the dye-release reaction, 1 μg of Mitrecin A was added to 200 μl of the standardized dye-labelled cell suspension. The mixtures were incubated at 37°C for 24 h with continual oscillation. The reactions were arrested by the addition of 25 μl of ethanol, and undigested substrate was removed by centrifugation at 5000 g for 5 min. A volume of 150 μl of supernatant for each reaction mixture was transferred to a Costar 3595 96-well flat-bottom microplate (Corning, Inc., Corning, NY) to quantify absorbance at 595 nm using a Synergy HT microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT). Increased absorbance by the free dye released from the labelled substrate into the reaction supernatant was directly related to Mitrecin A activity. The standardized dye-labelled cell suspension without Mitrecin A addition served as a negative control.
In the microslide agarose diffusion assay, microlitre quantities of bacterial cell substrate (heat-killed or viable) were added to 3 ml molten 0·6% agarose in PBS (45°C). The volume of each respective cell concentrate added was varied so that a 1 : 10 dilution of the final agarose suspension in PBS yielded an OD600 of 1·0. The agarose with substrate was dispensed to a glass microslide and allowed to solidify. Wells (4 mm diameter) were cut into the agarose. Mitrecin A (1 μg) in PBS at a final volume of 10 μl was placed into the wells. Lysozyme (1 μg) in PBS and bovine serum albumin (1 μg) in PBS were used as positive and negative controls, respectively. Slides were incubated for 24 h at 37°C in humidity chambers. After incubation, development of a zone of substrate hydrolysis radiating from the well indicated the presence of bacteriolytic activity.
Effects of temperature, salinity and pH on Mitrecin A antimicrobial activity
The thermal stability of purified histidine-tagged Mitrecin A was characterized by challenges over a range of temperatures (4, 26, 37, 55, 70 and 95°C) for 30 min. Residual activity of Mitrecin A (1 μg), after each respective temperature exposure period, was quantified using the dye-release assay in incubation buffer (25 mmol l−1 Tris-HCl, 10 mmol l−1 NaCl, pH 7·4). Halotolerance of Mitrecin A (1 μg) was assessed with the quantitative dye-release assay in the presence of varying NaCl concentrations (0–7·5%) in incubation buffer modified to reflect the respective saline concentrations. The stability of Mitrecin A (1 μg) against various pH values (pH 4, 5, 6, 7, 8, 9, 10 and 11) was measured as a change in activity within the quantitative dye-release assay. Appropriate pH was maintained using the following buffers in place of the incubation buffer: 100 mmol l−1 citrate buffer, pH 4·0 (Teknova); 100 mmol l−1 acetate buffer, pH 5·0 (Electron Microscopy Sciences, Hatfield, PA); 100 mmol l−1 citrate buffer, pH 6·0 (Teknova); 100 mmol l−1 sodium phosphate buffer, pH 7·0 (Teknova); 100 mmol l−1 sodium phosphate buffer, pH 8·0 (Teknova); and 100 mmol l−1 sodium phosphate dibasic buffer, pH 9·0 (Teknova), which was also adjusted to pH 10 and pH 11 using 1 mol l−1 NaOH.
Effect of Mitrecin A on the viability of Yersinia pseudotuberculosis
The residual viability of Yersinia pseudotuberculosis was assessed after incubation in the presence of increasing concentrations of Mitrecin A. Aliquots of 200 μl of the cell suspension were distributed into wells of a 48-well CELLSTAR cell culture plate (Greiner Bio-One GmbH, Germany). Mitrecin A was added to the wells at concentrations of 0, 0·015, 0·15, 0·375, 0·75, 1·5 mg ml−1. The reaction mixtures were sealed with polyethylene Titer-Tops film (Diversified Biotech, Dedham, MA) to prevent evaporation and incubated at 37°C with shaking for 16 h. After incubation, the residual viability of the Y. pseudotuberculosis cultures for each enzyme exposure was determined by cultivating on nutrient agar plates in triplicate.
Acknowledgments
This work was funded by The MITRE Corporation through the MITRE Innovation Program (Project 51MSR601-CA). The authors would like to thank Stephen Huffman, Ph.D., Alan Moore, Dolores Derrington, Carl Picconatto, Ph.D., Thomas McEntee, Ph.D. and Richard Doyle, Ph.D. for their support and review of this work. Streptomyces sp. strain 212 culture was kindly provided by Julie B. Olson, Ph.D. of the University of Alabama.
Conflict of interest
No conflict of interest is declared.
References
- Arthur M, Molinas C, Courvalin P. Sequence of the vanY gene required for production of a vancomycin-inducible D, D-carboxypeptidase in Enterococcus faecium BM4147. Gene. 1992;120:111–114. doi: 10.1016/0378-1119(92)90017-j. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Odintsov SG, Marcyjaniak M, Sabala I. Similar active sites in lysostaphins and D-Ala-D-Ala metallopeptidases. Protein Sci. 2004;13:854–861. doi: 10.1110/ps.03515704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briers Y, Schmelcher M, Loessner MJ, Hendrix J, Engelborghs Y, Volckaert G, Lavigne R. The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem Biophys Res Commun. 2009;383:187–191. doi: 10.1016/j.bbrc.2009.03.161. [DOI] [PubMed] [Google Scholar]
- Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster MR, Haug MC, Huwiler SG, Loessner MJ. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol Microbiol. 2011;81:1419–1432. doi: 10.1111/j.1365-2958.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- Farris MH, Duffy C, Findlay RH, Olson JB. Streptomyces scopuliridis sp. nov., a bacteriocin-producing soil streptomycete. Int J Syst Evol Microbiol. 2011;61:2112–2116. doi: 10.1099/ijs.0.023192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, Proenca D, Cantante C, Silva FA, Leandro C, Lourenco S, Milheirico C, de Lencastre H, et al. Novel chimerical endolysins with broad antimicrobial activity against methicillin-resistant Staphylococcus aureus. Microb Drug Resist. 2012;18:333–343. doi: 10.1089/mdr.2012.0025. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E, Garcia JL, Garcia P, Arraras A, Sanchez-Puelles JM, Lopez R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci USA. 1988;85:914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Martinez B, Rodriguez L, Rodriguez A. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int J Food Microbiol. 2010;141:151–155. doi: 10.1016/j.ijfoodmicro.2010.04.029. [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, et al. Isolation of antibiotics turbomycin a and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol. 2002;68:4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Huang E, Yuan C, Zhang L, Yousef AE. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl Environ Microbiol. 2012;78:3156–3165. doi: 10.1128/AEM.07782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Prasad Y. P-27/HP endolysin as antibacterial agent for antibiotic resistant Staphylococcus aureus of human infections. Curr Microbiol. 2011;63:39–45. doi: 10.1007/s00284-011-9939-8. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit–Biological Sequence Alignment Editor. Carlsbad, CA: Ibis Biosciences; 2011. [Google Scholar]
- Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Hermoso JA, Garcia JL, Garcia P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Higgins ML, Lechevalier MP. Poorly lytic bacteriophage from Dactylosporangium thailandensis (Actinomycetales) J Virol. 1969;3:210–216. doi: 10.1128/jvi.3.2.210-216.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger RD. Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult Sci. 2003;82:640–647. doi: 10.1093/ps/82.4.640. [DOI] [PubMed] [Google Scholar]
- Khan H, Flint S, Yu PL. Enterocins in food preservation. Int J Food Microbiol. 2010;141:1–10. doi: 10.1016/j.ijfoodmicro.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Growth and preservation of Streptomyces. In: Streptomyces P, editor. Genetics. Norwich, England: The John Innes Foundation; 2000. pp. 46–47. [Google Scholar]
- Kretzer JW, Lehmann R, Schmelcher M, Banz M, Kim KP, Korn C, Loessner MJ. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl Environ Microbiol. 2007;73:1992–2000. doi: 10.1128/AEM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachica RV, Genigeorgis C, Hoeprich PD. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl Microbiol. 1971;21:585–587. doi: 10.1128/am.21.4.585-587.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA Sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York, NY: John Wiley & Sons Ltd; 1991. pp. 115–175. [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, O’Brien E, Barrett AJ. MEROPS: the protease database. Nucleic Acids Res. 2002;30:343–346. doi: 10.1093/nar/30.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rubio L, Gutierrez D, Martinez B, Rodriguez A, Garcia P. Lytic activity of LysH5 endolysin secreted by Lactococcus lactis using the secretion signal sequence of bacteriocin Lcn972. Appl Environ Microbiol. 2012;78:3469–3472. doi: 10.1128/AEM.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Springs, NY: Cold Springs Harbor Laboratory Press; 2001. [Google Scholar]
- Schmelcher M, Shabarova T, Eugster MR, Eichenseher F, Tchang VS, Banz M, Loessner MJ. Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl Environ Microbiol. 2010;76:5745–5756. doi: 10.1128/AEM.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelcher M, Powell AM, Becker SC, Camp MJ, Donovan DM. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl Environ Microbiol. 2012;78:2297–2305. doi: 10.1128/AEM.07050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settanni L, Corsetti A. Application of bacteriocins in vegetable food biopreservation. Int J Food Microbiol. 2008;121:123–138. doi: 10.1016/j.ijfoodmicro.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Singh PK, Ashish C, Sharma V, Patil PB, Korpole S. Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS One. 2012;7:e31498. doi: 10.1371/journal.pone.0031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JS, Lee SJ, Jun SY, Yoon SJ, Kang SH, Paik HR, Kang JO, Choi YJ. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl Microbiol Biotechnol. 2010a;86:1439–1449. doi: 10.1007/s00253-009-2386-9. [DOI] [PubMed] [Google Scholar]
- Son JS, Jun SY, Kim EB, Park JE, Paik HR, Yoon SJ, Kang SH, Choi YJ. Complete genome sequence of a newly isolated lytic bacteriophage, EFAP-1 of Enterococcus faecalis, and antibacterial activity of its endolysin EFAL-1. J Appl Microbiol. 2010b;108:1769–1779. doi: 10.1111/j.1365-2672.2009.04576.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*–Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari K, Gupta RK. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol. 2012;32:108–132. doi: 10.3109/07388551.2011.562482. [DOI] [PubMed] [Google Scholar]
- Tolba M, Ahmed MU, Tlili C, Eichenseher F, Loessner MJ, Zourob M. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst. 2012;137:5749–5756. doi: 10.1039/c2an35988j. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Chamorro SA, Palou L, Del Rio MA, Perez-Gago MB. Antimicrobial edible films and coatings for fresh and minimally processed fruits and vegetables: a review. Crit Rev Food Sci Nutr. 2011;51:872–900. doi: 10.1080/10408398.2010.485705. [DOI] [PubMed] [Google Scholar]
- Walmagh M, Boczkowska B, Grymonprez B, Briers Y, Drulis-Kawa Z, Lavigne R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol. 2012;97:4369–4375. doi: 10.1007/s00253-012-4294-7. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen S, Recsei P. A dye release assay for determination of lysostaphin activity. Anal Biochem. 1988;171:141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]