Summary
Candida albicans and streptococci of the mitis group colonize the oral cavities of the majority of healthy humans. While C. albicans is considered an opportunistic pathogen, streptococci of this group are broadly considered avirulent or even beneficial organisms. However, recent evidence suggests that multi-species biofilms with these organisms may play detrimental roles in host homeostasis and may promote infection. In this review we summarize the literature on molecular interactions between members of this streptococcal group and C. albicans, with emphasis on their potential role in the pathogenesis of opportunistic oral mucosal infections.
Keywords: fungal–bacterial interactions, microbial synergy, mucosal infection, polymicrobial biofilms
Introduction
The bacterial genus Streptococcus consists of gram-positive organisms that are typically classified into α-, β-, and γ-hemolytic groups, based on the type or presence of blood agar hemolysis that they trigger (incomplete, complete or absent, respectively). The streptococci most frequently isolated from the oral cavity are either α-hemolytic (also known as viridans) or non-hemolytic and, with the exception of Streptococcus mutans, are broadly considered avirulent or even beneficial organisms. For example, certain oral streptococcal species can hinder the development of a cariogenic S. mutans biofilm (Kuramitsu et al., 2007).
Recent attention has been focused on a subgroup of these streptococci, known as the mitis group (MGS), since members of this group have been implicated in driving a pathogenic multi-species biofilm phenotype when they coaggregate with other bacterial or fungal species (Ramsey et al., 2011; Whitmore & Lamont, 2011; Xu et al., 2013b). The MGS are principally represented by the species Streptococcus gordonii, Streptococcus oralis, Streptococcus mitis, Streptococcus parasanguinis and Streptococcus sanguinis and comprise approximately 5% (Moore et al., 1982) to over 60% (Syed & Loesche, 1978) of the recoverable human oral microbiota, depending on the method of analysis. MGS colonize the majority of healthy humans (Alam et al., 2000) and are also credited with initiating community assembly in oral biofilms (reviewed in Whitmore & Lamont, 2011).
The ability of any microorganism to form an adhering biofilm on oral surfaces is essential for stable colonization of the oral cavity (Jakubovics & Kolenbrander, 2010). During biofilm growth, coaggregation of MGS with other organisms frequently confers a mutual advantage in biofilm formation. For example, other colonizers such as Actinomyces spp., Capnocytophaga spp., Eikenella spp., Haemophilus spp., Prevotella spp., Propionibacterium spp. and Veillonella spp., appear to benefit from the presence of MGS (Kolenbrander et al., 2002). Inter-Kingdom coaggregation interactions between MGS members and Candida albicans have also been extensively studied in vitro (Jenkinson et al., 1990) and there is now evidence for Candida–streptococcal biofilm interactions in vivo, both in humans and animal models (Zijnge et al., 2010; Xu et al., 2013b). Like MGS, C. albicans colonizes the oral cavity of the majority of healthy individuals (Odds, 1988; Wilkieson et al., 1991), so interactions between these organisms are likely to play a role not only during commensal colonization of oral surfaces, but also during the course of opportunistic infections.
Multi-species biofilm communities may play both beneficial and detrimental roles in host homeostasis. Within these communities, microorganisms use quorum-sensing molecules and other metabolites to communicate with each other, adjust their population density, change gene expression patterns, and continuously adapt to the biofilm microenvironment, or release planktonic cells and colonize new, distant sites. During these processes, cell–cell interactions in the community result in antagonism, synergy or mutualism. These events not only affect the behavior of microorganisms but also modulate host responses and the ability to trigger infection (Wright et al., 2013). As a consequence of the shared ecological niche and high frequency of oral carriage of oral streptococci and C. albicans, it is important to understand how these organisms modulate each other's capacity to interact with the host. This review summarizes recent evidence on this topic.
Common Colonization Sites in Oral Health and Disease
The human oral cavity contains a number of different potential habitats for oral microorganisms such as the keratinized and non-keratinized oral mucosa, dorsal and lateral tongue surfaces, subgingival crevices and teeth. The commensal microbiota at these sites have been traditionally regarded as important in maintaining oral health. Oral streptococci colonize both teeth and oral mucosal surfaces in healthy individuals (Frandsen et al., 1991). Streptococcus oralis and its closely related species S. sanguinis and S. mitis comprise 60–80% of the primary colonizers of clean tooth surfaces, depending on the method of analysis used (16S rRNA sequencing vs. culture) (Gibbons, 1989; Diaz et al., 2012a). Importantly, although streptococci had been traditionally regarded as early or late (as in the case of S. gordonii) tooth colonizers, recent 454 microbiome pyrosequencing analysis has revealed that MGS (particularly S. mitis) dominate in the buccal mucosa of healthy individuals (Diaz et al., 2012a). Furthermore, when C. albicans is present, MGS colonization and biofilm efficiency on mucosal surfaces increase, as shown in three-dimensional models of the human oral and esophageal mucosae, under conditions of salivary flow (Diaz et al., 2012b). These results were confirmed in a mouse model of Candida–streptococcal oral co-inoculation, where it was shown that S. oralis colonization of the oral and gastrointestinal tract is significantly augmented by the presence of C. albicans (Xu et al., 2013b). Similarly, in the healthy vaginal mucosa, group B streptococci have a greater probability of isolation when C. albicans is present (Monif & Carson, 1998).
Although C. albicans appears to favor streptococcal colonization of mucosal surfaces, the pioneer binding of MGS to oral surfaces has been proposed to directly reduce the opportunities of pathogens such as Candida species and Staphylococcus aureus to colonize (Wade, 2013). Indeed some oral streptococci produce small molecules with antibiotic-like activity that can inhibit growth of other organisms. For example, S. mutans, S. mitis, S. oralis, and S. sanguinis produce diffusible signal factors (Streptococcus diffusible signal factors), which can repress the yeast-to-hypha transition of C. albicans that is essential for mucosal colonization and virulence (Lo et al., 1997; Vilchez et al., 2010). In other instances, streptococci can serve as probiotics, as in the case of S. salivarius K12, which has probiotic properties that protect mice from severe candidiasis (Ishijima et al., 2012).
Similar to MGS, Candida species (principally represented by C. albicans) also colonize both hard (teeth, dentures) and soft (mucosal) surfaces (Campos et al., 2008; Zijnge et al., 2010). Interestingly, C. albicans isolates from dental plaque and tracheobronchial sites from the same mechanically ventilated intensive care patients are genetically identical (Heo et al., 2011), underscoring the potential for systemic fungal spread from this oral habitat. Although coaggregation of Candida and streptococci has been observed on human dental plaque (Zijnge et al., 2010), and on denture biofilms (Campos et al., 2008) in vivo the clinical or biological significance of these interactions is unknown. Candida albicans is frequently isolated in dental caries of children with high oral Candida carriage, but again whether it plays a direct pathogenic role or merely co-exists in these lesions with cariogenic viridans streptococci is not clear (Raja et al., 2010).
On oral mucosal surfaces C. albicans can trigger an inflammatory response associated with the development of candidiasis, a common oropharyngeal infection primarily afflicting immunosuppressed individuals (reviewed in Villar & Dongari-Bagtzoglou, 2008). Although the presence of C. albicans is required for the development of this infection, it is increasingly appreciated that oral candidiasis is a mixed fungal–bacterial mucosal biofilm-induced or denture biofilm-induced disease (Dongari-Bagtzoglou et al., 2009; Nett et al., 2010; Johnson et al., 2012; Nobile et al., 2012; Xu et al., 2013b). In fact two recently reported rodent models of denture stomatitis identified endogenous bacteria forming biofilm communities with C. albicans on denture surfaces (Nett et al., 2010; Johnson et al., 2012). Candida albicans biofilm formation is controlled by a complex transcriptional network essential for C. albicans adaptation to different host habitats, resistance to immune system, survival and growth (Dwivedi et al., 2011; Nobile et al., 2012).
In an experimental oropharyngeal candidiasis mouse model C. albicans formed complex polymicrobial biofilms on the dorsal surface of the tongue with indigenous bacterial cocci (Dongari-Bagtzoglou et al., 2009). The potential role of streptococci in the development of this lesion was recently explored experimentally in a mouse model where oral co-inoculation of C. albicans with S. oralis led to increased frequency and severity of biofilm lesions (Xu et al., 2013b). In humans, S. oralis and C. albicans are frequently co-isolated from the sputum of antibiotic-treated cystic fibrosis patients (Maeda et al., 2011). Furthermore, co-isolation of C. albicans and Streptococcus pneumoniae, which has a high degree of genetic relatedness to S. oralis (Johnston et al., 2010), was implicated as the cause of pulmonary infection (Yokoyama et al., 2011). However, MGS have not yet been implicated in the pathogenesis of oropharyngeal candidiasis in humans.
Molecular Mechanisms of Streptococcal–Candida Cell Interactions
Adhesive and coaggregation interactions
Streptococci bind directly to salivary proteins that are abundant in the pellicle covering teeth and mucosal surfaces, and this binding facilitates their initial colonization under salivary flow conditions. MGS members S. sanguinis, S. gordonii and S. oralis have the ability to coaggregate with many microorganisms on salivary pellicles (Kolenbrander et al., 2002), including C. albicans (Hsu et al., 1990; Jenkinson et al., 1990). Coaggregation of Candida with oral streptococci was suggested to be important for C. albicans colonization of oral surfaces (Jenkinson et al., 1990) but more recent evidence suggests that Candida facilitates oral biofilm accretion of MGS species that lack the ability to form robust mucosal biofilms, such as S. oralis (Diaz et al., 2012b; Xu et al., 2013b).
The dual binding ability to host surfaces and to other microorganisms promotes streptococcal multi-species communities in the oral cavity. Binding to the pleiomorphic fungus C. albicans can be both streptococcal species-specific and fungal morphotype-specific. Streptococcus sanguinis and S. gordonii adhere to starved C. albicans yeast cells and S. salivarius also shows some coaggregating ability to yeast, but S. mutans and Enterococcus faecalis are deficient (Jenkinson et al., 1990). In vitro, in the absence of nutrients, S. oralis cells bind to Candida germ tubes as visualized with multi-color fluorescence microscopy (Diaz et al., 2012b) and in the presence of nutrients, both S. oralis and S. gordonii cells form clusters along hyphae within a few hours (Bamford et al., 2009; Diaz et al., 2012b).
Streptococcus–Candida cell adhesion is physically mediated by a number of well-characterized cell wall surface proteins/receptors on both organisms. Streptococci have abundant surface cell wall-anchored protein adhesins discussed in detail elsewhere (Nobbs et al., 2009). Untill now, two families of proteins on the streptococcal cell surface have been found to be involved in C. albicans binding. Both protein families contain an LPXTG motif recognized by sortase A enzymes, tethering the related proteins to the peptidoglycan layer. These proteins have multiple functions, including binding to both human proteins and microorganisms. One family is the CshA protein family first identified in S. gordonii DL1 (McNab & Jenkinson, 1992). The CshA protein comprises 2508 amino acid residues containing three domains, leader sequence, non-repetitive region (NR), and amino acid repeat block region(R), and it forms fibrillar structures on the cell surface with adhesive and hydrophobic properties (McNab et al., 1999). Streptococcus gordonii DL1 cshA mutants are deficient in binding to immobilized human fibronectin and other oral bacterial and Candida cells and both NR and R domains of CshA protein are responsible for binding to Candida cells (Holmes et al., 1996). CshA-like proteins have also been found in S. oralis and S. sanguinis (Elliott et al., 2003). However, the binding mechanism of CshA is still not clear and no ligands of CshA on the Candida cell surface have been identified to date.
The second streptococcal adhesin family implicated in binding to C. albicans consists of the SspA and SspB proteins that belong to the antigen I/II polypeptide family. SspA and SspB precursors comprise 1575 and 1499 amino acid residues respectively and contain seven structural regions: signal peptide, N-terminal region, Ala-rich repeats, divergent region, Pro-rich repeats, C-terminal region, and cell-wall anchoring sequence (Jenkinson & Demuth, 1997). Interestingly, the divergent region shows sequence similarity with streptococcal glucan-binding protein C, which is involved in dextran-dependent aggregation of streptococci (Okamoto-Shibayama et al., 2006). The S. gordonii strain DL1 has both SspA and SspB proteins and the two genes are in one operon, but S. oralis only has an sspA gene (Vickerman et al., 2007; Reichmann et al., 2011). Both SspA/B proteins participate in S. gordonii binding to fungi, whereas binding of SspB protein to C. albicans requires C. albicans cell surface expression of protein ALS3 (agglutinin-like sequence 3), because S. gordonii cells fail to attach to Δals3/Δals3 Candida cells (Silverman et al., 2010). Heterologous expression of SspB and ALS3 proteins in Lactobacillus lactis and Streptococcus cerevisiae, respectively, confers coaggregation properties between the two organisms, which supports the idea that SspB and ALS3 proteins interact directly (Silverman et al., 2010).
The cell wall of the trimorphic fungus C. albicans contains proteins and carbohydrates that differ in yeast, pseudohyphal and hyphal forms and are also tightly regulated by the type and/or duration of the environmental stimuli that drive morphogenesis (Gopal et al., 1984; Ebanks et al., 2006). Different environmental stimuli may therefore lead to hyphae expressing different surface proteins or having different amounts or types of skeletal polysaccharides, depending on the specific environmental triggers (Gow & Hube, 2012). The best studied adhesins in C. albicans, belong to one of three families, the ALS, Hwp1 (Hyphal Wall Protein 1), and Iff/Hyr (Hyphally Regulated Protein 1) families, which are reviewed in detail elsewhere (de Groot et al., 2013). Some proteins of the ALS and Hwp1 families are involved in adhesion to both host epithelial cells and bacteria. The ALS family consists of eight large, cell wall surface GPI-anchored glycoproteins that show differential expression profiles in yeast and hyphae. ALS3, ALS1 and ALS5 proteins mediate recognition of S. gordonii cells, whereas ALS3 also mediates Staphylococcus aureus adhesion to C. albicans (Klotz et al., 2007; Silverman et al., 2010). Hwp1 and Eap1, which belong to the Hwp1 family, confer on yeast the ability to bind to S. gordonii when the proteins are heterologously expressed in S. cerevisiae, and the interactions do not depend on the S. gordonii CshA protein (Nobbs et al., 2010).
In addition to protein–protein types of adhesive interactions between C. albicans and oral streptococci, carbohydrate-mediated or extracellular polysaccharide (EPS)-mediated interactions may also play important roles in multi-species community assembly. The EPS is a major component of the biofilm matrix. Both oral streptococci and C. albicans produce EPS in the biofilm state, albeit with a different composition. The EPS of viridans streptococci is synthesized by multiple glucosyl-transferases (Gtfs) that exist widely in the streptococcal genus. GtfB is secreted by S. mutans and can bind to candidal yeast cells in an enzymatically active form, depositing α-glucans on the yeast cell surface (Gregoire et al., 2011). The newly synthesized α-glucans promote S. mutans binding to yeast cells and enhance S. mutans–C. albicans coaggregation (Gregoire et al., 2011). Similarly, GtfG-derived α-glucans play a role in promoting biofilm interactions between S. gordonii and C. albicans (Ricker et al., 2014).
Other streptococcal molecules that play a role in inter-species and inter-generic aggregation interactions are the coaggregation receptor polysaccharides (RPS). These are six, structurally distinct receptors for lectin-like adhesins mediating inter-species and intra-species streptococcal aggregation, or inter-generic interactions of streptococci with Actinomyces, and Veilonellae (Yoshida et al., 2006). For example, Yoshida et al. (2006) have shown that an RPS-negative S. oralis mutant fails to form mixed biofilms with an adhesin-expressing Actinomyces. Sequence analysis of human dental plaque organisms showed that RPS-positive streptococci clustered with S. oralis and RPS-negative clustered with S. gordonii (Hoshino et al., 2005), indicating species heterogeneity in the expression of these receptors within the MGS group. There is no information on the potential role of these receptors in inter-Kingdom interactions.
The major carbohydrate component of C. albicans EPS is β-glucans (Al-Fattani & Douglas, 2006). In C. albicans biofilms β-glucans are immunodetectable not only within the matrix but also on the hyphal surface (Dongari-Bagtzoglou et al., 2009). Since streptococci form ‘Corncob–like’ structures on hyphae in mixed biofilms both in vitro and in vivo (Bamford et al., 2009; Zijnge et al., 2010; Diaz et al., 2012b), this suggests that streptococci may benefit from the β-glucan-rich surface provided by hyphae in the biofilm growth state. It is therefore conceivable that glucan binding proteins of certain streptococci have both α- and β-glucan-binding properties that mediate such interactions, since these proteins are known to have multiple functions [for example glucan-binding protein D also has lipase activity (Shah & Russell, 2004)].
Inter-Kingdom signaling
During formation of polymicrobial biofilm communities streptococcal and Candida cells communicate by using their own ‘languages’, consisting of chemical signal molecules or metabolites that alter microbial behavior. Quorum-sensing systems are well-known communication systems in bacteria and fungi. Bacteria produce and secrete chemical signal molecules termed autoinducers to modulate cell behavior within communities (Miller & Bassler, 2001). There are at least two types of autoinducers, autoinducer 1 (AI-1) mediating intra-species communication, and autoinducer 2 (AI-2) serving as universal language involved in inter-species communication (Miller & Bassler, 2001; Vendeville et al., 2005). AI-2 is a byproduct of the activated methyl cycle and the luxS gene encodes the protein precursor responsible for its production (Vendeville et al., 2005). Streptococcal species possess a homologue of the luxS gene and a S. gordonii luxS mutant is unaffected in planktonic growth or monospecies biofilm formation (McNab et al., 2003). However, luxS mutants of S. gordonii fail to form mixed biofilms with Porphyromonas gingivalis (McNab et al., 2003). Along the same lines, the biofilm mass of C. albicans mixed with a S. gordonii luxS mutant is reduced, and interestingly C. albicans produces shorter hyphae, suggesting a role of this bacterial gene product in inter-Kingdom communication (Bamford et al., 2009).
Farnesol, the first quorum-sensing molecule identified in eukaryotes, accumulates abundantly in Candida biofilms to inhibit hyphal growth (Hornby et al., 2001). Farnesol also blocks yeast to hyphae transition induced by a variety of environmental cues such as serum, N-acetylglucosamine, and glucanase (Biswas et al., 2007; Sudbery, 2011; Xu et al., 2013a). As a consequence, farnesol downregulates a number of hyphal genes, including ALS3 and heatshock protein 90 (Uppuluri et al., 2007). AI-2 of S. gordonii relieves the repression of Candida hyphae and biofilm formation triggered by farnesol, further supporting the idea that C. albicans responds to this signal (Bamford et al., 2009). Hence by relieving farnesol's hyphal repression, AI-2 may indirectly increase hyphal virulence gene expression and increase fungal pathogenicity. On the other hand, farnesol can decrease the accumulation of EPS in S. mutans biofilms, without affecting bacterial cell viability, so potentially reducing the cariogenic potential of this organism (Koo et al., 2003).
Fatty acid diffusible signal factors (DSF) were recently identified as interspecies small signaling molecules that are structurally homologous to cis-11-methyl-2 dodecanoic acid (cis-DA). DSF were first identified in Xanthomonas campestris and were implicated in biofilm dispersal and full virulence of this organism (Barber et al., 1997; Dow et al., 2003). The synthesis of DSF is dependent on the RpfF protein while a two-component system (sensor kinase) RpfC/(regulator) RpfG is implicated in DSF perception. DSF of Stenotrophomonas maltophilia can be detected by Pseudomonas aeruginosa PA sensor kinase and influences dual-species biofilm structure, supporting a role in communication between the two species (Ryan et al., 2008). Similarly, these molecules play a role in inter-Kingdom communication since DSF purified from S. mutans cell culture supernatants inhibit yeast-to-hyphal transition of C. albicans and suppress HWP1 gene expression. Streptococcus mitis, S. oralis and S. sanguinis also produce DSF-type activity (Vilchez et al., 2010).
Streptococcus mutans produces another quorum-sensing molecule known as competence-stimulating peptide (CSP). CSP is encoded by the comC gene, which is under the regulation of ComD and a two-component regulatory system (Pestova et al., 1996). CSP not only inhibits Candida germ tube formation but also stimulates the hyphae-to-yeast transition (Jarosz et al., 2009). In conclusion, the net result of signaling between Candida and streptococci in mixed biofilms is not always the same but bound to be dictated by environmental conditions, population densities and the streptococcal species participating in these communities.
Metabolic interactions
Oral streptococci produce lactic acid as an end product of their carbohydrate fermentation process, which can contribute to a rapid decline in the environmental pH. Extracellular pH can modulate Candida hyphal growth, and in turn, biofilm formation. Candida albicans grows in the yeast form at acidic pH and in the hyphal form at alkaline pH (Calderone, 2002). PHR1 and PHR2 (pH-Response) are two pH-regulated genes in C. albicans that encode homologues of S. cerevisiae GAS1, predicted to be anchored to the plasma membrane (Fonzi, 1999). PHR1 expression is repressed at pH value <5.5 whereas PHR2 is repressed at pH > 6. Both PHR1 and PHR2 mutants exhibit a pH-conditional morphological defect (Saporito-Irwin et al., 1995; Muhlschlegel & Fonzi, 1997). Hence, in theory, by decreasing the environmental pH as a result of the fermentative process, streptococci may repress PHR1 gene expression and increase PHR2 expression leading to inhibition of the yeast-to-hyphae transition. However, under glucose-limiting conditions, C. albicans has the ability to neutralize the environmental pH and proceed with hyphal morphogenesis by excreting ammonia into the environment (Vylkova et al., 2011).
Viridans group streptococci generate hydrogen peroxide under standard laboratory growth conditions. In fact S. gordonii spent culture medium contains up to 0.15 mm H2O2. Genotoxic and oxidative stress mediated by streptococcal H2O2 can stimulate filamentous growth in C. albicans (Barnard & Stinson, 1996; Nasution et al., 2008). The H2O2 released by MGS may also increase catalase gene expression in C. albicans, which in turn may protect the organism from neutrophil oxidative killing, in a model similar to that proposed to exist in the pathogenic synergy between S. gordonii and Aggregatibacter actinomycetemcomitans (Ramsey et al., 2011).
As in any well-regulated community system with checks and balances, metabolic signals in polymicrobial communities can promote or repress activities of its member species, control population growth, and manipulate the community hierarchical structure with the establishment of ‘leader’ and ‘follower’ or ‘accessory’ species. The end result of these metabolic interactions for the microorganism community is a homeostatic state that extends the life of the community as a whole. For the host the end point is a beneficial, symbiotic or pathogenic relationship with the microbial community. The remainder of this review will concentrate on Candida–streptococcal interactions contributing to a pathogenic relationship with the host.
Virulence Models of Candida and Streptococcal Polymicrobial Infection
There is an abundance of in vivo models to study virulence and pathogenic mechanisms of C. albicans mono-infection. These include insects, fruit flies, nematodes, zebrafish, mice and rats (Cotter et al., 2000; Alarco et al., 2004; Hamamoto et al., 2004; Hanaoka et al., 2008; Meeker & Trede, 2008; Pukkila-Worley et al., 2009; Johnson et al., 2012; Solis & Filler, 2012). Larger mammals such as macaques, piglets, rabbits and guinea pigs have also been used (Fransen et al., 1984; Filler et al., 1991; Steele et al., 1999; Andrutis et al., 2000). Mice have been useful in the development of mucosal, intravenous and gastrointestinal dissemination models (MacCallum & Odds, 2005; Clemons et al., 2006; Koh et al., 2008; Dongari-Bagtzoglou et al., 2009) (also see review by MacCallum, 2012). Similarly, the virulence and pathogenetic mechanisms of an α-hemolytic Streptococcus, phylogenetically very closely related to some MGS (S. pneumoniae) have been studied in mouse, rat and rabbit models (Chiavolini et al., 2008). Mixed infections with this pathogen have also been modeled in vivo. For example, mouse models of pneumonia and otitis media have been used to study the synergistic effect of S. pneumoniae with non-typeable Haemophilus influenzae (Ratner et al., 2005; Ramsey & Whiteley, 2009). Rodent models have also been used to study the cariogenicity of S. mutans (Bowen et al., 1988, 1991), or the oral colonization capabilities of S. gordonii (Tanzer et al., 2001). However, with the exception of S. mutans, no other oral streptococcal species has shown virulence properties in a rodent model of single oral infection, even when animals were immunocompromised and inoculated with a high number of organisms (Xu et al., 2013b).
With the possible exception of virally triggered diseases, most oral infectious diseases are polymicrobial in nature or require ‘cooperation’ by multiple microbes, because they develop in oral habitats that harbor a vast number of different bacterial and fungal species. Despite this central axiom, animal models of oral polymicrobial diseases are scarce, and models of mixed infection have only recently begun to emerge. Because of the lack of significant virulence properties when inoculated on their own, the role of MGS as ‘accessories’ to established pathogens was recently tested in two mixed infection models. Ramsey and colleagues used a mouse abscess model to study co-infection with A. actinomycetemcomitans and S. gordonii (Ramsey et al., 2011). An oral co-infection mouse model has also been used to study alveolar bone loss caused by P. gingivalis in the presence of S. gordonii (Daep et al., 2011). In both studies this MGS species was implicated in enhanced pathogenicity of the primary oral pathogens (A. actinomycetemcomitans and P. gingivalis) and so the term ‘accessory pathogen’ was introduced to describe this organism (Whitmore & Lamont, 2011).
The first mixed Candida–bacterial (Escherichia coli) co-infection model was described in the 1950s (Gale & Sandoval, 1957) and has been followed by very few studies ever since. Recently, Peters & Noverr (2013) developed a murine model of peritonitis by co-infecting mice with C. albicans and Staphylococcus aureus. Using this model, the investigators found that co-infection of mice can lead to 40% mortality while single infection with either organism is non-lethal (Peters & Noverr, 2013). A murine oral Candida–streptococcal co-infection model was recently described by Dongari-Bagtzoglou and colleagues. In this model, it was shown that the MGS species S. oralis can significantly enhance C. albicans mucosal pathogenicity in mice immunocompromised with cortisone (Xu et al., 2013b). This resembles the significant enhancement in P. aeruginosa pathogenicity in a rodent lung co-infection model where MGS on their own show no virulence attributes (Duan et al., 2003). Finally, in a fruit fly model of polymicrobial gastrointestinal infection oral streptococci, including S. oralis and its close phylogenetic relative S. mitis, were grouped into three distinct pathogenicity groups: virulent, avirulent and synergistic, i.e. streptococci that alone are not pathogenic but in combination with a pathogen can significantly enhance pathogenicity (Sibley et al., 2008b). Collectively most animal models of mixed infection have so far consistently demonstrated that, although MGS lack significant virulence properties on their own, they can promote the virulence of established bacterial and fungal oral opportunistic pathogens.
Mechanisms of Synergistic Virulence in Candida and Streptococcal Polymicrobial Infections
Microorganisms can interact to increase each other's pathogenicity in four principal ways: (i) modulation of host responses; (ii) increased virulence; (iii) environmental alterations; and (iv) metabolic interactions. Below, we present a comprehensive discussion of evidence of each of these mechanisms as they pertain to C. albicans-streptococcal interactions.
Mixed biofilm growth and virulence gene expression
The mechanisms of synergistic virulence of Strepto-coccus and Candida are still not fully understood. One early mechanism of synergy may be at the level of colonization, whereby one species helps others to colonize certain oral surfaces more efficiently. Although most MGS streptococci preferentially colonize tooth surfaces, when a perturbation of the oral microbial flora is present they may colonize mucosal sites in higher numbers. For example, in lactobacilli-free and streptococci-free mice, when orally inoculated, S. gordonii colonizes the palatal and tongue surfaces in higher numbers compared with in mice with intact oral microbiota (Loach et al., 1994). Similarly the introduction of C. albicans to the oral cavity of mice facilitates colonization of mucosal surfaces by S. oralis (Xu et al., 2013b). Hence, C. albicans creates favorable conditions for mucosal colonization and biofilm growth of these bacteria, in line with recent evidence showing that introduction of C. albicans in the gastrointestinal tract of antibiotics-treated mice leads to a preferential re-growth of enterococci (Mason et al., 2012). Because increased S. oralis mucosal colonization in the presence of C. albicans led to an increased size and frequency of oral lesions (Xu et al., 2013b), it is conceivable that like many opportunistic pathogens, a critical mass of this species, reached in the presence of fungal organisms, is needed to induce pathology. These results dispute the long-held belief that these members of the commensal bacterial microbiota protect the host against mucosal candidiasis (Liljemark & Gibbons, 1973). Similarly, C. albicans promoted Staphylococcus aureus burdens and pathology in a peritoneal co-infection model (Peters & Noverr, 2013).
A second mechanism of synergy at the biofilm accretion level is when one species modulates the structure of a polymicrobial biofilm to form certain architectural patterns, which may promote biofilm pathogenicity. One such example is the filamentous architecture of P. aeruginosa within biofilms mixed with S. maltophilia, which is not found in P. aeruginosa mono-species biofilms. This structural change was linked to an increased tolerance of P. aeruginosa to polymyxins (Ryan et al., 2008). In C. albicans–S. oralis mixed biofilms, C. albicans hyphae obtain a more uniform vertical orientation against the biofilm substratum surface, both in vitro and in vivo (Fig. 1). Based on the well-accepted role of hyphal organisms in mucosal invasion (Gow & Hube, 2012), it is tempting to speculate that such an orientation against the oral mucosal surface may be more conducive to tissue pathology and systemic dissemination, as seen in this co-infection model (Xu et al., 2013b).
Figure 1.
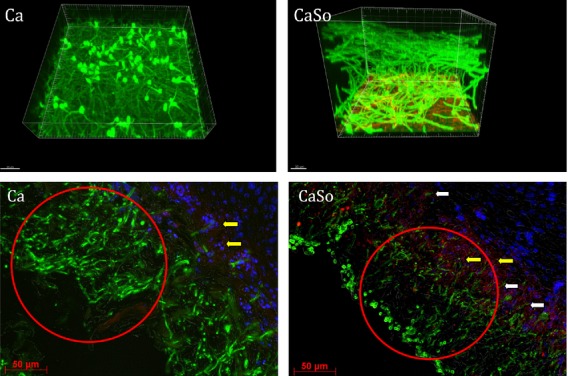
Streptococcus oralis modifies Candida albicans biofilm structure. Upper panel: C. albicans SC5314 biofilms were grown with (CaSo) or without (Ca) S. oralis 34 on a glass surface under static conditions at 37°C. After 20 h, C. albicans and S. oralis 34 were labeled with a fluorescein isothiocyanate-conjugated anti-C. albicans polyclonal antibody (shown in green) and a S. oralis-specific fluorescence in situ hybridization probe labeled with Alexa 546 (shown in red), then observed by confocal scanning laser microscopy. Note the orientation of the hyphae toward the substratum of the dual biofilm. Lower panel: Overlay images of mouse tissue sections showing C. albicans biofilms (green) growing on the oral mucosal surface of animals infected with C. albicans SC5314 alone (Ca), or C. albicans SC5314 plus S. oralis So34 (CaSo). Stained in red are neutrophils infiltrating the oral mucosa. Mucosal cells are counterstained with the nucleic acid stain Hoechst 33258 (blue). Sections were stained as described elsewhere (Xu et al., 2013b). Note the difference in hyphal orientation toward the mucosal surface in single (Ca) and mixed (CaSo) infection, associated with more localized mucosal invasion (white arrows), as well as more pronounced neutrophil infiltration (yellow arrows) in the latter.
Finally, in polymicrobial biofilms gene expression patterns of organisms can be mutually modulated, which can have an important impact on the virulence of opportunistic pathogens. For example, proteomic analysis of in vitro Candida–Staphylococcus co-culture biofilms showed that 27 proteins of C. albicans and Staphylococcus aureus are significantly upregulated (Peters et al., 2010). Similarly, genome-wide transcriptional analysis shows that about 4% of genes in the P. aeruginosa genome are modulated by the presence of MGS strains isolated from cystic fibrosis patients, and that a large portion of upregulated genes are involved in P. aeruginosa pathogenesis (Duan et al., 2003). Using Drosophila as a surrogate host for polymicrobial infection it was shown that nearly half of the MGS strains isolated from human sputum have pathogenic synergy with P. aeruginosa, by altering Pseudomonas gene expression without affecting its growth (Sibley et al., 2008a). Since MGS species have been shown to promote Candida yeast to hyphae transition under certain in vitro conditions (Bamford et al., 2009) (Fig. 2), it is plausible that several virulence genes involved in mucosal invasion are upregulated during these interactions.
Figure 2.
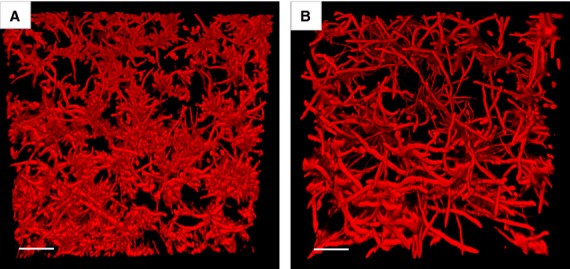
Streptococcus gordonii stimulates yeast to hypha transition in Candida albicans. The C. albicans biofilms were grown with or without S. gordonii DL1 expressing green fluorescent protein under flow conditions at 37°C. After 6 h the biofilms were visualized by confocal scanning laser microscopy. The growth medium contained Calcofluor white (1 μg ml−1) to fluorescently label C. albicans (shown in red). (A) An underneath image of a C. albicans biofilm grown without S. gordonii, and (B) an underneath image of a dual species biofilm of C. albicans with S. gordonii. In panel B the S. gordonii component has been subtracted using Volocity computer software. (Scale bars, 50 μm.)
Influence on host response through pattern recognition receptors
One principal way by which microorganisms can interact to increase the pathogenicity each of the other is by modulation of host responses. Even with the introduction of more pathogenic species, multiple bacterial species from the commensal microbiota can contribute to mucosal inflammation (Jergens et al., 2007; Hajishengallis et al., 2011) and there are several examples of co-infections where infection with one organism influences the innate host response to the other (Jamieson et al., 2010; Vylkova et al., 2011). One possible molecular mechanism of such interactions is a unilateral or reciprocal upregulation of pattern recognition receptors in the oral mucosa, which drive inflammatory responses to otherwise commensal or opportunistic organisms. It has been postulated that commensal oral microorganisms alone do not trigger host inflammatory responses because a critical density of such receptors is not present on oral epithelia to transmit inflammatory signals. This is supported by the fact that, even when given in high infectious doses, S. oralis did not induce an inflammatory response in the oral mucosa (Xu et al., 2013b).
When S. oralis is orally co-inoculated with C. albicans an exaggerated mucosal inflammatory response ensues (Xu et al., 2013b). The majority of the immune regulatory genes upregulated in the co-infected animals involve genes in the general categories of chemotaxis response, neutrophilic response, cytokine activity and phagocytosis. Interestingly, strong induction of multiple neutrophil-activating cytokines (interleukin-17C, Chemokine (C-X-C motif) ligand 1/CXCL1, macrophage inflammatory protein-2/MIP-2, tumor necrosis factor, interleukin-1α, interleukin-1β) with concomitant increased neutrophilic infiltration was observed in this model (Fig. 1). Synergistic virulence mediated by an exaggerated host inflammatory response was also observed in mice co-infected peritoneally with Staphylococcus aureus and C. albicans, with increased expression levels of proinflammatory cytokines that augment a neutrophilic response, including interleukin-6, granulocyte colony-stimulating factor, keratinocyte-derived chemokine, monocyte chemoattractant protein-1, and macrophage inflammatory protein-1. In addition, the cyclooxygenase pathway was implicated in synergistic virulence in this model (Peters & Noverr, 2013).
Gram-positive bacteria-triggered epithelial inflammation is principally regulated by Toll-like receptor 2 (TLR2) (Aderem & Ulevitch, 2000), however, TLR2 plays a secondary role in bacterial clearance, at least in chronic models of infection (Knapp et al., 2004; Burns et al., 2006). In an oral infection model, it was shown that the TLR2 oral mucosal expression levels increased in mice infected with C. albicans and that S. oralis can signal directly via this receptor to activate a neutrophilic response (Xu et al., 2013b). Oral mucosal epithelial cells normally express low levels of TLR2, but can be stimulated to express higher levels by C. albicans (Zhang et al., 2004). Hence, a model of synergy was suggested whereby infection with C. albicans increases both the biomass of S. oralis and the TLR2 expression to critical levels required for mucosal proinflammatory signaling by this otherwise commensal organism. This was supported by the fact that in TLR2−/− co-infected mouse proinflammatory gene expression and polymorphonuclear cell infiltration were partly attenuated (Xu et al., 2013b). TLR2-dependent epithelial innate immune response genes also augmented the neutrophilic influx in an alveolar mucosa co-infection model with oral streptococci (Sibley et al., 2008a).
Similarly, respiratory co-infection with H. influenzae and S. pneumoniae in mice leads to increased production of nuclear factor-κB in mouse lung tissue and this synergistic effect is dependent on TLR2 because nuclear factor-κB is not increased in TLR2−/− mice (Lim et al., 2008). The TLR2 expression in the alveolar mucosa is upregulated by S. pneumoniae, but can be amplified further by co-infection with S. pneumoniae with H. influenzae, as seen in the oral co-infection model with C. albicans and S. oralis (Xu et al., 2013b). Interestingly, TLR2 induction by S. pneumoniae and/or H. influenzae in murine lung tissues is dependent on the TLR4 receptor (Lim et al., 2008).
Not all oral streptococcal species are involved in increased virulence of primary pathogens or in exaggerated epithelial inflammatory responses. In fact, S. salivarius strain K12 antagonizes P. aeruginosa-induced interleukin-8 secretion from human bronchial epithelial cells, suggesting a role for commensal streptococci in down-regulating epithelial cell inflammatory responses in the nasopharynx (Cosseau et al., 2008). Hence, there is a great variability in the involvement of oral streptococcal species in oral health and disease, with some species having probiotic properties and others acting as accessories in infectious disease pathogenesis.
Influence on epithelial barrier integrity and invasion
Invasion of epithelial surfaces allows organisms to reach the bloodstream, with potential subsequent escape into deep organs. Gram-positive commensals exert the opposite effects on mucosal epithelial barriers depending on their population size, ranging from promotion of homeostasis in low numbers (Rakoff-Nahoum et al., 2004), to deleterious effects in high numbers (Clarke et al., 2011). Although the resident bacterial biota can play an important protective role against the invasion of pathogens in the lower gastrointestinal tract (Falk et al., 1998), certain members of the MGS promote C. albicans breaching of the mucosal barrier in organotypic constructs in vitro (Diaz et al., 2012b) and facilitate systemic dissemination of fungal cells in vivo (Xu et al., 2013b).
E-cadherin plays a crucial role in epithelial cell tight junctions and in the maintenance of the stratified oral mucosal tissue architecture. Although barrier function regulation in oral epithelium is not well defined, E-cadherin protein integrity is important in different oral models of infection (Katz et al., 2002; Dongari-Bagtzoglou et al., 2009). In addition to causing extensive epithelial cell damage via cytoplasmic invasion (Zhu & Filler, 2010), C. albicans uses a paracellular pathway of mucosal invasion via protease-mediated E-cadherin degradation (Villar et al., 2007), in agreement with the reduced E-cadherin protein levels in human oral candidiasis biopsies (Leigh et al., 2004). During the invasion process, hyphae bind to E–cadherin on epithelial cells via the hyphal-specific protein ALS3. Subsequently, hyphae secrete aspartic proteinases that, apart from hydrolyzing tight junction proteins, bind to integrins on the epithelial surface and induce apoptosis, further expediting the breach of the mucosal barrier (Villar et al., 2007; Parnanen et al., 2010; Liu & Filler, 2011; Wu et al., 2013). This process may be further enhanced or expedited by the synergistic effect of MGS members that can promote yeast to hyphal transition (Bamford et al., 2009).
Epithelial cells are the direct targets of bacterial cytolysins, including Pneumolysin (Ply). Ply is a major toxin of S. pneumoniae that belongs to cholesterol-dependent cytolysins that attack cholesterol-rich membranes to form large oligomeric transmembrane pores on cell membranes and kill target cells (Morgan et al., 1994). Increased colonization of the mouse upper respiratory tract of S. pneumoniae and H. influenzae is dependent on Ply (Ratner et al., 2005). It has been suggested that peptidoglycan of H. influenzae, a key mediator of inflammation in this model, can access the epithelial cytoplasm via Ply-induced pores to trigger cytoplasmic inflammatory signaling (Ratner et al., 2005). Sequence analysis suggests that the S. mitis genome contains putative virulence factor genes encoding pneumolysin and autolysin that are also found in virulent S. pneumoniae strains (Whatmore et al., 2000). Also, an S. oralis clinical isolate from endocarditis produces sialidase (Byers et al., 2000), an enzyme that is thought to facilitate breach of mucosal barriers and provide nutritional support to this organism. Hence it is not entirely surprising that these streptococci can disseminate systemically and cause sepsis, especially when the mucosa is injured or disrupted by cytotoxic chemotherapy (Khan & Wingard, 2001). Surprisingly, although the biomass of S. oralis 34 on the oral mucosa increases in the presence of C. albicans, bacterial breach of the mucosa and systemic dissemination is not promoted (Xu et al., 2013b). This is probably attributed to the large strain to strain and species to species genetic variability in MGS.
Lethal co-infection of mice with the pandemic H1N1 virus (Mex09) and S. pneumoniae is associated with severe lung epithelial cell damage. Co-infection with Mex09 and S. pneumoniae decreases the expression of a number of genes associated with tissue repair or remodeling and cell differentiation, including KGF and Sftpa1 (Kash et al., 2011). Along similar lines, in S. oralis 34 and C. albicans co-infected mice more severe oral lesions were accompanied by downregulation of a large number of epithelial cell structure genes, including several cytokeratins, (Xu et al., 2013b). Hence streptococcal synergy in tissue pathology and invasion may also be mediated by a compromise in host structural repair pathways.
Conclusions and Future Directions
Studies of synergistic or antagonistic roles of the oral microbiota in oral health and disease are still in their infancy, despite decades of pioneering, productive research by oral microbiologists. To begin with, animal or complex organotypic in vitro models of oral polymicrobial infections are scarce and new ones are urgently needed. One intriguing aspect of microbial commensalism is defining the conditions under which a commensal relationship with the host may be transformed to pathogenic. Dysbiotic changes in the commensal microbiota quantity or composition, accompanied by a distorted mucosal immunological or inflammatory response, appear to play central roles in the development of several polymicrobial infections in the upper and lower gastrointestinal tract. It is possible that a two-step process is needed by ‘ordinary commensals’ to trigger disease either as accessories to primary pathogens, or as primary opportunistic pathogens themselves: (i) increase in colonization efficiency at oral sites not abundantly colonized in health; and (ii) local mucosal alterations that favor initiation of inflammatory signaling cascades that are not conducive to microbial clearance.
Due to their abundance and plethora of oral habitats, oral streptococci of the MGS are bound to play important roles in both health and disease (Table1). We propose that the interactions of C. albicans with members of this group serve as a prototype of the interactions between opportunistic pathogens with commensals and their influence on oral mucosal homeostasis. A wide range in disease manifestations of oral candidiasis (pseudomembranous, erythematous, hyperplastic) has not been adequately explained by differences in Candida virulence or host factors. We therefore further propose that differences in clinical manifestations of this infection may be explained by interactions of this organism with the commensal streptococcal biota that occupies each site. Streptococcus pneumoniae, a non-oral member of the MGS, shares an extremely high degree of genetic homology with both S. mitis and S. oralis (Kawamura et al., 1995). Despite the close genetic relationships between these species, while S. pneumoniae can be a severe respiratory pathogen, S. oralis and other MGS members have only been implicated in opportunistic bloodstream infections. This major phenotypic difference is intriguing because a whole genome microarray analysis showed that S. oralis hybridized to 83% of pneumococcal virulence genes (Johnston et al., 2010). Based on existing evidence we propose that C. albicans can trigger a dysbiotic change in S. oralis by promoting colonization and growth at mucosal sites that would normally not be a primary habitat for this organism in health. Whether this dysbiotic change is merely a result of favorable coaggregation interactions between these microbes or is secondary to epithelial changes induced by the primary pathogen, C. albicans, is still incompletely understood. Inter-Kingdom signaling, metabolic interactions and local environmental changes are all likely to play a role in the development of this polymicrobial mucosal biofilm infection. Modulation of host responses by oral streptococci and increased Candida virulence may lead to a pathogenic synergy of these microorganisms in the oral mucosa. As the infection progresses there is a possible fluctuation of the role and contribution of each organism in the infectious process, with one or the other playing central or accessory roles in host responses or tissue pathology, at different disease stages. More investigations are needed into the inter-Kingdom communication systems that have a bearing on virulence gene expression patterns when these organisms co-inhabit new sites. In addition to partaking in dysbiosis and inflammatory changes, MGS and C. albicans may regulate pattern recognition receptor expression to multiple other microorganisms that also occupy these sites. Future studies should focus on defining the molecular mechanisms of these complex microbial interactions with the host.
Table 1.
Summary of molecular interactions between oral streptococci and Candida albicans
| Streptococcal molecules | Functional interaction with Candida | References |
|---|---|---|
| CshA | Binds to unidentified ligand(s) on C. albicans to promote coaggregation | Holmes et al., 1996; McNab et al., 1999 |
| SspA/SspB | Bind to hyphae-specific ALS3 cell wall protein of C. albicans to promote coaggregation | Jenkinson & Demuth, 1997; Silverman et al., 2010 |
| α-glucans | Synthesized by GtfB or GtfG to promote mixed biofilm accretion with C. albicans | Gregoire et al., 2011; Ricker et al., 2014 |
| Autoinducer-2 | Relieves farnesol repression of C. albicans hyphal growth | Bamford et al., 2009 |
| Competence-stimulating peptide | Inhibits germ tube formation and stimulates hyphae-to-yeast transition | Jarosz et al., 2009 |
| Streptococcus diffusible signal factors | Inhibit yeast-to-hyphae transition | Vilchez et al., 2010 |
| Lactic acid | Decreases pH potentially affects pH- response gene expression | Calderone, 2002; Vylkova et al., 2011 |
| H2O2 | Oxidative and genotoxic stress, promotes filamentous growth | Barnard & Stinson, 1996; Nasution et al., 2008 |
Acknowledgments
The authors would like to acknowledge funding by NIH/NIDCR (Grants DE013986 to ADB and DE016690 to HFJ), as well as Takanori Sobue and Lindsay Dutton for providing some of the confocal images.
References
- Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- Alam S, Brailsford SR, Adams S, et al. Genotypic heterogeneity of Streptococcus oralis and distinct aciduric subpopulations in human dental plaque. Appl Environ Microbiol. 2000;66:3330–3336. doi: 10.1128/aem.66.8.3330-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarco AM, Marcil A, Chen J, Suter B, Thomas D, Whiteway M. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J Immunol. 2004;172:5622–5628. doi: 10.4049/jimmunol.172.9.5622. [DOI] [PubMed] [Google Scholar]
- Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- Andrutis KA, Riggle PJ, Kumamoto CA, Tzipori S. Intestinal lesions associated with disseminated candidiasis in an experimental animal model. J Clin Microbiol. 2000;38:2317–2323. doi: 10.1128/jcm.38.6.2317-2323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CE, Tang JL, Feng JX, et al. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- Barnard JP, Stinson MW. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect Immun. 1996;64:3853–3857. doi: 10.1128/iai.64.9.3853-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Pearson SK, Young DA. The effect of desalivation on coronal and root surface caries in rats. J Dent Res. 1988;67:21–23. doi: 10.1177/00220345880670010301. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Schilling K, Giertsen E, et al. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4606–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- Byers HL, Tarelli E, Homer KA, Beighton D. Isolation and characterisation of sialidase from a strain of Streptococcus oralis. J Med Microbiol. 2000;49:235–244. doi: 10.1099/0022-1317-49-3-235. [DOI] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington, D.C: ASM Press; 2002. [Google Scholar]
- Campos MS, Marchini L, Bernardes LA, Paulino LC, Nobrega FG. Biofilm microbial communities of denture stomatitis. Oral Microbiol Immunol. 2008;23:419–424. doi: 10.1111/j.1399-302X.2008.00445.x. [DOI] [PubMed] [Google Scholar]
- Chiavolini D, Pozzi G, Ricci S. Animal models of Streptococcus pneumoniae disease. Clin Microbiol Rev. 2008;21:666–685. doi: 10.1128/CMR.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TB, Francella N, Huegel A, Weiser JN. Invasive bacterial pathogens exploit TLR-mediated downregulation of tight junction components to facilitate translocation across the epithelium. Cell Host Microbe. 2011;9:404–414. doi: 10.1016/j.chom.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons KV, Gonzalez GM, Singh G, et al. Development of an orogastrointestinal mucosal model of candidiasis with dissemination to visceral organs. Antimicrob Agents Chemother. 2006;50:2650–2657. doi: 10.1128/AAC.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosseau C, Devine DA, Dullaghan E, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host–microbe homeostasis. Infect Immun. 2008;76:4163–4175. doi: 10.1128/IAI.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol. 2000;27:163–169. doi: 10.1111/j.1574-695X.2000.tb01427.x. [DOI] [PubMed] [Google Scholar]
- Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Dupuy AK, Abusleme L, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol. 2012a;27:182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012b;80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS ONE. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- Dwivedi P, Thompson A, Xie Z, et al. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS ONE. 2011;6:e16218. doi: 10.1371/journal.pone.0016218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebanks RO, Chisholm K, McKinnon S, Whiteway M, Pinto DM. Proteomic analysis of Candida albicans yeast and hyphal cell wall and associated proteins. Proteomics. 2006;6:2147–2156. doi: 10.1002/pmic.200500100. [DOI] [PubMed] [Google Scholar]
- Elliott D, Harrison E, Handley PS, et al. Prevalence of Csh-like fibrillar surface proteins among mitis group oral streptococci. Oral Microbiol Immunol. 2003;18:114–120. doi: 10.1034/j.1399-302x.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler SG, Crislip MA, Mayer CL, Edwards JE., Jr Comparison of fluconazole and amphotericin B for treatment of disseminated candidiasis and endophthalmitis in rabbits. Antimicrob Agents Chemother. 1991;35:288–292. doi: 10.1128/aac.35.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen J, Van Cutsem J, Vandesteene R, Janssen PA. Histopathology of experimental systemic candidosis in guinea-pigs. Sabouraudia. 1984;22:455–469. doi: 10.1080/00362178485380731. [DOI] [PubMed] [Google Scholar]
- Frandsen EV, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- Gale D, Sandoval B. Response of mice to the inoculations of both Candida albicans and Escherichia coli I. The enhancement phenomenon. J Bacteriol. 1957;73:616–624. doi: 10.1128/jb.73.5.616-624.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ. Bacterial adhesion to oral tissues: a model for infectious diseases. J Dent Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- Gopal PK, Shepherd MG, Sullivan PA. Analysis of wall glucans from yeast, hyphal and germ-tube forming cells of Candida albicans. J Gen Microbiol. 1984;130:3295–3301. doi: 10.1099/00221287-130-12-3295. [DOI] [PubMed] [Google Scholar]
- Gow NA, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, et al. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 2011;77:6357–6367. doi: 10.1128/AEM.05203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013;12:470–481. doi: 10.1128/EC.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto H, Kurokawa K, Kaito C, et al. Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob Agents Chemother. 2004;48:774–779. doi: 10.1128/AAC.48.3.774-779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka N, Takano Y, Shibuya K, Fugo H, Uehara Y, Niimi M. Identification of the putative protein phosphatase gene PTC1 as a virulence-related gene using a silkworm model of Candida albicans infection. Eukaryot Cell. 2008;7:1640–1648. doi: 10.1128/EC.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SM, Sung RS, Scannapieco FA, Haase EM. Genetic relationships between Candida albicans strains isolated from dental plaque, trachea, and bronchoalveolar lavage fluid from mechanically ventilated intensive care unit patients. J Oral Microbiol. 2011;3:6332. doi: 10.3402/jom.v3i0.6362. DOI: 10.3402/jom.v3i0.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, McNab R, Jenkinson HF. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin–receptor interactions. Infect Immun. 1996;64:4680–4685. doi: 10.1128/iai.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby JM, Jensen EC, Lisec AD, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Fujiwara T, Kilian M. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J Clin Microbiol. 2005;43:6073–6085. doi: 10.1128/JCM.43.12.6073-6085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LY, Minah GE, Peterson DE, et al. Coaggregation of oral Candida isolates with bacteria from bone marrow transplant recipients. J Clin Microbiol. 1990;28:2621–2626. doi: 10.1128/jcm.28.12.2621-2626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima SA, Hayama K, Burton JP, et al. Effect of Streptococcus salivarius K12 on the in vitro growth of Candida albicans and its protective effect in an oral candidiasis model. Appl Environ Microbiol. 2012;78:2190–2199. doi: 10.1128/AEM.07055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis. 2010;16:729–739. doi: 10.1111/j.1601-0825.2010.01701.x. [DOI] [PubMed] [Google Scholar]
- Jamieson AM, Yu S, Annicelli CH, Medzhitov R. Influenza virus-induced glucocorticoids compromise innate host defense against a secondary bacterial infection. Cell Host Microbe. 2010;7:103–114. doi: 10.1016/j.chom.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jergens AE, Wilson-Welder JH, Dorn A, et al. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut. 2007;56:934–940. doi: 10.1136/gut.2006.099242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CC, Yu A, Lee H, Fidel PL, Jr, Noverr MC. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun. 2012;80:1736–1743. doi: 10.1128/IAI.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Hinds J, Smith A, van der Linden M, Van Eldere J, Mitchell TJ. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J Clin Microbiol. 2010;48:2762–2769. doi: 10.1128/JCM.01746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Walters KA, Davis AS, et al. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio. 2011;2:e00172–11. doi: 10.1128/mBio.00172-11. DOI: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Yang QB, Zhang P, et al. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun. 2002;70:2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- Khan SA, Wingard JR. Infection and mucosal injury in cancer treatment. J Natl Cancer Inst Monogr. 2001;3:1–36. doi: 10.1093/oxfordjournals.jncimonographs.a003437. [DOI] [PubMed] [Google Scholar]
- Klotz SA, Gaur NK, De Armond R, et al. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol. 2007;45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- Knapp S, Wieland CW, van ‘t Veer C, et al. Toll-like receptor 2 plays a role in the early inflammatory response to murine pneumococcal pneumonia but does not contribute to antibacterial defense. J Immunol. 2004;172:3132–3138. doi: 10.4049/jimmunol.172.5.3132. [DOI] [PubMed] [Google Scholar]
- Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Hayacibara MF, Schobel BD, et al. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J Antimicrob Chemother. 2003;52:782–789. doi: 10.1093/jac/dkg449. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JE, Shetty K, Fidel PL., Jr Oral opportunistic infections in HIV-positive individuals: review and role of mucosal immunity. AIDS patient care STDs. 2004;18:443–456. doi: 10.1089/1087291041703665. [DOI] [PubMed] [Google Scholar]
- Liljemark WF, Gibbons RJ. Suppression of Candida albicans by human oral streptococci in gnotobiotic mice. Infect Immun. 1973;8:846–849. doi: 10.1128/iai.8.5.846-849.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Ha U, Sakai A, et al. Streptococcus pneumoniae synergizes with nontypeable Haemophilus influenzae to induce inflammation via upregulating TLR2. BMC Immunol. 2008;9:40. doi: 10.1186/1471-2172-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. 2011;10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Loach DM, Jenkinson HF, Tannock GW. Colonization of the murine oral cavity by Streptococcus gordonii. Infect Immun. 1994;62:2129–2131. doi: 10.1128/iai.62.5.2129-2131.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM. Hosting infection: experimental models to assay Candida virulence. Int J Microbiol. 2012;2012:363764. doi: 10.1155/2012/363764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Elborn JS, Parkins MD, et al. Population structure and characterization of viridans group streptococci (VGS) including Streptococcus pneumoniae isolated from adult patients with cystic fibrosis (CF) J Cyst Fibros. 2011;10:133–139. doi: 10.1016/j.jcf.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Mason KL, Downward JR, Mason KD, et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R, Jenkinson HF. Gene disruption identifies a 290 kDa cell-surface polypeptide conferring hydrophobicity and coaggregation properties in Streptococcus gordonii. Mol Microbiol. 1992;6:2939–2949. doi: 10.1111/j.1365-2958.1992.tb01753.x. [DOI] [PubMed] [Google Scholar]
- McNab R, Forbes H, Handley PS, Loach DM, Tannock GW, Jenkinson HF. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J Bacteriol. 1999;181:3087–3095. doi: 10.1128/jb.181.10.3087-3095.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker ND, Trede NS. Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Monif GR, Carson HJ. Female genital tract bacterial coisolates with Candida albicans in patients without clinical vaginitis. Infect Dis Obstet Gynecol. 1998;6:52–56. doi: 10.1002/(SICI)1098-0997(1998)6:2<52::AID-IDOG4>3.0.CO;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, et al. Bacteriology of experimental gingivitis in young adult humans. Infect Immun. 1982;38:651–667. doi: 10.1128/iai.38.2.651-667.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PJ, Hyman SC, Byron O, Andrew PW, Mitchell TJ, Rowe AJ. Modeling the bacterial protein toxin, pneumolysin, in its monomeric and oligomeric form. J Biol Chem. 1994;269:25315–25320. [PubMed] [Google Scholar]
- Muhlschlegel FA, Fonzi WA. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasution O, Srinivasa K, Kim M, et al. Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot Cell. 2008;7:2008–2011. doi: 10.1128/EC.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Marchillo K, Spiegel CA, Andes DR. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 2010;78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell. 2010;9:1622–1634. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Activity of cilofungin (LY121019) against Candida species in vitro. J Antimicrob Chemother. 1988;22:891–897. doi: 10.1093/jac/22.6.891. [DOI] [PubMed] [Google Scholar]
- Okamoto-Shibayama K, Sato Y, Yamamoto Y, Ohta K, Kizaki H. Identification of a glucan-binding protein C gene homologue in Streptococcus macacae. Oral Microbiol Immunol. 2006;21:32–41. doi: 10.1111/j.1399-302X.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Parnanen P, Meurman JH, Samaranayake L, Virtanen I. Human oral keratinocyte E-cadherin degradation by Candida albicans and Candida glabrata. J Oral Pathol Med. 2010;39:275–278. doi: 10.1111/j.1600-0714.2009.00866.x. [DOI] [PubMed] [Google Scholar]
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Peters BM, Noverr MC. Candida albicans–Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun. 2013;81:2178–2189. doi: 10.1128/IAI.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. Microbial interactions and differential protein expression in Staphylococcus aureus–Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell. 2009;8:1750–1758. doi: 10.1128/EC.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja M, Hannan A, Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010;44:272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner AJ, Lysenko ES, Paul MN, Weiser JN. Synergistic proinflammatory responses induced by polymicrobial colonization of epithelial surfaces. Proc Natl Acad Sci U S A. 2005;102:3429–3434. doi: 10.1073/pnas.0500599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann P, Nuhn M, Denapaite D, et al. Genome of Streptococcus oralis strain Uo5. J Bacteriol. 2011;193:2888–2889. doi: 10.1128/JB.00321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker A, Vikerman MM, Dongari-Bagtzoglou A. S. gordonii glucosyltransferase promoters biofilm interactions with C. albicans. J Oral Microbiol. 2014:6. doi: 10.3402/jom.v6.23419. doi: 10.3402/jom.v6.23419. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Garcia BF, et al. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- Saporito-Irwin SM, Birse CE, Sypherd PS, Fonzi WA. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DS, Russell RR. A novel glucan-binding protein with lipase activity from the oral pathogen Streptococcus mutans. Microbiology. 2004;150:1947–1956. doi: 10.1099/mic.0.26955-0. [DOI] [PubMed] [Google Scholar]
- Sibley CD, Duan K, Fischer C, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008a;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A. 2008b;105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele C, Ratterree M, Fidel PL., Jr Differential susceptibility of two species of macaques to experimental vaginal candidiasis. J Infect Dis. 1999;180:802–810. doi: 10.1086/314964. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Syed SA, Loesche WJ. Bacteriology of human experimental gingivitis: effect of plaque age. Infect Immun. 1978;21:821–829. doi: 10.1128/iai.21.3.821-829.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM, Baranowski LK, Rogers JD, Haase EM, Scannapieco FA. Oral colonization and cariogenicity of Streptococcus gordonii in specific pathogen-free TAN:SPFOM(OM)BR rats consuming starch or sucrose diets. Arch Oral Biol. 2001;46:323–333. doi: 10.1016/s0003-9969(00)00126-6. [DOI] [PubMed] [Google Scholar]
- Uppuluri P, Mekala S, Chaffin WL. Farnesol-mediated inhibition of Candida albicans yeast growth and rescue by a diacylglycerol analogue. Yeast. 2007;24:681–693. doi: 10.1002/yea.1501. [DOI] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Vickerman MM, Iobst S, Jesionowski AM, Gill SR. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007;189:7799–7807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez R, Lemme A, Ballhausen B, et al. Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF) ChemBioChem. 2010;11:1552–1562. doi: 10.1002/cbic.201000086. [DOI] [PubMed] [Google Scholar]
- Villar CC, Dongari-Bagtzoglou A. Immune defence mechanisms and immunoenhancement strategies in oropharyngeal candidiasis. Expert Rev Mol Med. 2008;10:e29. doi: 10.1017/S1462399408000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect Immun. 2007;75:2126–2135. doi: 10.1128/IAI.00054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2:e00055–00011. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, Efstratiou A, Pickerill AP, et al. Genetic relationships between clinical isolates of Streptococcus pneumoniae Streptococcus oralis, and Streptococcus mitis: characterization of “Atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkieson C, Samaranayake LP, MacFarlane TW, Lamey PJ, MacKenzie D. Oral candidosis in the elderly in long term hospital care. J Oral Pathol Med. 1991;20:13–16. doi: 10.1111/j.1600-0714.1991.tb00880.x. [DOI] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Downs D, Ghosh K, et al. Candida albicans secreted aspartic proteases 4-6 induce apoptosis of epithelial cells by a novel Trojan horse mechanism. FASEB. 2013;27:2132–2144. doi: 10.1096/fj.12-214353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Nobile CJ, Dongari-Bagtzoglou A. Glucanase induces filamentation of the fungal pathogen Candida albicans. PLoS ONE. 2013a;8:e63736. doi: 10.1371/journal.pone.0063736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Thompson A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2013b;16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Sasaki J, Matsumoto K, et al. A necrotic lung ball caused by co-infection with Candida and Streptococcus pneumoniae. Infect Drug Resist. 2011;4:221–224. doi: 10.2147/IDR.S24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Palmer RJ, Yang J, Kolenbrander PE, Cisar JO. Streptococcal receptor polysaccharides: recognition molecules for oral biofilm formation. BMC Oral Health. 2006;6(Suppl 1):S12. doi: 10.1186/1472-6831-6-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li J, Jia X, Wu Y. The expression of toll-like receptor 2 and 4 mRNA in local tissues of model of oropharyngeal candidiasis in mice. J Huazhong Univ Sci Technolog Med Sci. 2004;24:639–641. doi: 10.1007/BF02911380. [DOI] [PubMed] [Google Scholar]
- Zhu W, Filler SG. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 2010;12:273–282. doi: 10.1111/j.1462-5822.2009.01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MB, Degener JE, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]


