Abstract
Founder or isolated populations have advantages for genetic studies due to decreased genetic and environmental heterogeneity. However, whereas longer range linkage disequilibrium (LD) in these populations is expected to facilitate gene localization, extensive LD may actually limit the ability for gene discovery. The North American Hutterite population is one of the best characterized young founder populations and members of this isolate have been the subjects of our studies of complex traits, including fertility, asthma and cardiovascular disease, for >20 years. Here, we directly assess the patterns and extent of global LD using single nucleotide polymorphism (SNP) genotypes with minor allele frequencies (MAFs) ≥5% from the Affymetrix GeneChip® Mapping 500K array in 60 relatively unrelated Hutterites and 60 unrelated Europeans (HapMap CEU). Although LD among some marker pairs extends further in the Hutterites than in Europeans, the pattern of LD and minor allele frequencies are surprisingly similar. These results indicate that 1) identifying disease genes should be no more difficult in the Hutterites than in outbred European populations, 2) the same common susceptibility alleles for complex diseases should be present in the Hutterites and outbred European populations, and 3) imputation algorithms based on HapMap CEU should be applicable to the Hutterites.
Introduction
Finding and characterizing variation in the human genome that confers risk for common diseases remains a major challenge in human genetics. The advantages of isolated and founder populations for genetic mapping studies have long been recognized with respect to Mendelian diseases [de la Chapelle 1993; Peltonen 2000], and more recently with respect to common diseases [Heutink and Oostra 2002; Peltonen, et al. 2000]; also see Wright, Carothers et al. [Wright, et al. 1999] for a review of the importance of population choice for study design [Lander and Schork 1994; Sheffield, et al. 1998]. Both reduced genetic heterogeneity due to the small number of founders and the increased linkage disequilibrium (LD) due to recent ancestry facilitated the discovery of many rare Mendelian disease genes in founder populations, such as the Finns (e.g., [Nikali, et al. 1995; Sankila, et al. 1987]), Amish (e.g., [Polymeropoulos, et al. 1996; Ruiz-Perez, et al. 2000]), Hutterites (e.g. [Boycott, et al. 2008; Weiler, et al. 1998]), Ashkenazi Jews (e.g., [Anderson, et al. 2001; Slaugenhaupt, et al. 2001]), Sardinians (e.g., [Verhoeven, et al. 2001]), and Icelanders (e.g., [Gulcher, et al. 1997]). Although these same features, in addition to the generally more uniform lifestyle shared among members of these groups, should also enhance our ability to discover common risk alleles for complex diseases [Heutink and Oostra 2002; Lander and Schork 1994; Newman, et al. 2004], the more extensive LD in these populations may limit the ability to move from gene localization to gene discovery.
Recent technological advances have allowed the incorporation of LD-based genome-wide association studies into the arsenal of mapping tools in many laboratories. Numerous platforms are available that generate dense single nucleotide polymorphism (SNP) genotypes (300,000 to 1 million), and the HapMap [Frazer, et al. 2007] provides the framework for both interpreting the results of genome-wide association studies and imputing genotypes for SNPs not represented on the platforms [Browning and Browning 2007; Marchini, et al. 2007; Nicolae 2006; Servin and Stephens 2007]. However, the applicability of the HapMap resource for interpreting results of genome-wide association studies and for imputing genotypes in isolated populations has not been extensively evaluated (see Bonnen et al. 2006 [Bonnen, et al. 2006] for an exception).
We have focused our genetic mapping studies in the Hutterites of South Dakota. The Hutterites are an Anabaptist religious group established in the Tyrolean Alps in 1528. The more than 1,000 Hutterites in our studies are related to each other in a 13-generation pedigree with 62 founders, The average inbreeding coefficient is approximately 3%, equivalent to that of 1 ½ cousins (first cousins once removed) [Ober, et al. 1998]. Hutterites live communally on large farms, called colonies, resulting in a remarkably similar environment among individuals both within and between colonies and reduces confounding effects of environmental variation. This isolate is one of the best characterized young founder populations and has been the subject of our genetic studies for more than 20 years [Abney, et al. 2002; Gallego Romero and Ober 2008; Newman, et al. 2004; Ober, et al. 2001; Ober, et al. 1987; Ober, et al. 2000; Weiss, et al. 2006].
Here, we present the results of the first genome-wide comparison of LD between the Hutterites and unrelated Europeans (HapMap CEU) using genotype data from the Affymetrix Mapping GeneChip® 500K mapping array. Our results illustrate that the differences in patterns of LD and minor allele frequencies (MAFs) between these two populations are quite subtle and indistinguishable in much of the genome. We conclude that identifying complex diseases genes should be no more challenging in the Hutterites compared to outbred European populations, common susceptibility alleles for complex diseases that are present in outbred European populations should also be present in the Hutterites, and imputation algorithms based on HapMap CEU should be applicable to the Hutterites.
Materials and Methods
For these studies, we selected 60 Hutterites (30 females and 30 males) who are not related to one another as first-degree relatives. For an outbred European comparison group, HapMap CEU, we utilized genotypes for the parents of 30 trios to represent 60 unrelated individuals (30 females and 30 males).
Genotypes for SNPs from the Affymetrix GeneChip ® Mapping 500K Array that were present on both the early access and commercial chip (N=428,867) and had a MAF >5% (N=315,493) in the Hutterites were subject to quality control (QC) checks (for details on our QC checks and SNP composition, see [Ober, et al. 2008]). Briefly, SNPs with Hardy-Weinberg equilibrium P <0.001 (N=5,080), more than five Mendelian errors (N=11,359), and call rates <90% (N=3,617) were further excluded, yielding a final set of 295,437 SNPs. Genotypes for all autosomal QC SNPs in the Hutterites were obtained for the HapMap (CEU) to assess global LD.
In addition to assessments of global (genome-wide) LD, we also investigated the LD pattern in eight discrete regions. First, we selected the longest and shortest q arms (2q and 21q, respectively) in the human genome. Next, we selected four 500 Kb genomic regions on the basis of recombination rate and gene density, and one region each on Xp and Xq from the dataset of Conrad et al. [Conrad, et al. 2006], to represent the most extreme of the first two categories (highest and lowest recombination rates and gene densities) and the two X chromosome arms.
Results
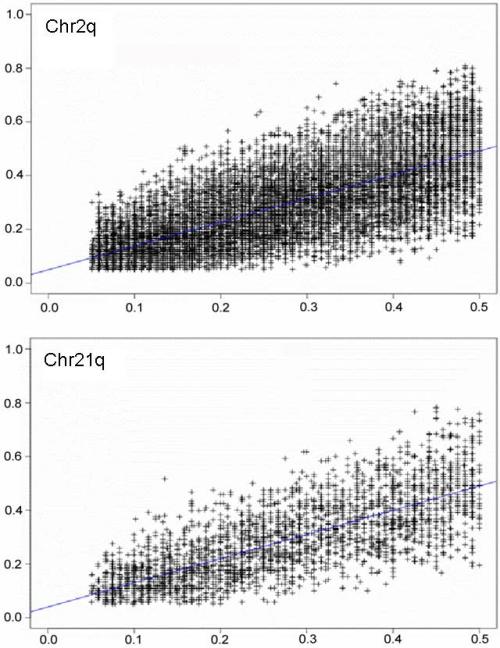
We first examined MAFs in the Hutterites and CEU by comparing the longest (chromosome 2q) and shortest (chromosome 21q) chromosome q-arm. Allele frequencies for SNPs on 2q and 21q are highly correlated in the Hutterites and CEU (r=0.646 and 0.656, respectively), but show wide variation at each MAF (Figure 1). A similar pattern of scatter to that in Figure 1 was seen in simulated data sets where genotypes were generated from populations with identical MAFs (data not shown).
Figure 1.
Comparison of minor allele frequencies between Hutterites and CEU samples on chr2q and chr21q. MAFs in Hutterites (N=60) are plotted on the X axis, and CEU (N=60) appears on the Y axis. Only SNPs with MAF≥5% are included.
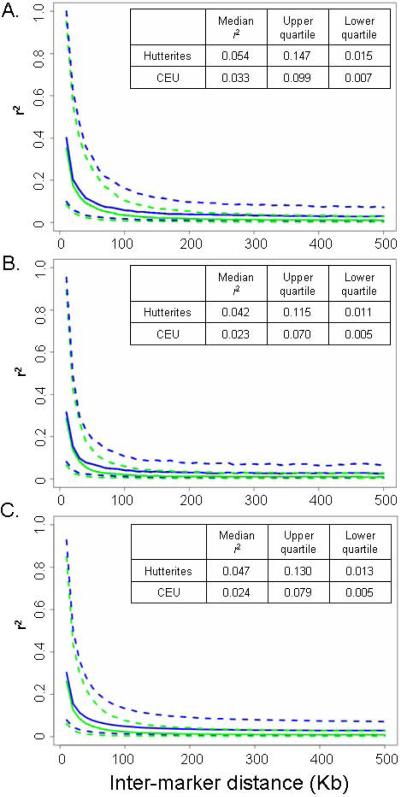
We next investigated patterns of LD between the two populations by comparing chromosomes 2q and 21q as well as the entire genome, for all SNPs within 500Kb windows (Figure 2). Median r2 values at distances greater than 100 Kb are similar for the two populations (solid lines in Figure 2), although LD extends over longer distances in the Hutterites at r2<0.20 (genome-wide median r2=0.047 in the Hutterites vs. 0.024 in CEU; see inset tables, Figure 2).
Figure 2.
Comparison of LD (measured using r2) in CEU (blue lines) and Hutterites (green lines). Solid lines correspond to median values and dotted to upper and lower quartiles. Inter-marker distance (Kb) is shown on the X axis. Pairwise LD values between markers on chromosomes 2q (A), 21q (B), and the whole genome (C), are plotted. r2 values in inset tables are calculated based on all SNP pairs in 10 Kb windows.
The percentages of SNP pairs with strong LD (r2≥0.8) show minor differences in the two populations, even at large inter-marker distances (Table 1). The number of SNP pairs with r2≥0.8 at distances up to 10kb is remarkably similar for more common SNPs (82 and 79% for MAF between 31-40% and 83 and 80% for MAF between 41-50% in Hutterites and CEU, respectively) as well as less frequent SNPs (i.e., MAFs of 5-10%) (89 and 85% in Hutterites and CEU, respectively). Even at distances up to 500kb, the percentage of SNP pairs showing either r2≥0.8 or r2=1 is very similar at MAFs greater than 10%.
Table 1.
Counts and percentages of autosomal SNP pairs showing perfect (r2=1) or strong (r2≥0.8) LD at various inter-marker distances (IMD) in the Hutterites and CEU. For each MAF bin, only SNPs with corresponding MAFs in both populations are included.
| MAF | 5-10% | |||||||
|---|---|---|---|---|---|---|---|---|
| IMD (kb) | r2=1 | r2≥0.8 | ||||||
| HT | CEU | HT | CEU | |||||
| Count | % | Count | % | Count | % | Count | % | |
| ≤10 | 2,653 | 82.73 | 2,332 | 72.72 | 2,867 | 89.40 | 2,735 | 85.28 |
| 10-20 | 1,195 | 68.80 | 895 | 51.53 | 1,315 | 75.71 | 1,177 | 67.76 |
| 20-50 | 1,733 | 57.19 | 1,163 | 38.38 | 2,001 | 66.04 | 1,688 | 55.71 |
| 50-100 | 1,077 | 36.52 | 714 | 24.21 | 1,294 | 43.88 | 1,045 | 35.44 |
| 100-200 | 692 | 16.34 | 375 | 8.86 | 889 | 21.00 | 648 | 15.30 |
| 200-300 | 201 | 5.74 | 78 | 2.23 | 275 | 7.86 | 113 | 3.23 |
| 300-400 | 76 | 2.36 | 28 | 0.87 | 99 | 3.07 | 38 | 1.18 |
| 400-500 | 45 | 1.50 | 8 | 0.27 | 78 | 2.60 | 19 | 0.63 |
| MAF | 11-20% | |||||||
|---|---|---|---|---|---|---|---|---|
| IMD (kb) | r2=1 | r2≥0.8 | ||||||
| HT | CEU | HT | CEU | |||||
| Count | % | Count | % | Count | % | Count | % | |
| ≤10 | 11,887 | 69.07 | 10,193 | 59.23 | 13,631 | 79.20 | 13,141 | 76.36 |
| 10-20 | 4,736 | 49.40 | 3,422 | 35.69 | 5,841 | 60.93 | 5,307 | 55.36 |
| 20-50 | 6,355 | 32.49 | 4,163 | 21.28 | 8,372 | 42.80 | 7,127 | 36.43 |
| 50-100 | 3,667 | 15.35 | 2,220 | 9.29 | 5,414 | 22.67 | 4,127 | 17.28 |
| 100-200 | 2,578 | 6.60 | 1,480 | 3.79 | 3,859 | 9.89 | 2,702 | 6.92 |
| 200-300 | 1,224 | 3.34 | 787 | 2.15 | 1,799 | 4.91 | 1,153 | 3.15 |
| 300-400 | 705 | 2.04 | 538 | 1.55 | 980 | 2.83 | 677 | 1.95 |
| 400-500 | 545 | 1.61 | 489 | 1.44 | 748 | 2.21 | 565 | 1.67 |
| MAF | 21-30% | |||||||
|---|---|---|---|---|---|---|---|---|
| IMD (kb) | r2=1 | r2≥0.8 | ||||||
| HT | CEU | HT | CEU | |||||
| Count | % | Count | % | Count | % | Count | % | |
| ≤10 | 19,123 | 69.10 | 16,024 | 57.90 | 22,243 | 80.38 | 21,537 | 77.82 |
| 10-20 | 7,370 | 49.30 | 5,170 | 34.59 | 9,306 | 62.26 | 8,495 | 56.83 |
| 20-50 | 9,421 | 31.26 | 5,956 | 19.76 | 12,977 | 43.06 | 11,114 | 36.87 |
| 50-100 | 5,221 | 14.27 | 2,968 | 8.11 | 8,049 | 22.00 | 6,309 | 17.24 |
| 100-200 | 3,201 | 5.35 | 1,746 | 2.92 | 5,170 | 8.65 | 3,753 | 6.28 |
| 200-300 | 1,354 | 2.41 | 814 | 1.45 | 2,135 | 3.81 | 1,399 | 2.49 |
| 300-400 | 720 | 1.35 | 541 | 1.02 | 1,083 | 2.03 | 716 | 1.34 |
| 400-500 | 552 | 1.07 | 492 | 0.96 | 805 | 1.56 | 617 | 1.20 |
| MAF | 31-40% | |||||||
|---|---|---|---|---|---|---|---|---|
| IMD (kb) | r2=1 | r2≥0.8 | ||||||
| HT | CEU | HT | CEU | |||||
| Count | % | Count | % | Count | % | Count | % | |
| ≤10 | 25,439 | 69.47 | 21,200 | 57.90 | 29,885 | 81.62 | 29,055 | 79.35 |
| 10-20 | 9,477 | 49.00 | 6,552 | 33.88 | 12,241 | 63.30 | 11,243 | 58.14 |
| 20-50 | 11,816 | 30.87 | 7,282 | 19.02 | 16,786 | 43.85 | 14,544 | 37.99 |
| 50-100 | 6,326 | 13.68 | 3,425 | 7.41 | 10,351 | 22.39 | 8,141 | 17.61 |
| 100-200 | 3,719 | 4.94 | 1,949 | 2.59 | 6,292 | 8.36 | 4,800 | 6.37 |
| 200-300 | 1,465 | 2.10 | 822 | 1.18 | 2,350 | 3.37 | 1,564 | 2.24 |
| 300-400 | 721 | 1.09 | 541 | 0.81 | 1,102 | 1.66 | 735 | 1.11 |
| 400-500 | 552 | 0.86 | 492 | 0.76 | 812 | 1.26 | 618 | 0.96 |
| MAF | 41-50% | |||||||
|---|---|---|---|---|---|---|---|---|
| IMD (kb) | r2=1 | r2≥0.8 | ||||||
| HT | CEU | HT | CEU | |||||
| Count | % | Count | % | Count | % | Count | % | |
| ≤10 | 30,168 | 69.79 | 24,942 | 57.70 | 35,678 | 82.54 | 34,752 | 80.40 |
| 10-20 | 10,877 | 49.10 | 7,374 | 33.29 | 14,218 | 64.18 | 13,108 | 59.17 |
| 20-50 | 13,467 | 30.57 | 8,123 | 18.44 | 19,645 | 44.59 | 17,078 | 38.77 |
| 50-100 | 7,058 | 13.47 | 3,798 | 7.25 | 11,982 | 22.86 | 9,526 | 18.17 |
| 100-200 | 4,011 | 4.75 | 2,072 | 2.45 | 7,127 | 8.44 | 5,481 | 6.49 |
| 200-300 | 1,526 | 1.96 | 838 | 1.07 | 2,598 | 3.33 | 1,727 | 2.21 |
| 300-400 | 756 | 1.02 | 542 | 0.73 | 1,229 | 1.65 | 867 | 1.17 |
| 400-500 | 583 | 0.81 | 492 | 0.69 | 881 | 1.23 | 687 | 0.96 |
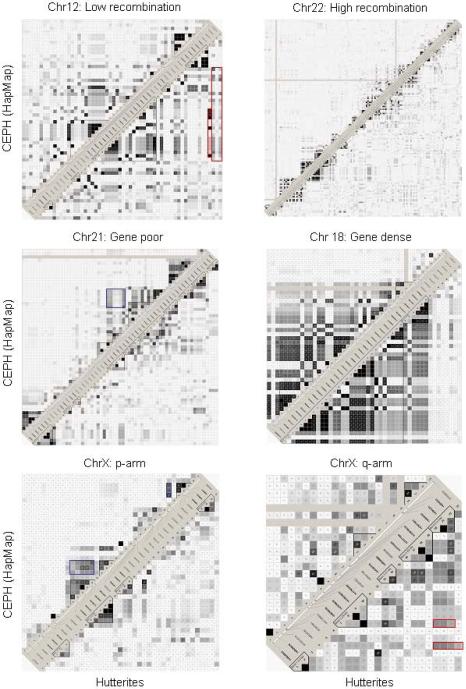
Six discrete regions representing different chromosomal features were selected for comparison between the Hutterite and CEU (Figure 3; for details regarding each region see Table S1). Similar to the genome-wide patterns discussed above, patterns of LD in the six regions are remarkably similar between the two populations. While there are some instances of stronger LD between distant marker pairs in the Hutterites (for example, the q-arm of chromosome X and the region of low recombination on chromosome 12; outlined in red), there are also examples of stronger LD between marker pairs in the CEU sample (gene-poor region of chromosome 21 and p-arm of chromosome X, outlined in blue). These results suggest that while a particular pair of markers may show differences in LD between populations, overall, the CEU and Hutterite populations are not characterized by large scale differences in the distribution and extent of LD. Thus, patterns of LD in the HapMap CEU samples should be informative for imputing genotypes and patterns of LD in the Hutterites, for the majority of variation in the genome.
Figure 3.
Comparisons of LD between SNPs in 500 Kb regions in 60 CEU and 60 Hutterite individuals. Hutterite data appear on the lower right and CEU on the upper left of each panel. Examples of regions that show greater LD in Hutterites or CEU are boxed in red or blue, respectively (see Results). Solid gray lines extending the length of the region in the HapMap samples indicate missing data. The genomic coordinates for the six regions can be found in Table S1.
DISCUSSION
Despite the extreme bottleneck in the Hutterite population and the small number of founding genomes that have given rise to the current population [Hostetler 1985], the allele frequency spectrum and pattern of LD are remarkably similar to modern Europeans. This may be partially due to the rapid population expansion following their arrival in North America: the population grew from about 116 individuals when they settled in Russia in the 1700's to an estimated 40,000-50,000 members living today in North America [Hostetler 1985].
Simulation studies predicted that rapid population growth following a founding event can reduce LD in a population [Slatkin 1994], and unexpected decays of LD over relatively short distances have been attributed to rapid population growth in other relatively young genetic isolates [Katoh, et al. 2002; Laan and Paabo 1997]. In addition, high-frequency polymorphisms, which we focused on in this study, are likely to be ancient in origin and to have been present in more than one ancestor and on multiple haplotypes at the time of the founding. Thus, as suggested by simulations, recent demographic events may have little impact on the extent of LD between common SNPs [Kruglyak 1999; Pritchard and Przeworski 2001]. This is supported by empirical studies of LD in populations with different demographic histories but of shared origins [Dunning, et al. 2000; Eaves, et al. 2000; Pardo, et al. 2009; Taillon-Miller, et al. 2000; Xing, et al. 2008].
Only one other recent study has examined genome-wide patterns of LD in a founder population, the Kosrae of Micronesia [Bonnen, et al. 2006]. In that study, pairwise LD between and MAFs for ~110,000 SNPs were compared between 30 Kosraen trios (KOS) and 30 trios from each HapMap population (CEU, YRI, JPT, CHB). Allele frequency distributions were similar for SNPs with MAFs ≥15%, although the proportion of SNP pairs with r2 > 0.8 was higher in the KOS than in the HapMap samples for SNPs with MAF > 0.15, using the (then) current technology could still cover 78% of the SNPs in the genome with high (r2 > 0.8) efficiency [Bonnen, et al. 2006]. Thus, similar to the results of our studies [and the other reports in this issue], we conclude that while differences between isolated and outbred populations may exist on a local scale, global patterns LD, as well as MAF distributions, are likely to be similar for common SNPs across populations with very different demographic histories.
In conclusion, we suggest that common susceptibility alleles for complex diseases will be shared by the Hutterites and outbred European populations (for examples, see [Newman, et al. 2004; Ober, et al. 2008; Weiss, et al. 2005] and that identifying these risk variants should be no more difficult in the Hutterites than in outbred European populations. Lastly, the similar patterns of LD in the Hutterites and HapMap CEU further suggest that imputation algorithms based on LD patterns in HapMap CEU individuals are applicable to the Hutterites. On the other hand, the homogeneous environment that results from their communal lifestyle should reduce environmental and lifestyle heterogeneity, and may facilitate gene discovery for complex diseases in the Hutterites in smaller sample sizes than those required for gene discovery in outbred individuals.
Supplementary Material
Acknowledgments
GRANT SUPPORT: R01 HL085197.
References
- Abney M, Ober C, McPeek MS. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet. 2002;70(4):920–34. doi: 10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68(3):753–8. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnen PE, Pe'er I, Plenge RM, Salit J, Lowe JK, Shapero MH, Lifton RP, Breslow JL, Daly MJ, Reich DE. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38(2):214–7. doi: 10.1038/ng1712. others. [DOI] [PubMed] [Google Scholar]
- Boycott KM, Parboosingh JS, Chodirker BN, Lowry RB, McLeod DR, Morris J, Greenberg CR, Chudley AE, Bernier FP, Midgley J. Clinical genetics and the Hutterite population: a review of Mendelian disorders. Am J Med Genet A. 2008;146A(8):1088–98. doi: 10.1002/ajmg.a.32245. others. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38(11):1251–60. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A. Disease gene mapping in isolated human populations: the example of Finland. J Med Genet. 1993;30(10):857–65. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Durocher F, Healey CS, Teare MD, McBride SE, Carlomagno F, Xu CF, Dawson E, Rhodes S, Ueda S. The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet. 2000;67(6):1544–54. doi: 10.1086/316906. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA. The genetically isolated populations of Finland and sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet. 2000;25(3):320–3. doi: 10.1038/77091. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Ober C. CFTR mutations and reproductive outcomes in a population isolate. Hum Genet. 2008;122(6):583–8. doi: 10.1007/s00439-007-0432-1. [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Jonsson P, Kong A, Kristjansson K, Frigge ML, Karason A, Einarsdottir IE, Stefansson H, Einarsdottir AS, Sigurthoardottir S. Mapping of a familial essential tremor gene, FET1, to chromosome 3q13. Nat Genet. 1997;17(1):84–7. doi: 10.1038/ng0997-84. others. [DOI] [PubMed] [Google Scholar]
- Heutink P, Oostra BA. Gene finding in genetically isolated populations. Hum Mol Genet. 2002;11(20):2507–15. doi: 10.1093/hmg/11.20.2507. [DOI] [PubMed] [Google Scholar]
- Hostetler JA. History and relevance of the Hutterite population for genetic studies. Am J Med Genet. 1985;22(3):453–62. doi: 10.1002/ajmg.1320220303. [DOI] [PubMed] [Google Scholar]
- Katoh T, Mano S, Ikuta T, Munkhbat B, Tounai K, Ando H, Munkhtuvshin N, Imanishi T, Inoko H, Tamiya G. Genetic isolates in East Asia: a study of linkage disequilibrium in the X chromosome. Am J Hum Genet. 2002;71(2):395–400. doi: 10.1086/341608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22(2):139–44. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Laan M, Paabo S. Demographic history and linkage disequilibrium in human populations. Nat Genet. 1997;17(4):435–8. doi: 10.1038/ng1297-435. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Newman DL, Hoffjan S, Bourgain C, Abney M, Nicolae RI, Profits ET, Grow MA, Walker K, Steiner L, Parry R. Are common disease susceptibility alleles the same in outbred and founder populations? Eur J Hum Genet. 2004;12(7):584–90. doi: 10.1038/sj.ejhg.5201191. others. [DOI] [PubMed] [Google Scholar]
- Nicolae DL. Testing untyped alleles (TUNA)-applications to genome-wide association studies. Genet Epidemiol. 2006;30(8):718–27. doi: 10.1002/gepi.20182. [DOI] [PubMed] [Google Scholar]
- Nikali K, Suomalainen A, Terwilliger J, Koskinen T, Weissenbach J, Peltonen L. Random search for shared chromosomal regions in four affected individuals: the assignment of a new hereditary ataxia locus. Am J Hum Genet. 1995;56(5):1088–95. [PMC free article] [PubMed] [Google Scholar]
- Ober C, Abney M, McPeek MS. The genetic dissection of complex traits in a founder population. Am J Hum Genet. 2001;69(5):1068–79. doi: 10.1086/324025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Bombard A, Dhaliwal R, Elias S, Fagan J, Laffler TG, Martin AO, Rosinsky B. Studies of cystic fibrosis in Hutterite families by using linked DNA probes. Am J Hum Genet. 1987;41(6):1145–51. [PMC free article] [PubMed] [Google Scholar]
- Ober C, Hyslop T, Elias S, Weitkamp LR, Hauck WW. Human leukocyte antigen matching and fetal loss: results of a 10 year prospective study. Hum Reprod. 1998;13(1):33–8. doi: 10.1093/humrep/13.1.33. [DOI] [PubMed] [Google Scholar]
- Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–91. doi: 10.1056/NEJMoa0708801. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67(5):1154–62. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo L, Bochdanovits Z, de Geus E, Hottenga JJ, Sullivan P, Posthuma D, Penninx BW, Boomsma D, Heutink P. Global similarity with local differences in linkage disequilibrium between the Dutch and HapMap-CEU populations. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L. Positional cloning of disease genes: advantages of genetic isolates. Hum Hered. 2000;50(1):66–75. doi: 10.1159/000022892. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1(3):182–90. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Ide SE, Wright M, Goodship J, Weissenbach J, Pyeritz RE, Da Silva EO, Ortiz De Luna RI, Francomano CA. The gene for the Ellis-van Creveld syndrome is located on chromosome 4p16. Genomics. 1996;35(1):1–5. doi: 10.1006/geno.1996.0315. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69(1):1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Ide SE, Strom TM, Lorenz B, Wilson D, Woods K, King L, Francomano C, Freisinger P, Spranger S. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat Genet. 2000;24(3):283–6. doi: 10.1038/73508. others. [DOI] [PubMed] [Google Scholar]
- Sankila EM, de la Chapelle A, Karna J, Forsius H, Frants R, Eriksson A. Choroideremia: close linkage to DXYS1 and DXYS12 demonstrated by segregation analysis and historical-genealogical evidence. Clin Genet. 1987;31(5):315–22. doi: 10.1111/j.1399-0004.1987.tb02815.x. [DOI] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3(7):e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Stone EM, Carmi R. Use of isolated inbred human populations for identification of disease genes. Trends Genet. 1998;14(10):391–6. doi: 10.1016/s0168-9525(98)01556-x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics. 1994;137(1):331–6. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68(3):598–605. doi: 10.1086/318810. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet. 2000;25(3):324–8. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- Verhoeven K, Villanova M, Rossi A, Malandrini A, De Jonghe P, Timmerman V. Localization of the gene for the intermediate form of Charcot-Marie-Tooth to chromosome 10q24.1-q25.1. Am J Hum Genet. 2001;69(4):889–94. doi: 10.1086/323742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler T, Greenberg CR, Zelinski T, Nylen E, Coghlan G, Crumley MJ, Fujiwara TM, Morgan K, Wrogemann K. A gene for autosomal recessive limb-girdle muscular dystrophy in Manitoba Hutterites maps to chromosome region 9q31-q33: evidence for another limb-girdle muscular dystrophy locus. Am J Hum Genet. 1998;63(1):140–7. doi: 10.1086/301925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Lester LA, Gern JE, Wolf RL, Parry R, Lemanske RF, Solway J, Ober C. Variation in ITGB3 is associated with asthma and sensitization to mold allergen in four populations. Am J Respir Crit Care Med. 2005;172(1):67–73. doi: 10.1164/rccm.200411-1555OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat Genet. 2006;38(2):218–22. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23(4):397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Watkins WS, Zhang Y, Tolpinrud W, Jorde LB. HapMap tagSNP transferability in multiple populations: general guidelines. Genomics. 2008;92(1):41–51. doi: 10.1016/j.ygeno.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.