Abstract
Purpose
Inter-individual differences in quantitative expression could underlie a propensity for lung cancer. To determine precise individual gene expression signatures on a lung compartment-specific basis, we investigated the expression of carcinogen metabolism genes encoding cytochromes P450 (CYP) 1B1, 2A13, glutathione S-transferase (GST) P1, and a tumor suppressor gene p16 in laser capture microdissected samples of human alveolar compartment (AC) and bronchial epithelial compartment (BEC) lung tissue from 62 smokers and non-smokers.
Experimental Design
Tobacco exposure was determined by plasma nicotine, cotinine, and smoking history. Precise mRNA expression was determined using our RNA-specific qRT-PCR strategy, and correlated with detailed demographic and clinical characteristics.
Results
Several correlations of mRNA expression included: (a) CYP1B1 in AC (positively with plasma nicotine level, P = 0.008; plasma cotinine level, P = 0.001); (b) GSTP1 in AC (positively with plasma cotinine level, P = 0.003); (c) GSTP1 in BEC (negatively with smoke dose, P = 0.043; occupational risk, P = 0.019). CYP2A13 was rarely expressed in AC, and not expressed in BEC. p16 expression was not correlated with any measured factor. For each gene, subjects showed expression that was individually concordant between these compartments. No clear association of mRNA expression with lung cancer risk was observed in this pilot analysis.
Conclusions
The association between lung mRNA expression and tobacco exposure implies that gene-tobacco interaction is a measurable quantitative trait, albeit with wide inter-individual variation. Gene expression tends to be concordant for alveolar and bronchial compartments for these genes in an individual, controlling for proximate tobacco exposure.
Keywords: Lung carcinoma, gene expression, laser capture microdissection, lung epithelial cells, tobacco smoking
INTRODUCTION
The gene expression analysis of clinical tissue specimens has provided patterns of gene-environmental interplay associated with cancer, and suggests a broad range of potential candidate diagnostic and prognostic biomarkers of human cancer. Several reports have used gene expression profiling to discriminate primary lung cancers from noncancerous lung (1–5). Given that cigarette smoke creates a field of injury throughout the airway, several groups have applied gene expression profiles to identify smoking-associated markers in cytologically normal human airway epithelial cells brushed at bronchoscopy from smokers and nonsmokers (5–9). A recent study performed gene expression microarrays on 135 fresh frozen tissue samples of adenocarconoma and paired noninvolved lung tissue from current, former and never smokers (10). In these surgical resection studies, however, the inherent heterogeneity of primary tissues with an admixture of various cells might be a significant confounder affecting the outcome and interpretation of molecular studies (11).
Laser capture microdissection (LCM) entails microscopically visualizing the cells of interest from a stained tissue section, which can obtain pure populations of cells from a section of complex, heterogeneous tissue (11), thus, overcoming some of the challenges of tissue complexity. The human bronchial epithelial compartment (BEC) lines the bronchial and bronchiolar airways of the lung, and plays a key role in regulating the entry and metabolism of inhaled foreign compounds, or xenobiotics. The BEC represents a primary defense against inhaled pollutants, including polycyclic aromatic hydrocarbon (PAH) procarcinogens and other organic chemicals (12). On the other hand, the cellular origins of peripheral lung adenocarcinomas are thought to be type II alveolar cells in the alveolar compartment (AC) or bronchial alveolar junction (13–15). To obtain more anatomically precise information of gene expression profiles of lung carcinogenesis, it would be useful to precisely assess the differential expression between human AC and BEC lung compartments, using the LCM technique.
Several groups have tried to compare the differential gene expression patterns in AC and BEC, employing LCM and microarray analysis strategies (16–19). Although microarray data offers a global assessment of genetic alterations, dynamic range and accuracy is compressed when analyzing genes individually, as compared to quantitative PCR. Moreover, there have been considerable differences in identified genes between the reports, and the roles of specific genes have still to be elucidated. In this study, we applied an LCM technique combined with a quantitative RNA-specific reverse transcription-PCR (RT-PCR) strategy (20, 21) to determine precise individual expression phenotypes for a candidate set of known carcinogenesis-relevant genes in the primary BEC and AC from 62 subject donors. The selection of candidate target genes was based on their known importance for metabolism of tobacco carcinogens and/or susceptibility to lung cancer, and their known or hypothesized levels in human lung.
Cytochrome P450 (CYP) 1B1 is polycyclic aromatic hydrocarbons (PAH) inducible, and the most abundantly expressed phase I oxidizing carcinogen metabolism gene in human lung (22). It serves as an indicator both for exposure to procarcinogens, and its cds variants have been hypothesized to confer susceptibility to cancer (23). CYP2A13, a functional member of the human CYP2A gene subfamily, has been reported to be selectively expressed in the respiratory tract (24), and its protein has been detected in adult human lung (25, 26). CYP2A13-catalyzed alpha-hydroxylation is an important step in the activation of the tobacco-specific carcinogens, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and (S)-N'-nitrosonornicotine (26, 27). Since our previous studies showed that CYP1A1, one of the most highly studied genes in the literature, is minimally expressed in human lung (21, 22), it was not investigated in this study. The glutathione S-transferase (GST) P1 is the major metabolic phase II enzyme in nonmalignant human lung, as evaluated by mRNA and protein expression (21, 28) and by activity (29). Knockout of GSTP1 in mice has been reported to lead to a significant increase in both carcinogen-induced and spontaneous tumorigenesis (30–32). p16 is a cell cycle regulatory pathway gene, commonly referred to as a tumor suppressor gene, and has been observed as silenced or abnormal in approximately 70% of non-small cell lung cancer (33). Therefore, the expression signature of these four candidate target genes (CYP1B1, CYP2A13, GSTP1 and p16) was explored, and analyzed in a multivariate model for probing gene-environment interactions associated with lung carcinoma.
MATERIALS AND METHODS
Patient Recruitment and Sample Collection
This study was comprised of 62 consenting individuals undergoing lung resectional surgery for clinically suspected carcinoma. As previously described (21, 28), recruits were part of the pulmonary medicine or thoracic surgery practices at Albany Medical Center and were otherwise destined for bronchoscopy or lung resectional surgery for clinical indications, under a protocol approved by Albany Medical Center and New York State Department of Health institutional review boards. After informed consent was obtained, the subjects were interviewed by a trained research nurse. The information on mainstream tobacco exposure history (type, amount, duration, and quitting date), environmental tobacco smoke exposure history, preexisting lung diagnoses (chronic obstructive pulmonary disease and interstitial lung disease), dietary history; occupational exposure history to asbestos, silica, or other known carcinogens, family history of cancer, and medication history were obtained pre-diagnosis/pre-operatively by direct interview.
Additionally, 15 ml blood sample for each subject was collected, stored briefly at room temperature, and the plasma fraction was frozen and banked at −180°C for plasma nicotine and cotinine analysis. Lung tissue samples were snap-frozen in liquid nitrogen within 15 minutes of surgical resection, and stored in a −180°C tissue bank until analyzed. The assigned clinical surgical pathologist confirmed the diagnosis of lung cancer, per clinical routine, and classified the samples as adenocarcinoma, squamous cell carcinoma, mixed non-small cell carcinoma, or benign process of lung, according to the 1999 World Health Organization (WHO) histological classification of lung and pleural tumours.
Laser-Capture Microdissection of Lung Tissue
Immediately prior to snap-freezing in liquid nitrogen, the freshly-resected malignant or nonmalignant lung tissues were embedded in Tissue-Tek® optimal cutting temperature (OCT) medium (Sakura Finetek USA, Inc., Torrance, CA) in the “frozen section” room, immediately adjacent to the operating room. The frozen malignant or nonmalignant lung tissue was subjected to frozen microtome sectioning at −25°C, using a refrigerated cryostat, and the sections mounted on uncoated, cooled glass slides. The sections were immediately stained with hematoxylin, and were dehydrated by a series of 70% ethanol (30 s), 95% ethanol (30 s), 100% ethanol (30 s) and xylene (5 minutes) treatments. The OCT material was removed after sectioning during the rapid alcohol fixation and dehydration process, prior to immediately placing the section on the stage for microdissection. Once air-dried, desired cells were microdissected onto CapSure™ transfer films (Arcturus Engineering, Mountain View, CA) using Pixcell IIe LCM system (Arcturus Engineering, Mountain view, CA) for each sample. Distinguishing these two lung tissue compartments (alveolar AC vs. bronchial BEC) could be performed with confidence in these snap-frozen samples, given the quality of realtime images guiding microdissection.
We performed sampling of four areas of a frozen tissue section, using 200 pulses/area (each 30 µm pulse yielding ∼2 to 4 cells), so that each cap (of a 500 µl tube containing a RNA extraction buffer, as described below) contained 1,600 to 3,200 cells for qualitative mRNA-PCR. The Arcturus Capsure™ cap was placed, immediately after tissue microdissection, in a 500 µl microfuge tube preloaded with 100 µl of RLT/β-ME buffer (Qiagen, Valencia, CA), inverted to submerse the tissue, and stored in −80°C until use.
RNA Extraction and Quantitative Real Time RT-PCR
Total RNA was extracted from the microdissected samples using a RNAeasy Mini kit (Qiagen, Valencia, CA) as described previously (28). Reverse transcription was performed on a scale of 20 µl using SuperScript™ II Reverse Transcriptase (Life Technologies, Inc., Invitrogen, Grand Island, NY) with our universal tagged RT primer (20, 21). Briefly, RNA (about 100 ng), 1 µl universal RT primer (100 µM) and 2 µl dNTPs (10 mM) were combined in a PCR reaction tube, and RNase/DNase-free water was added to make up the final volume to 13 µl. The RNA was denatured at 65°C for 5 minutes and then at 4°C for 5 minutes. The following were added: 4 µl 5 x SuperScript™ II First-Strand buffer (Invitrogen); 2 µl DTT (0.1 M). After 6 µl of the master mix/RT reaction was aliquoted, incubation at 42°C for 2 minutes was followed by the addition of 1 µl SuperScript™ II RT polymerase to each tube, and each tube was individually mixed gently by pipetting up and down, followed by incubation at 42°C for 60 minutes and then at 70°C for 15 minutes. All RT steps were performed in a block thermal cycler (ABI 9700, Applied Biosystem, CA).
Quantitation of cDNA for CYP1B1, CYP2A13, GSTP1, and p16 by realtime PCR was performed in the ABI Prism 7500HT sequence detection system (Applied Biosystems), using the previously published RNA-specific qRT-PCR strategy (20, 21). cDNA-specific amplification (versus genomic DNA amplification) was inherent to the strategy, and verified using RT-omission controls (20). A primer pair for each gene was designed by using online Primer 3.0 software based on the published sequences as following: CYP1B1, 5’-GCC ACT ATC ACT GAC ATC T-3’ (sense) and 5’-CTT GCC TCT TGC TTC TTA TT-3’ (antisense); CYP2A13, 5’- ACC TGG TGA TGA CCA CCC-3’ (sense) and 5’- CGT GGA TCA CTG CCT CTG-3’ (antisense); GSTP1, 5’-TCT CCT TCG CTG ACT ACA AC-3’ (sense) and 5’-AAC GAG ACG ACG ACA GAC-3’ (antisense); p16, 5’-GGA AGG TCC CTC AGA CAT CC-3’ (sense) and 5’-AGG CTG CGA GGC TCG CAA G-3’ (antisense); and GAPDH, 5’-AGC CCC AGC AAG AGC ACA A-3’ (sense) and 5’-AAC GAG ACG ACG ACA GAC-3’ (antisense). Real-time PCR was performed using a 96-well optical plate (Applied Biosystem), and with GoTaq Flexi DNA polymerase kit (Promega, Madison, WI). In each well, the following were combined: 5 µl of the 5 x Colorless GoTaq Flexi buffer, 1.5 µl of 25 mM MgCl2, 14 µl of RNase/DNase-free H2O; 1.6 µl of dNTPs (2.5 mM each triphosphate nucleotide); 1.25 µl SYBR Green dye (1:10,000) (Applied Biosystem); 0.4 µl GoTaq Flexi DNA Polymerase; 0.25 µl of 50 µM each primer pair; and 1 µl template cDNA from the RT reaction. PCR conditions included a denaturing cycle at 95°C for 30 s; 5 cycles of 95°C for 30 s, 66°C for 30 s and 72°C for 30 s, followed by 50 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Melting analysis for one cycle was as follows: 95°C for 15 s; 60°C for 60 s; and then slow up-ramp for a continuous acquisition to 95°C. After the reaction, PCR products underwent electrophoresis on a 1.5% agarose-ethidium bromide gel, and visualized under UV light.
For each target gene and the internal and external target reference gene, PCR was performed in triplicate for each subject’s alveolar and bronchial sample, in duplicate for both positive controls (transcript specific vector cDNA and a mixed cDNA pool, below), and an H2O blank as negative control in parallel. For scaling expression to any RNA degradation, the expression level of each gene was normalized to RNA content for each sample by using validated GAPDH transcript sequence as an internal “housekeeping” control transcript, in parallel uniplex reactions. For positive controls, transcript-specific positive parallel controls for run-to-run variability included quantitated cDNA from vectors containing the appropriate target sequence insert. Another transcript-specific positive control was cDNA derived from total RNA of a set of standard cell lines, chosen according to the characteristic expression patterns. For CYP1B1 and GAPDH, total RNA from dioxin-stimulated MCF-7 breast cancer cell line was used for a positive external control. For CYP2A13 and GSTP1, a positive external control was from human lung tissue derived total RNA (Clontech Laboratories, Inc., Mountain View, CA), while for p16, total RNA from NHBE cell lines was used. All positive control cDNAs were synthesized from 2.5 µg total RNA.
Plasma Nicotine and Cotinine Analysis by Gas Chromatograpy/Mass Spectrometry
Plasma nicotine and cotinine levels were measured using an isotope dilution-high performance liquid chromatography/electrospray ionization tandem mass spectrometric method at the Wadsworth Center for Labs and Research, New York State Department of Health, as described previously (28, 34). The limits of detection for nicotine and cotinine were 0.25 and 0.05 µg/l plasma, respectively. Therefore, we considered the plasma nicotine level < 0.25 µg/l and cotinine level < 0.05 µg/l as 0 µg/l, when we performed data analysis using the plasma nicotine/cotinine levels as continuous variables.
Data Analysis
All successive replicate experimental RT-PCR on any given sample displayed appropriate negative and positive controls for that trial; otherwise the trial was not included in the data analysis. For quantitative analysis, the ratio of expression of target gene t in sample s relative to positive control samples c is given by
, where r labels the reference gene used for normalization (35, 36). All mRNA expression data of CYP1B1, GSTP1 and p16 levels in BEC and AC were log10 transformed for statistical analysis. Since only six of 62 AC samples expressed CYP2A13, it was excluded from further quantitative data analysis.
In the initial univariate analyses, endogenous variables (age, gender, histology, family history of lung cancer) and exposure variables (plasma nicotine, cotinine, smoking status, smoking dose, consumption of fruit & vegetable, and occupation risk) were assessed individually as nine independent variables for their correlation with the individual mRNA expression of CYP1B1, GSTP1, and p16 in bronchial epithelial and alveolar cells, using Spearman’s correlation coefficient. When univariate comparison of independent variable to the outcome variable (gene expression) displayed P ≤ 0.05, the independent variable was taken to further explore the correlation of the gene expression between the bronchial epithelial and alveolar cells in the multivariate models.
Paired BEC and AC were compared employing the Wilcoxon signed-rank test to determine the differential expression in paired samples. The Mann-Whitney test (for two categories) and Kruskal-Wallis test (for more than two categories) were used for testing the differences in median gene expression across the categories investigated. Odds ratios (OR) and their 95% confidence intervals (95% CI) measuring the associations of our gene expression data, as well as other risk factors, with risk for lung carcinoma were estimated using multivariable logistical regression models, employing the PHREG procedure of the statistical software package SAS 9.1 (SAS Institute, Inc., Cary, NC). For these evaluations, considering the Bonferroni correction for multiple testing, the standard significance level of 5% was adjusted to α = 0.05/ n, where n is the number of variables in the model for each gene.
RESULTS
The clinical and demographic characteristics of study participants are listed in Table 1. As described in Table 1, precise smoking status and exposure levels were verified by assay of plasma nicotine and cotinine level. Plasma nicotine and cotinine were undetectable in never smokers, while the highest level of plasma and nicotine and cotinine was observed in current smokers (40.8 ± 12.3 and 137.4 ± 15.7 µg/l, respectively). There were no significant differences between smoke status with reference to age at diagnosis, fruit and vegetable consumption, occupation risk, preexisting lung disease, family history of lung cancer or histological parameters.
Table 1.
Clinical and demographic characteristics of study participants
| Characteristics | Never smokers (N=10) |
Former smokers (N=36) |
Current smokers (N=16) |
P Value |
|---|---|---|---|---|
| Age at diagnosis (yr), mean ± se§ | 53.3 ± 4.6 | 63.3 ± 1.8 | 55.3 ± 3.2 | 0.075* |
| Gender, males/ females | 6/4 | 19/17 | 5/11 | 0.241† |
| Smoking dose (pack-yr), mean ± se | 0 | 53.8 ± 6.4 | 43.7 ± 9.3 | <0.001* |
| Plasma nicotine (µg/l), mean ± se | 0 | 1.2 ± 0.8 | 40.9 ± 12.3 | <0.001* |
| Plasma cotinine (µg/l), mean ± se | 0 | 8.8 ± 2.1 | 137.4 ± 15.7 | <0.001* |
| Family history of lung cancer, yes/no | 1/9 | 10/26 | 2/13 | 0.456† |
|
Fruits & Vegetable consumption (estimated servings/day), mean ± se |
2.9 ± 0.6 | 2.3 ± 0.2 | 2.7 ± 0.6 | 0.682* |
| Occupation risk, yes/no | 3/7 | 7/29 | 1/14 | 0.368† |
| Preexisting lung disease, yes/ no | 1/9 | 11/25 | 3/12 | 0.385† |
|
Histological diagnosis, AdC/ SqC/ mixed/ benign of lung |
2/1/0/7 | 7/10/12/7 | 4/2/5/5 | 0.062† |
Definition of abbreviations: se = standard error; AdC = adenocarcinoma; SqC = squamous cell carcinoma.
Kruskal Wallis test
Fisher’s exact test.
Typical microdissections for AC and BEC are depicted in Figure 1. Univariate correlations between the clinical and demographic characteristics and the mRNA expression of CYP1B1, GSTP1 and p16 in AC and BEC are presented in Table 2. CYP1B1 gene expression in AC was positively correlated with the plasma nicotine and cotinine levels (r = 0.336, P value = 0.008; and r = 0.413, P value = 0.001, respectively). Additionally, GSTP1 gene expression in AC was positively correlated with the plasma cotinine level (r = 0.366, P value = 0.003), and was marginally positively correlated with the plasma nicotine level (r = 0.241, P value = 0.054) and marginally negatively with the family history of lung cancer (r = −0.248, P value = 0.052). GSTP1 expression in BEC had a moderate inverse correlation with smoke dose (pack-yrs) (r = −0.334, P value = 0.043) and occupational exposure to lung carcinogens (r = −0.385, P value = 0.019). p16 gene expression in BEC and AC was not correlated with any clinical or demographic characteristic investigated.
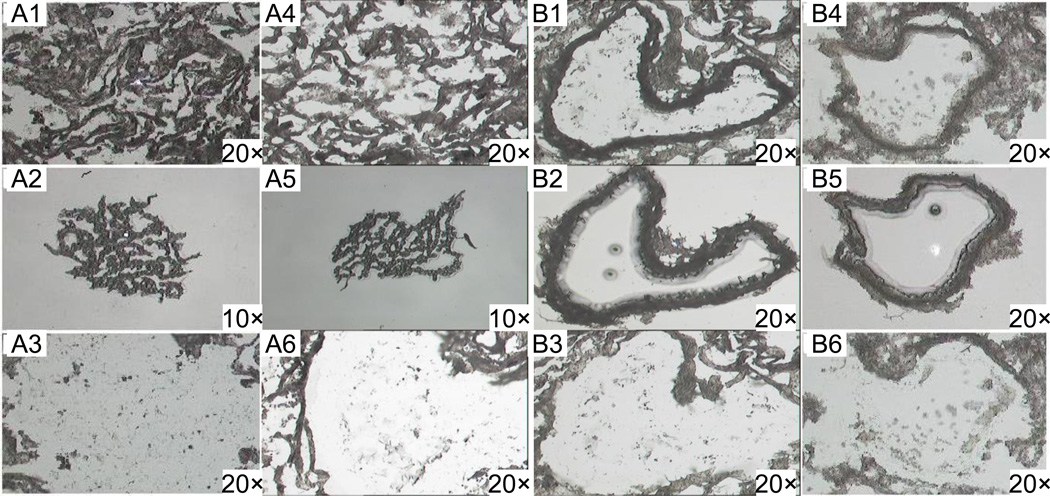
Figure 1.
LCM from hematoxylin and eosin-stained sections of alveolar (A1-A6) and bronchial epithelial (B1-B6) cells from human lung tissue. Histological sections before capture (A1, A4 and B1, B4); captured cells on transfer film (A2, A5 and B2, B5); and histological sections after capture (A3, A6 and B3, B6).
Table 2.
Spearman’s correlation coefficients for the association between mRNA expression of CYP1B1, GSTP1 and p16 in alveolar compartment and bronchial epithelial compartment, and each investigated characteristic of the study subjects
| Characteristics | Gene expression in AC§ |
Gene expression in BEC |
||||
|---|---|---|---|---|---|---|
| CYP1B1 | GSTP1 | P16 | CYP1B1 | GSTP1 | P16 | |
| Age at diagnosis (per yr) | −0.110 | −0.164 | 0.100 | −0.030 | −0.100 | −0.072 |
| Gender (males/ females) | −0.174 | −0.124 | −0.231 | −0.156 | −0.168 | −0.226 |
|
Smoke status (never/ former/ current smokers) |
0.093 | 0.198 | 0.220 | 0.182 | 0.102 | −0.060 |
| Smoke dose (per pack-yr) | 0.063 | 0.028 | −0.066 | 0.030 | −0.334* | −0.069 |
| Plasma nicotine (per µg/l) | 0.336‡ | 0.241† | 0.024 | 0.143 | −0.022 | −0.068 |
| Plasma cotinine (per µg/l) | 0.413‡ | 0.366‡ | 0.045 | 0.103 | 0.052 | −0.068 |
|
Fruit & vegetable intake (per serving/ day) |
−0.088 | −0.219 | −0.124 | −0.083 | 0.009 | 0.142 |
| Occupation risk (yes/ no) | −0.132 | −0.055 | −0.010 | −0.204 | −0.385* | −0.028 |
| Preexisting lung disease (yes/ no) | −0.052 | −0.042 | 0.106 | −0.041 | −0.117 | 0.047 |
|
Family history of lung cancer (yes/ no) |
−0.095 | −0.248† | −0.169 | 0.079 | −0.184 | −0.157 |
|
Histological diagnosis (AdCs/ SqCs/ mixed/ benign of lung) |
−0.058 | 0.130 | 0.076 | 0.156 | −0.085 | −0.019 |
Definition of abbreviations: AdC = adenocarcinoma; SqC = squamous cell carcinoma; AC = Alveolar Cells; BEC = Bronchial Epithelial cells.
P < 0.01
P < 0.05
P = 0.05.
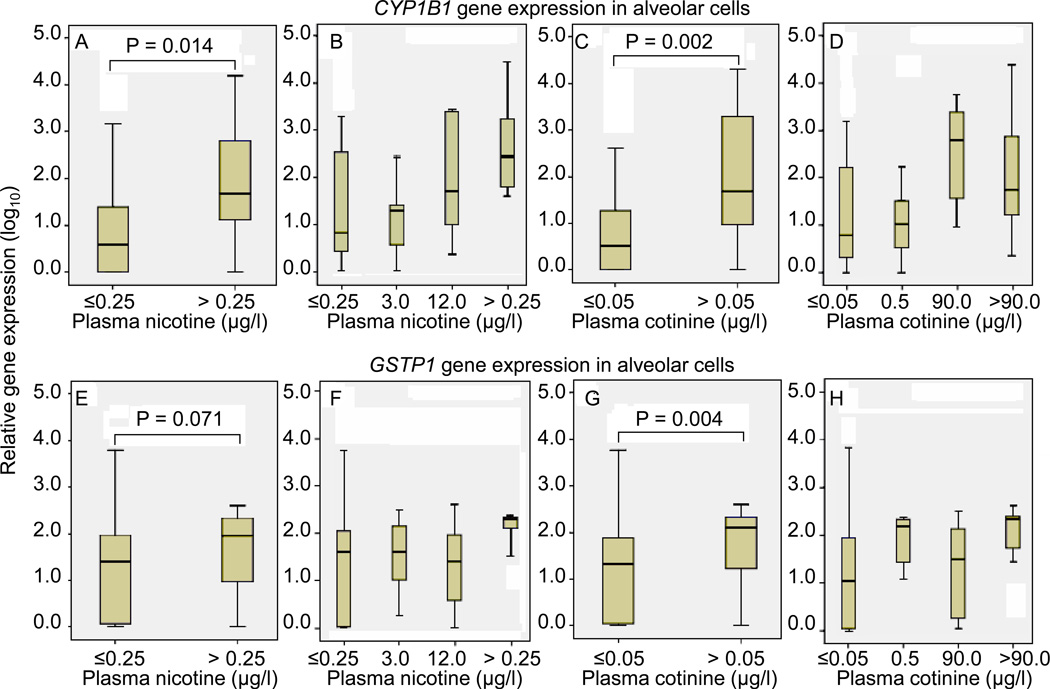
Differences in median gene expression across categorized plasma nicotine and cotinine levels are depicted in Figure 2, using the cut-points of 0.25 µg/l and 0.05 µg/l, the respective assay limits of detection. Overall, CYP1B1 and GSTP1 gene expression in AC was significantly higher in the subjects with plasma cotinine level > 0.05 µg/l, compared to the subjects with plasma cotinine level ≤ 0.05 µg/l (P value = 0.002 and 0.004, respectively) (Figure 2C and G). Similarly, subjects with the plasma nicotine level > 0.25 µg/l had a higher CYP1B1 gene expression in AC, compared to the subjects with plasma nicotine level ≤ 0.25 µg/l (P value = 0.014) (Figure 2A). However, the expression difference based on plasma nicotine biomarker was not significant for GSTP1 gene expression in AC (P value = 0.071) (Figure 2E), albeit it was positive for the longer half-life cotinine (P = 0.004). Furthermore, the categorization of tobacco exposure is further resolved (Figure 2B, D, F and H), and the dose-dependent response with plasma nicotine level was only found for CYP1B1 gene expression in AC (Figure 2B). No differential expression in BEC across the categories of plasma nicotine and cotinine level was found for CYP1B1 and GSTP1 gene expression, nor for p16 gene expression in either AC or BEC (data not shown).
Figure 2.
Box plots for the relative mRNA expression of CYP1B1 and GSTP1 in microdissected alveolar cells (AC) associated with the plasma nicotine and cotinine levels. The Mann-Whitney test was used for testing the differences in median gene expression across the categories of the plasma nicotine and cotinine levels. The vertical axis is log10, such that a range from 0 – 4 relative units represents a 10,000-fold range. Figure 2C, D, and G, H resolve the tobacco exposure further, depicting the significant variance in expression in CYP1B1 and GSTP1, respectively, in finer categories of plasma nicotine and cotinine.
Based on the above univariate analysis, we assessed the intra-individual correlations between AC and BEC compartments in mRNA expression of CYP1B1, GSTP1 and p16, adjusting by plasma nicotine and cotinine levels, pack-years, occupational risk, and family history of lung cancer (Table 3). We observed a strong positive correlation between AC and BEC for CYP1B1 gene expression (r = 0.679, P value < 0.001). A positive correlation between the gene expression in AC and BEC for GSTP1 and p16 was also present (r = 0.364, P value = 0.04 and r = 0.380, P value = 0.03, respectively). No correlation between different genes in either AC or BEC compartments was found.
Table 3.
Spearman’s correlation coefficients adjusted by plasma nicotine and cotinine levels, pack-years, occupational risk and family history of lung cancer, for the association between alveolar compartment and bronchial epithelial compartment in mRNA expression of CYP1B1, GSTP1 and p16
| Gene expression in AC § |
Gene expression in BEC |
||||||
|---|---|---|---|---|---|---|---|
| CYP1B1 | GSTP1 | P16 | CYP1B1 | GSTP1 | P16 | ||
| Gene expression in AC |
CYP1B1 | — | |||||
| GSTP1 | 0.257 | — | |||||
| P16 | 0.252 | 0.322 | — | ||||
| Gene expression in BEC |
CYP1B1 | 0.679‡ | 0.124 | 0.115 | — | ||
| GSTP1 | 0.030 | 0.364* | 0.075 | 0.225 | — | ||
| P16 | 0.120 | −0.018 | 0.380* | 0.299 | 0.274 | — | |
Definition of abbreviations: AC = Alveolar Cells; BEC = Bronchial Epithelial cells.
P < 0.001
P < 0.05.
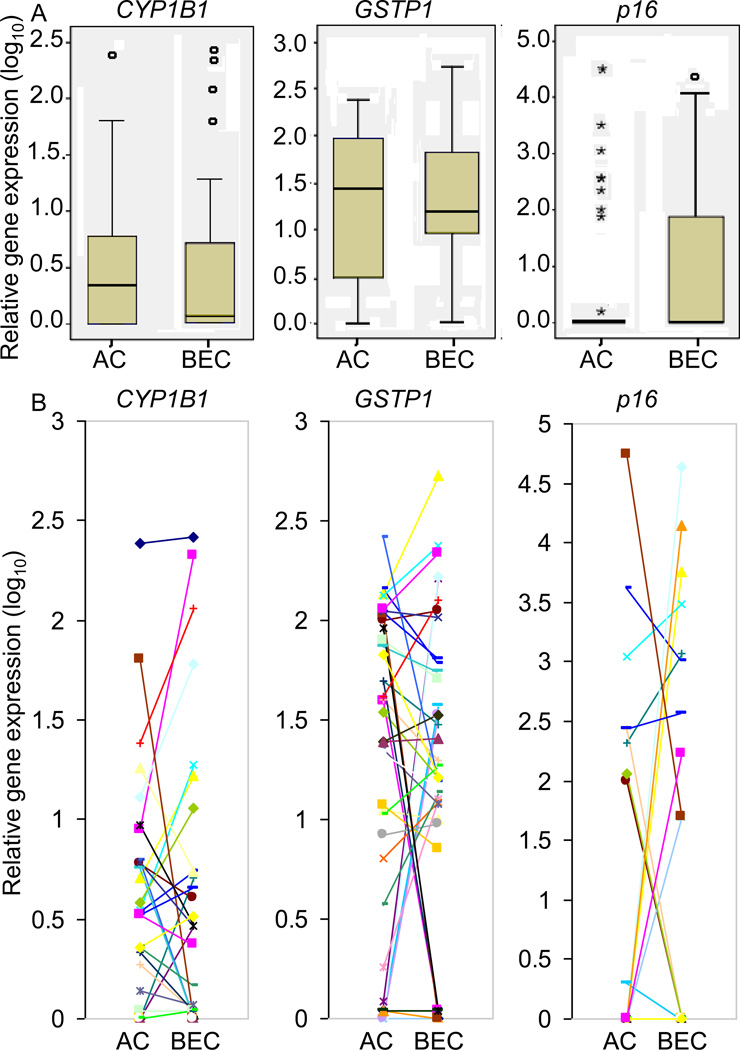
Additionally, the differential expression in microdissected AC and BEC was estimated using the Wilcoxon signed-rank test in 38 paired samples. There was no significant difference between AC and BEC for mean expression of each of the expressed genes (of the three expressed genes CYP1B1, GSTP1 and p16) (Figure 3A). Individual gene expression patterns suggested a concordance between both compartments (i. e. AC and BEC) with exceptions (Figure 3B).
Figure 3.
The relative mRNA expression levels of CYP1B1, GSTP1 and p16 in alveolar compartment (AC) and bronchial epithelial compartment (BEC) from paired tissues from 38 subjects. Paired AC and BEC were compared employing the Wilcoxon signed-rank test to determine the differential expression in the paired samples. No overall significant differences were found for the relative mRNA expression of CYP1B1, GSTP1 and p16 between microdissected AC and BEC (P value > 0.05, top panels). However, individual expression patterns (bottom panels) showed ordinal concordance between alveolar and bronchial compartments, for most individuals, particularly for CYP1B1 and GSTP1, with readily apparent exceptions.
Table 4 depicts the lack of obvious association of incident lung carcinoma with the gene expression signature of CYP1B1, GSTP1 and p16 in AC and BEC in a pilot case-control analysis. Age-adjusted association of GSTP1 gene expression in microdissected AC with incident lung carcinoma showed borderline significance, and this association was stronger when adjusted by age at diagnosis and smoke dose, or adjusted by age at diagnosis, smoke dose, and family history of lung cancer. However, there was marginal association between the GSTP1 gene expression signature in AC, and the risk of lung carcinoma after adjusting for multiple testing. No other gene expression signatures showed significant association with incident lung carcinoma.
Table 4.
Adjusted odds ratios and their 95% confidence intervals for the association of lung cancer case-control status with mRNA expression levels of CYP1B1, GSTP1 and p16 in alveolar compartment and bronchial epithelial compartment
| Gene expression in AC § |
Gene expression in BEC |
|||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| CYP1B1 | ||||||
| Crude | 0.92 | 0.63–1.33 | 0.65 | 2.02 | 0.51–8.01 | 0.32 |
| Age adjusted | 0.95 | 0.61–1.50 | 0.84 | 1.73 | 0.33–9.16 | 0.52 |
| Age + smoking dose | 0.83 | 0.46–1.48 | 0.52 | 2.17 | 0.20–23.34 | 0.52 |
| Age + smoking dose + FH | 0.84 | 0.46–1.52 | 0.56 | 2.71 | 0.20–36.51 | 0.45 |
| GSTP1 | ||||||
| Crude | 1.50 | 0.83–2.70 | 0.18 | 0.80 | 0.31–2.07 | 0.65 |
| Age adjusted | 2.06 | 0.96–4.41 | 0.06 | 0.58 | 0.16–2.05 | 0.39 |
| Age + smoking dose | 3.05 | 0.99–9.35 | 0.05 | 0.86 | 0.19–3.90 | 0.85 |
| Age + smoking dose + FH | 4.19 | 1.24–14.12 | 0.02 | 1.76 | 0.24–12.95 | 0.58 |
| p16 | ||||||
| Crude | 0.98 | 0.62–1.54 | 0.91 | 0.92 | 0.56–1.48 | 0.72 |
| Age adjusted | 0.92 | 0.50–1.69 | 0.79 | 0.91 | 0.52–1.59 | 0.74 |
| Age + smoking dose | 1.18 | 0.58–2.43 | 0.65 | 1.06 | 0.52–2.16 | 0.87 |
| Age + smoking dose + FH | 1.36 | 0.61–3.00 | 0.45 | 1.08 | 0.50–2.31 | 0.85 |
Definition of abbreviations: OR = Odds ratios; CI = Confidence intervals; AC = Alveolar Cells; BEC = Bronchial Epithelial cells; FH = family history of lung cancer.
DISCUSSION
In this study, we have demonstrated that the quantitative mRNA expression signature of CYP1B1 and GSTP1 in AC, but not BEC, is strongly associated with biomarkers of tobacco exposure. Additionally, we have shown an intra-individual correlation between AC and BEC compartments for the mRNA expression signature for each studied gene, suggesting regulatory signatures may be individual donor-specific. Notably, there was no significant difference between AC and BEC in the mean expression of these four studied genes, suggesting that at a group level, carcinogen metabolism in one compartment may be somewhat mirrored in the other. We did not observe clear association of a quantitative gene expression signature of this geneset with the risk of lung cancer, accounting for other confounding factors.
For CYP1B1, the smoking-related induction of this bioactivating enzyme is consistent with previous reported findings (37–38). Tobacco-induced expression of CYP1B1 in human lung has been suggested to vary widely inter-individually (21, 39); we observed ranges of 1,000 to >10,000-fold differences, even when stratified by smoke exposure biomarkers nicotine and cotinine, where the respective half-lives of 2–24 hours (40–43) are compatible with transcription kinetics. The responsible regulatory factors for such variability are not yet elucidated, nor are the downstream (adduct and mutation) consequences as yet.
This level of precision in human exposure-gene expression correlation has not, to our knowledge, been reported before. We saw suggestions of CYP1B1 tobacco-responsiveness only in the alveolar compartment, a somewhat surprising phenomena, given the bronchogenic nature of many malignancies, but cannot exclude responsiveness in the bronchial epithelium given the limits of resolution and possibility of admixture of other cells when microdissecting bronchial walls of frozen sections. Additionally, given that there were fewer samples available with bronchial epithelium (38 total, with 10 controls and 28 cases); there were clear statistical power limitations.
Previous studies showed CYP2A13 gene is expressed in liver and a number of extrahepatic tissues, including nasal mucosa, lung, trachea, brain, mammary gland, prostate, testis, and uterus (24). We only found expression of CYP2A13 transcript in six of 62 AC samples; while acknowledging the hypothetical possibility of sensitivity limits of our anchored RNA-specific RT-PCR assay, our findings exclude high levels of CYP2A13 expression in either AC or BEC compartments.
GSTs appeared to be modestly tobacco-responsive in the alveolar compartment, uniquely. Gene expression of GSTP1 in alveoli, alveolar macrophages, and respiratory bronchioles is more abundant than that of GSTM and other GSTs (21, 44). It is therefore possible that GSTP1 may play an important role in local detoxification of xenobiotics in the lung, but is mostly non-inducible to tobacco exposure, which we have observed in multiple in vitro studies (28, 45, 46). If true, this is biologically problematic, as this phase I versus phase II imbalance potentially favors mutagenesis, rather than mutagen quenching. A recent animal GSTP1 knockout study indicates that GSTP1 plays a key role in vivo in determining susceptibility to lung cancer following exposure to chemical carcinogens (32).
p16 has been reported to be silenced early in lung carcinogenesis (47, 48). It is an inhibitor of CDK4 and CDK6 and negatively regulates cyclin D-dependent phosphorylation of Rb gene product, thereby inhibiting cell cycle progression from G1 to S-phase by sequestration of E2F. We reasoned that low expressers might be at higher risk of lung cancer. However no significant p16 expression differences were found with any of the lung cancer risk factors, nor with case-control status. This finding still remains compatible with epigenetic silencing of the gene as one step in carcinogenesis, which was not tested in this study.
There are several possible reasons why we did not find a signature that predicted case or control status of the donor. First, the pathways affected by tobacco smoke are numerous, interconnected, and potentially redundant, so a small candidate gene set such as ours, while precise, may lose etiologic signatures. Indeed, tobacco smoke is a complex mixture of compounds, each of which is likely to have multiple targets. Second, tobacco use clusters with other human behaviors, which, if these are not accounted for, leads to confounding and biased or misclassified estimates of association. Thus, the gene expression profile associated with smoking may also be associated with a pattern of other unmeasured behaviors (e.g., higher alcohol intake, lower exercise) that by themselves may influence gene expression. Third, we cannot exclude the possibility that the null relationships found in this pilot case-control analysis between the gene expression of CYP1B1, CYP2A13, GSTP1, and p16 with lung cancer were due to the small sample size. At the current sample size (19 controls and 43 cases for AC, and 10 controls and 28 cases for BEC), the analyses were expect to have less than 35% power at the nominal significance level of 0.05 to detect the observed differences of CYP1B1, GSTP1, and p16 gene expression between cases and controls.
In the current study, we examined the CYP1B1, CYP2A13, GSTP1 and p16 gene expression in microdissected AC and BEC, rather than in whole lung tissue. Tissue microdissection is important for more precise molecular analysis, to reduce the number of contaminating non-target cells. The presence of discordant expression results between AC and BEC compartments, particularly for GSTP1 and p16, underscore this point. Earlier studies (16–19) have often been lacking information on potential confounders or availability of paired AC and BEC samples for the distinction of gene changes involved in lung carcinogenesis, from those representing a transient smoking effect. We attempted to overcome these latter pitfalls with detailed covariate information (e.g., age, sex, occupation exposure risk, previous lung disease and family history of lung cancer, diet) and biochemical validation of the tobacco smoking exposure.
As clinical specimens deteriorate, they and the attendant transcripts do so at variable rates that are difficult to measure, such that mRNA measurements must include valid internal reference transcript controls, allowing one to scale the target transcript amount to that of the internal reference, each assumed to degenerate at similar rates. Further, because many of the internal standard mRNA “housekeeper” transcripts (β-actin, GAPDH, 36B4, others) are highly homologous to (pseudogene) sequences in contaminating genomic DNA in the tissue RNA extract, their measurement can be overestimated inadvertently in many clinical extracts. For these reasons, we went to considerable lengths to use a reference RNA-specific qRT-PCR strategy (20, 21), and employed both validated internal (GAPDH) and external reference (pooled quantitated total RNA, and quantitated insert-containing cDNA insert containing vector) controls, to bolster our ability to precisely quantitate expression.
In sum, this exposure-coupled gene expression profiling study provides further insight into lung compartment-specific patterns of expression of genes known to be involved in tobacco carcinogen and oxidant metabolism in lung carcinogenesis. The data suggest that widely varying expression signatures exist, there is tobacco-responsiveness on a gene- and compartment-specific basis, and concordance of expression between the two main lung epithelial compartments is common. The regulatory determinants of the wide inter-individual differences in expression of this and other gene sets warrant further study.
Translational Relevance.
Little is known about compartment-specific inter-individual differences in quantitative expression in the lung, and this is true for carcinogen and oxidant metabolism pathways that may underlie the development of lung phenotypes, such as cancer. A precisely-executed study, entailing both plasma biomarkers of proximate tobacco exposure, laser microdissected alveolar and bronchial compartments, and RNA-specific real-time qRT-PCR, demonstrated: (a) Inter-individual variation is very wide even within exposure strata; (b) Quantitative expression signatures tend to be characteristic of an individual, and manifest in both alveolar and bronchial compartments; (c) Gene × Tobacco interaction is a measurable, individually unique quantitative trait in the lung. This study provides further translational insight into lung compartment-specific patterns of expression of tobacco carcinogen and oxidant metabolism genes in lung carcinogenesis.
ACKNOWLEDGMENTS
We acknowledge the Wadsworth Center analytic chemistry core at the New York State Department of Health (Albany, NY) for nicotine and cotinine assays. The cooperation of surgeons Riivo Ilves (Albany Medical College Hospital, Albany, NY) and Darroch Moores (St. Peter’s Hospital, Albany, NY), the respective pathology departments at the two Albany hospitals, and the invaluable aide of Albany research nurses Angela Sheehan, Ann Venezia, and Kathy Mokhiber are also acknowledged.
Sources of Support: This work was supported by NIH-R01 CA 10618 (to SD Spivack), NIH-R21 CA 94714 (to SD Spivack) and Prevent Cancer Foundation (Research Fellowship, to XL Tan)
REFERENCES
- 1.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singhal S, Miller D, Ramalingam S, Sun SY. Gene expression profiling of non-small cell lung cancer. Lung Cancer. 2008;60:313–324. doi: 10.1016/j.lungcan.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacroix L, Commo F, Soria JC. Gene expression profiling of non-small-cell lung cancer. Expert Rev Mol Diagn. 2008;8:167–178. doi: 10.1586/14737159.8.2.167. [DOI] [PubMed] [Google Scholar]
- 5.Beane J, Spira A, Lenburg ME. Clinical impact of high-throughput gene expression studies in lung cancer. J Thorac Oncol. 2009;4:109–118. doi: 10.1097/JTO.0b013e31819151f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Liu G, Lenburg ME, Spira A. Comparison of smoking-induced gene expression on Affymetrix Exon and 3'-based expression arrays. Genome Inform. 2007;18:247–257. [PubMed] [Google Scholar]
- 8.Carolan BJ, Harvey BG, De BP, Vanni H, Crystal RG. Decreased expression of intelectin 1 in the human airway epithelium of smokers compared to nonsmokers. J Immunol. 2008;181:5760–5767. doi: 10.4049/jimmunol.181.8.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 10.Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie JW, Gannot G, Tangrea MA, et al. Molecular profiling of cancer. Toxicol Pathol. 2004;32(Suppl 1):67–71. doi: 10.1080/01926230490430728. [DOI] [PubMed] [Google Scholar]
- 12.Willey JC, Coy E, Brolly C, et al. Xenobiotic metabolism enzyme gene expression in human bronchial epithelial and alveolar macrophage cells. Am J Respir Cell Mol Biol. 1996;14:262–271. doi: 10.1165/ajrcmb.14.3.8845177. [DOI] [PubMed] [Google Scholar]
- 13.Have-Opbroek AA, Benfield JR, van Krieken JH, Dijkman JH. The alveolar type II cell is a pluripotential stem cell in the genesis of human adenocarcinomas and squamous cell carcinomas. Histol Histopathol. 1997;12:319–336. [PubMed] [Google Scholar]
- 14.Otto WR. Lung epithelial stem cells. J Pathol. 2002;197:527–535. doi: 10.1002/path.1160. [DOI] [PubMed] [Google Scholar]
- 15.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Miura K, Bowman ED, Simon R, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking- and prognosis-related molecular profiles. Cancer Res. 2002;62:3244–3250. [PubMed] [Google Scholar]
- 17.Kobayashi K, Nishioka M, Kohno T, et al. Identification of genes whose expression is upregulated in lung adenocarcinoma cells in comparison with type II alveolar cells and bronchiolar epithelial cells in vivo. Oncogene. 2004;23:3089–3096. doi: 10.1038/sj.onc.1207433. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura N, Kobayashi K, Nakamoto M, et al. Identification of tumor markers and differentiation markers for molecular diagnosis of lung adenocarcinoma. Oncogene. 2006;25:4245–4255. doi: 10.1038/sj.onc.1209442. [DOI] [PubMed] [Google Scholar]
- 19.Rohrbeck A, Neukirchen J, Rosskopf M, et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurteau GJ, Spivack SD. mRNA-specific reverse transcription-polymerase chain reaction from human tissue extracts. Anal Biochem. 2002;307:304–315. doi: 10.1016/s0003-2697(02)00058-1. [DOI] [PubMed] [Google Scholar]
- 21.Spivack SD, Hurteau GJ, Fasco MJ, Kaminsky LS. Phase I and II carcinogen metabolism gene expression in human lung tissue and tumors. Clin Cancer Res. 2003;9:6002–6011. [PubMed] [Google Scholar]
- 22.Spivack SD, Hurteau GJ, Reilly AA, Aldous KM, Ding X, Kaminsky LS. CYP1B1 expression in human lung. Drug Metab Dispos. 2001;29:916–922. [PubMed] [Google Scholar]
- 23.Roos PH, Bolt HM. Cytochrome P450 interactions in human cancers: new aspects considering CYP1B1. Expert Opin Drug Metab Toxicol. 2005;1:187–202. doi: 10.1517/17425255.1.2.187. [DOI] [PubMed] [Google Scholar]
- 24.Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- 25.Zhu LR, Thomas PE, Lu G, et al. CYP2A13 in human respiratory tissues and lung cancers: an immunohistochemical study with a new peptide-specific antibody. Drug Metab Dispos. 2006;34:1672–1676. doi: 10.1124/dmd.106.011049. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, D'Agostino J, Wu H, et al. CYP2A13: variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J Pharmacol Exp Ther. 2007;323:570–578. doi: 10.1124/jpet.107.127068. [DOI] [PubMed] [Google Scholar]
- 27.Jalas JR, Hecht SS, Murphy SE. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem Res Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 28.Spivack SD, Hurteau GJ, Jain R, et al. Gene-environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004;64:6805–6813. doi: 10.1158/0008-5472.CAN-04-1771. [DOI] [PubMed] [Google Scholar]
- 29.Coles B, Yang M, Lang NP, Kadlubar FF. Expression of hGSTP1 alleles in human lung and catalytic activity of the native protein variants towards 1-chloro-2,4-dinitrobenzene, 4-vinylpyridine and (+)-anti benzo[a]pyrene-7,8-diol-9,10-oxide. Cancer Lett. 2000;156:167–175. doi: 10.1016/s0304-3835(00)00458-4. [DOI] [PubMed] [Google Scholar]
- 30.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci USA. 1998;95 doi: 10.1073/pnas.95.9.5275. 5275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gate L, Majumdar RS, Lunk A, Tew KD. Influence of glutathione S-transferase pi and p53 expression on tumor frequency and spectrum in mice. Int J Cancer. 2005;113:29–35. doi: 10.1002/ijc.20540. [DOI] [PubMed] [Google Scholar]
- 32.Ritchie KJ, Henderson CJ, Wang XJ, et al. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res. 2007;67:9248–9257. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- 33.Wright GS, Gruidl ME. Early detection and prevention of lung cancer. Curr Opin Oncol. 2000;12:143–8. doi: 10.1097/00001622-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Davis RA. The determination of nicotine and cotinine in plasma. J Chromatogr Sci. 1986;24:134–141. doi: 10.1093/chromsci/24.4.134. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lampe JW, Stepaniants SB, Mao M, et al. Signatures of environmental exposures using peripheral leukocyte gene expression: tobacco smoke. Cancer Epidemiol Biomarkers Prev. 2004;13:445–453. [PubMed] [Google Scholar]
- 39.Willey JC, Coy EL, Frampton MW, et al. Quantitative RT-PCR measurement of cytochromes p450 1A1, 1B1, and 2B7, microsomal epoxide hydrolase, and NADPH oxidoreductase expression in lung cells of smokers and nonsmokers. Am J Respir Cell Mol Biol. 1997;17:114–124. doi: 10.1165/ajrcmb.17.1.2783. [DOI] [PubMed] [Google Scholar]
- 40.Kyerematen GA, Taylor LH, deBethizy JD, Vesell ES. Pharmacokinetics of nicotine and 12 metabolites in the rat. Application of a new radiometric high performance liquid chromatography assay. Drug Metab Dispos. 1988;16:125–129. [PubMed] [Google Scholar]
- 41.Jarvis MJ, Russell MA, Benowitz NL, Feyerabend C. Elimination of cotinine from body fluids: implications for noninvasive measurement of tobacco smoke exposure. Am J Public Health. 1988;78:696–698. doi: 10.2105/ajph.78.6.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foth H, Looschen H, Neurath H, Kahl GF. Nicotine metabolism in isolated perfused lung and liver of phenobarbital- and benzoflavone-treated rats. Arch Toxicol. 1991;65:68–72. doi: 10.1007/BF01973505. [DOI] [PubMed] [Google Scholar]
- 43.Green JT, Evans BK, Rhodes J, et al. An oral formulation of nicotine for release and absorption in the colon: its development and pharmacokinetics. Br J Clin Pharmacol. 1999;48:485–493. doi: 10.1046/j.1365-2125.1999.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantlay AM, Smith CA, Wallace WA, Yap PL, Lamb D, Harrison DJ. Heterogeneous expression and polymorphic genotype of glutathione S-transferases in human lung. Thorax. 1994;49:1010–1014. doi: 10.1136/thx.49.10.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han W, Pentecost BT, Pietropaolo RL, Fasco MJ, Spivack SD. Estrogen receptor alpha increases basal and cigarette smoke extract-induced expression of CYP1A1 and CYP1B1, but not GSTP1, in normal human bronchial epithelial cells. Mol Carcinog. 2005;44:202–211. doi: 10.1002/mc.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cauchi S, Han W, Kumar SV, Spivack SD. Haplotype-environment interactions that regulate the human glutathione S-transferase P1 promoter. Cancer Res. 2006;66:6439–6448. doi: 10.1158/0008-5472.CAN-05-4457. [DOI] [PubMed] [Google Scholar]
- 47.Gazzeri S, Gouyer V, Vour'ch C, Brambilla C, Brambilla E. Mechanisms of p16 INK4A inactivation in non small-cell lung cancers. Oncogene. 1998;16:497–504. doi: 10.1038/sj.onc.1201559. [DOI] [PubMed] [Google Scholar]
- 48.Groeger AM, Caputi M, Esposito V, et al. Independent prognostic role of p16 expression in lung cancer. J Thorac Cardiovasc Surg. 1999;118:529–535. doi: 10.1016/s0022-5223(99)70192-3. [DOI] [PubMed] [Google Scholar]