Abstract
Rationale
Though treatment of the prematurely born infant breathing with assistance of a mechanical ventilator has much advanced in the past decades, predicting extubation outcome at a given point in time remains challenging. Numerous studies have been conducted to identify predictors for extubation outcome; however, the rate of infants failing extubation attempts has not declined.
Objective
To develop a decision-support tool for the prediction of extubation outcome in premature infants using a set of machine learning algorithms
Methods
A dataset assembled from 486 premature infants on mechanical ventilation was used to develop predictive models using machine learning algorithms such as artificial neural networks (ANN), support vector machine (SVM), naïve Bayesian classifier (NBC), boosted decision trees (BDT), and multivariable logistic regression (MLR). Performance of all models was evaluated using area under the curve (AUC).
Results
For some of the models (ANN, MLR and NBC) results were satisfactory (AUC: 0.63–0.76); however, two algorithms (SVM and BDT) showed poor performance with AUCs of ~0.5.
Conclusion
Clinician's predictions still outperform machine learning due to the complexity of the data and contextual information that may not be captured in clinical data used as input for the development of the machine learning algorithms. Inclusion of preprocessing steps in future studies may improve the performance of prediction models.
Keywords: Premature infant, mechanical ventilation, extubation, prediction, machine learning
Introduction/Background
Though treatment of the prematurely born infant breathing with assistance of a mechanical ventilator has much advanced in the past decades, predicting extubation outcome at a given point in time remains challenging. Numerous studies have been conducted to identify predictors for extubation outcome; however, the rate of infants failing extubation attempts has not declined [1–5].
After promising results using Artificial Neural Networks (ANN) to determine the most important predictors for extubation success that resulted in an ANN predicting which infant would succeed an extubation attempt with 85% accuracy [6], our team used similar methods attempting to further improve the previously achieved results. The goal of this study was to develop a decision-support tool using a heterogeneous set of machine learning algorithms for the determination of whether or not a given infant should be extubated at a given time point. Algorithms included artificial neural networks (ANN), support vector machines (SVM), naïve Bayesian classifiers (NBC), boosted decision trees (BDT), and multivariable logistic regression (MLR). The intent of this study was to use the individual prediction from its different algorithms to determine an overall prediction providing better generalization and performance in the combined results compared to the individual predictions [7]. It was hypothesized that providing a large amount of data would enable a set of algorithms to return predictions for unseen data with a high level of accuracy.
Methods
Data collection
After receiving approval from the local IRB (HR#18064) in a first step, 682 potentially eligible babies born at the Medical University of South Carolina (MUSC) between January 2005 and September 2009 were identified from the MUSC Perinatal Information System (PINS) database.
Infants were identified to be potentially eligible on the basis of having been mechanically ventilated and having a diagnosis of RDS. In a second step, a trained data abstractor with more than twenty years experience as a neonatal intensive care nurse accessed each infant's medical record to collect study specific variables, including demographic characteristics of the infant (such as age in days, gender, race/ethnicity, gestational age, birth weight and weight at extubation), clinical characteristics (such as Apgar scores at 1 and 5 minutes, heart rate, respiratory rate, blood pressure), medication (maternal: betamethasone and infants: surfactant, saline [given for hypotension as decided by the clinical team], methylxanthines), ventilator information (including time to intubation from birth, time from last blood gas until extubation, type of ventilator, and ventilator settings at extubation and at the last time point prior to extubation, blood gas values prior to, at and after extubation), whether the extubation was successful or failed, and type of ventilatory support infants received after extubation within 48 and 72 hours.
Following clinical guidelines for ventilatory management of the preterm infant, clinicians, nurses and respiratory therapists worked in concert to wean ventilatory and oxygen support (see Appendix I). The decision to extubate was made by the clinical team based on the set of criteria specified in the guidelines. When these criteria were met by the preterm infant, the infant was extubated to either nasal CPAP, nasal cannula or room air, the type of post-extubation support being dictated by the work of breathing, gestational age, and the oxygen requirements of that infant at the time of extubation.
Study sample
Infants were included in the study if they were born prematurely; had a birth weight between 500 and 2000 grams; had a primary diagnosis of RDS confirmed radiographically; and were intubated and managed on a ventilator within 6 hours after birth. Infants were excluded if they had chromosomal, surgical or congenital anomalies; had comorbidities such as pulmonary hypertension; were on high frequency ventilation; or had life support withdrawn without a previous attempt to extubate.
Statistical methods
Means, standard deviations and proportions are reported to describe the study sample. For comparison between the group of infants who failed their first attempt to extubate and the group of infants who were extubated successfully, t-tests were used for continuous variables and chi-square tests for categorical variables.
Machine learning
A heterogeneous set of algorithms to predict extubation outcome was chosen. Such a set allows for better generalization and performance of the combined prediction compared to individual results since each of these algorithms provides different strengths through their diverse mathematical approaches. In addition, these algorithms allow for nonlinear relationships between variables without the need to explicitly specify them. Missing data were imputed based on weight category and variable type using mean or median.
Algorithms used in this study included multivariable logistic regression (MLR), artificial neural networks (ANNs), support vector machines (SVM), naïve Bayesian classifier (NBC), and bagged decision trees (BDT).
Multivariable Logistic Regression (MLR) is used to predict some event from several independent variables using a logistic function. This function can take inputs with any value from negative infinity to positive infinity and produces an output between 0 and 1, expressed as a probability. An advantage of this method is its ease of interpretation [8].
Artificial neural networks (ANNs) are modeled after the brain by using layers of so-called neurons that are connected within and between layers. In a simplified form, ANNs use multiple logistic regression models in parallel and in sequence. Advantages of ANNs are that relationships between variables do not have to be pre-specified and can be non-linear as ANNs learn from data [9]. ANNS have been known as “black boxes” that are difficult to interpret; however, in recent years software provides measures of sensitivity indicating the importance of individual variables in the ANN model used for prediction of a given outcome [6].
Similarly, Support Vector Machines (SVM) are used to assign events to one of two classes (e.g. infant failed/did not fail extubation). SVMs can be thought of as representing points in a more dimensional space where the two classification categories are clearly separated by a gap between the categories. This gap is called a hyperplane (or a set of hyperplanes), which is positioned in a way that both classes of points have the largest possible distance from the hyperplane. SVMs have simple geometric explanations and are less prone to overfitting [10].
The Naïve Bayesian Classifier (NBC) is a simple, probabilistic method to classify data using the Bayes theorem. With this theorem, the probability to classify an event into a class (A) given a set of parameters (B) equals the probability for a class multiplied by the probability for a set of variables given a certain class divided by the probability for the set of variables:
This classifier assumes that all variables are independent from each other. NBCs can be trained on relatively small data sets and perform well despite the naïve design [11].
Traditionally, decision trees (DTs) were created manually as a tree-like structure by branching into alternatives for each subsequent variable and expected values for each alternative, then were calculated. DTs are easy to interpret and understand and require little data; however, they are prone to produce very different results for small differences in data. Bagging, which is a bootstrapping method, i.e. samples repeatedly from the same data set with replacement, is used as a method to improve accuracy of the DTs and reduce the susceptibility to small disturbances in the data. In the Bagged Decision Tree (BDT) method a set of DTs is generated with differing subsets of data. The final classification result of the set of trees is determined through the average over all individual trees [12]. A detailed description of these algorithms including their parameters as well as the feature selection procedure employed and number of features included can be found in Mueller et al. [13].
For the purpose of this study, all algorithms were developed using MATLAB (Version R2009b, Copyright 1984–2009, The MathWorks Inc.) with 100 data sets that were created through resampling. For this process we repeatedly (100 times) randomly split the total sample into 2/3 vs. 1/3 for training and validation data and applied each of the algorithms to each dataset. The median performer for each algorithm among the 100 applications was determined using the area under the curve (AUC) obtained from Receiver Operating Characteristic (ROC) curves. Similarly, performance of all algorithms was compared using AUCs from training and validation data.
In addition to the main data set, several different sub data sets were used as described above. These subsets were created based on: a) birth weight (<1000g vs. ≥1000g and 500g–999g vs. 1000g–1499g vs. 1500g–1999g); b) birth year (2006–2007 vs. 2008–2009); c) use of weaning protocol (yes vs. no); d) correlation - variables that were highly correlated were excluded (for example birth weight vs. current weight, lag time from last blood gas to extubation vs. lag time between last two blood gases, HCO3 vs. BE); e) Principal Components Analysis - variables were excluded if loadings were below 0.4, 0.35 and 0.3; and f) tests for statistical significance (t-tests/chi-square tests) – variables were excluded if p was found to be above 0.1 (table 1). For these subsets the same resampling procedure was used as for the original data set described above.
Table 1.
Demographic and clinical characteristics of complete study population by extubation outcome.
| Succeeded extubation (n=427) | Failed extubation (n=59) | p-value | |
|---|---|---|---|
| Gestational age (weeks) | 28.8 ± 2.4 | 27.2 ± 2.3 | <0.0001 |
| Birth weight (grams) | 1212.8 ± 350.8 | 929.2 ± 325.6 | <0.0001 |
| Weight at extubation (grams) | 1238.2 ± 352.8 | 948.6 ± 322.9 | <0.0001 |
| Gender (Females) | 51.5% (220/427) | 49.2% (29/59) | 0.733 |
| Race/Ethnicity | 0.844 | ||
| White | 46.5% (226/427) | 52.5% (31/59) | |
| African-American or Black | 39.3% (168/427) | 39.0% (23/59) | |
| Hispanic | 6.8% (29/427) | 6.8% (4/59) | |
| Asian | 0.9% (4/427) | 1.7% (1/59) | |
| APGAR at 1 minute | 5.1 ± 2.4 | 4.8 ± 2.3 | 0.291 |
| APGAR at 5 minutes | 8.1 ± 9.0 | 6.9 ± 2.0 | 0.013 |
| FiO2(%) | 23.7 ± 4.7 | 26.0 ± 5.9 | 0.006 |
| Rate (ventilated breaths per minute) | 21.0 ± 5.4 | 21.4 ± 6.9 | 0.604 |
| Peak Inspiratory Pressure (PIP; cm H20) | 15.3 ± 1.2 | 14.9 ± 1.6 | 0.113 |
| Positive End Expiratory Pressure (PEEP; cm H20) | 4.2 ± 0.5 | 4.2 ± 0.5 | 0.377 |
| Mean Airway Pressure (MAP; cm H20) | 6.2 ± 0.9 | 6.2 ± 0.9 | 0.694 |
| Inspiratory time (seconds) | 0.39 ± 0.04 | 0.37 ± 0.06 | 0.043 |
| I:E ratio | 3.0 ± 2.0 | 3.1 ± 2.2 | 0.708 |
| Tidal Volume (VT; mL) | 5.1 ± 2.9 | 3.7 ± 1.9 | <0.0001 |
| Minute volume (mL) | 0.5 ± 0.3 | 0.3 ± 0.2 | <0.0001 |
| Pressure support (cm H20) | 3.6 ± 2.8 | 3.7 ± 2.8 | 0.746 |
| pH | 7.35 ± 0.06 | 7.33 ± 0.04 | 0.0003 |
| PaCO2 | 41.2 ± 7.2 | 43.6 ± 6.9 | 0.014 |
| PaO2 | 49.9 ± 18.1 | 47.3 ± 13.8 | 0.191 |
| SaO2 | 96.3 ± 3.3 | 94.6 ± 3.5 | 0.0002 |
| HCO3 | 22.8 ± 3.4 | 23.0 ± 3.4 | 0.668 |
| Base excess | −2.8 ± 2.7 | −3.1 ± 3.4 | 0.556 |
| Over-ventilated (PCO2<35) | 12.7% (54/427) | 8.5% (5/59) | 0.522 |
| Balanced pattern* | 92.0% (393/427) | 91.5% (54/59) | 0.802 |
| Heart rate (beats per minute) | 145.3 ± 13.5 | 152.1 ± 15.2 | 0.0003 |
| Blood pressure (mm Hg) | 42.1 ± 9.7 | 39.6 ± 8.5 | 0.057 |
| Rate ratio (spontaneous breathing / ventilatory rate) | 2.1 ± 1.0 | 1.8 ± 1.0 | 0.097 |
| Maternal betamethasone (yes) | 67.7% (348/514) | 77.6% (52/67) | 0.067 |
| Surfactant (# of dosages prior to extubation) | 1.6 ± 1.0 | 2.1 ± 1.4 | 0.004 |
| Saline bolus (yes) | 12.9% (55/427) | 6.8% (4/59) | 0.208 |
| Methylxanthines (yes) | 67.2% (287/427) | 83.1% (49/59) | 0.014 |
| Lag time (from last blood gas analysis to extubation) | 146.3 ± 206.8 | 225.0 ± 266.3 | 0.033 |
| Weaning protocol† followed (yes) | 56.7% (242/427) | 59.3% (35/59) | 0.700 |
Balanced pattern of ventilator settings. For example, the infant may have weaned to room air (i.e. FiO2 = 21%) but is still requiring a high peak inspiratory pressure to maintain oxygenation.
Weaning protocol included in Appendix I
The goal of this study was to provide clinicians with a decision-support tool for the prediction of extubation outcome in artificially ventilated premature infants using a set of heterogeneous machine learning algorithms.
Results
Data on 682 potentially eligible infants were obtained from the PINS database from January 2006 to September 2009. Of those, 196 infants were excluded for the following reasons: 95 preterm infants had a birth weight greater than 2000 grams, 47 infants were intubated post 6 hours after birth; 5 infants did not receive mechanical ventilation; 7 infants required surgical intervention; 4 infants had a diagnosis of congenital anomaly(ies); 20 infants had support withdrawn prior to the first extubation attempt; 5 infants were extubated from ventilators different than SIMV (HFOV or HFJV); and medical records of 13 infants were incomplete and data were unobtainable.
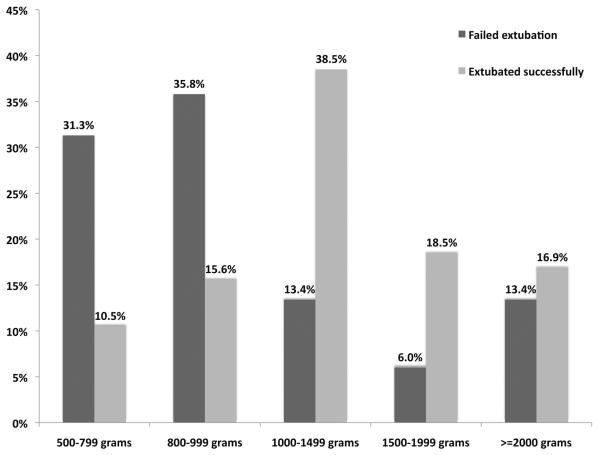
Of the remaining 486 infants, ventilator and blood gas data were obtained from the medical record. Of those, 59 (12.1%) infants failed extubation, 53% were White, 49% were Male; mean gestational age was 28.6 weeks, mean birth weight was 1178 grams. Infants failing extubation were born on average 1.5 weeks earlier compared to infants who were extubated successfully (gestational age 27.2 ± 2.3 vs. 28.8 ± 2.4, p<0.0001). Consequently, birth weight for infants who failed their first extubation attempt was lower (929 ± 326 grams) compared to infants who were extubated successfully (1213 ± 351 grams; p<0.0001). Figure 1 depicts distributions of birth weight for infants who were extubated successfully versus those who failed. Among ventilator settings, tidal volume (VT) immediately prior to extubation was statistically significantly higher in infants who succeeded their first extubation attempt than in infants who failed (5.1 ± 2.9 vs. 3.7 ± 1.9; p<0.0001). PaCO2, pH and SaO2 were statistically significantly different between the two groups: pH and SaO2 were higher, while PaCO2 was lower for infants succeeding their first extubation compared to those who failed (table 1, p<0.05). Infants who failed extubation received more dosages of surfactant prior to extubation compared to infants who did not fail (2.1 ± 1.3 vs. 1.6 ± 1.1; p=0.01). Time between last blood gas analysis and extubation was statistically significantly shorter in the group that was extubated successfully compared to the group that failed extubation (142 +/− 200 min vs. 211 +/− 254 min, p=0.04).
Figure 1.
Distribution of birth weight categories by extubation outcome
In Table 2, findings after extubation are reported. Of all 486 infants included in this study, 59 (12.1%) infants failed extubation within 48 hours and 69 (14.0%) failed within 72 hours. One infant was extubated unintentionally and needed reintubation. Among infants who had failed the first extubation attempt, 17% succeeded in a second attempt within 72 hours of the first. Among the infants who were considered extubated successfully at 48 hours, only 10 (2.3%) ultimately needed reintubation within 72 hours.
Table 2.
Characteristics at or after extubation
| Succeeded extubation (n=427) | Failed extubation (n=59) | p-value | |
|---|---|---|---|
| Extubated | 87.9% (427/486) | 12.1% (59/486) | - |
| Extubated unintentionally | 0 | 1.7% (1/59) | - |
| Escalation of ventilatory support within 48 hours of extubation | 13.2% (56/424)* | 98.3% (58/59) | <0.0001 |
| Extubation success at 72 hours post extubation | 97.7% (417/427) | 17.0 (10759)† | <0.0001 |
| Highest level of support prior to extubation: | 0.100 | ||
| SIMV | 95.7% (404/422)* | 89.8% (53/59) | |
| HFOV/HFJV | 4.3% (18/422)* | 10.2% (6/59) | |
| Number of days at highest level of ventilatory support (mean ± std) | 2.6 ± 5.4 | 3.3 ± 4.7 | 0.358 |
| Age at extubation (days) (mean ± std) | 3.2 ± 8.0 | 4.6 ± 8.8 | 0.213 |
| Day of life regained birth weight (mean ± std) | 9.6 ± 5.0 | 7.9 ± 5.4 | 0.016 |
| Extubated to: | 0.204 | ||
| Room air | 5.0% (21/422)* | 0 | |
| Nasal cannula | 11.4% (48/422)* | 10.2% (6/59) | |
| CPAP | 83.7% (353/422)* | 89.8% (53/59) |
Information not available for several infants
After additional extubation attempt
Thirteen percent of infants in the group who extubated successfully required escalated ventilatory support, i.e., reintubation with increased FiO2 and PIP within 48 hours of extubation compared to 98% in the group that failed (p<0.0001). More infants in the group that failed extubation were extubated to CPAP and no infants were extubated to room air compared to the group that was extubated successfully, though these differences were not statistically significant (p=0.2). Number of days at highest level of ventilatory support was similar between the groups (p=0.4); while approximately 10% of infants who failed extubation had HFOV or HFJV as highest level of ventilatory support prior to extubation compared to only 4% of infants who did not fail (p=0.1). Reasons for extubation failure were primarily apnea of prematurity, increased work of breathing, marked increase of O2 requirements and CO2 retention (table 3).
Table 3.
Reasons for extubation failure (more than one possible)*
| Reasons for failure: | N=56 % (n) |
|---|---|
| Apnea of prematurity | 48.2 (27) |
| Recurrence of RDS | 5.4 (3) |
| Respiratory failure† | 7.1 (4) |
| CO2 retention | 14.3 (8) |
| Frequent desaturations‡ | 7.1 (4) |
| Marked increase in O2 requirements | 14.3 (8) |
| Increased work of breathing | 35.7 (20) |
| Pulmonary interstitial emphysema (PIE) | 1.8 (1) |
| Bradycardia | 1.8 (1) |
| Pneumothorax | 1.8 (1) |
Information not available for several infants
Acute respiratory failure defined as PCO2>55 along with increased work of breathing (tachypnea, costal and/or subcostal retractions) and increasing FiO2 requirement above 50% to maintain saturations of at least 88% or higher
Frequent desaturations defined as > 1/hour
Machine learning
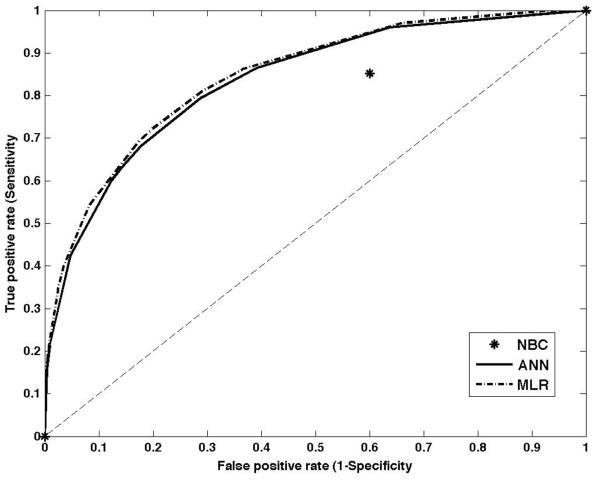
Table 4 reports performance of the different prediction methods as measured by the area under the curve (AUC) determined from the Receiver Operating Characteristic (ROC) curves using the full data set for training and validation. Figure 2 displays results using the validation set for three methods: for ANN and MLR methods ROC curves are depicted; for NBC a single prediction point (except 0 and 1) is displayed. As shown in table 4 several algorithms performed with high accuracy for the training data. The high accuracy for the training sets results from overfitting of the algorithms to the available data. When models are overfitting to data, the methods predict the known outcomes in the training data set extremely well but at the same time generalizability is decreased which means that the methods exhibit diminished capability to predict outcomes for the validation set or future data. This reduced generalizability is reflected in the poor performance in the validation data with two of the methods resulting in AUCs close to 0.5, i.e., with a 50/50 chance to predict the correct outcome. Only ANN and MLR showed satisfactory performance for the validation data with MLR having slightly higher AUC than ANN. None of the methods performed above 0.8, which would be considered minimally acceptable performance for this population.
Table 4.
Performance of algorithms as measured by the area under the curve (AUC) for dataset including all variables (median AUC from 100 resampling steps).
| Algorithm | Training | Validation |
|---|---|---|
| ANN | 0.930 | 0.753 |
| BDT | 1.000 | 0.513 |
| MLR | 0.880 | 0.762 |
| NBC | 0.610 | 0.626 |
| SVM | - | 0.493 |
Figure 2.
ROC curves for validation for ANN, MLR and NBC (full set)
Regardless of sub-sets of the full data used for model development such as sets based on birth weight or year of use of weaning protocol, sets without highly correlated variables, sets created though use of Principal Components Analysis and combinations of these criteria, all methods consistently exhibited low performance (results not shown). The only subset that showed satisfactory performance (AUC=0.78) comprised of only those variables that showed statistically significant differences when comparing infants who failed extubation to infants that were extubated successfully. For this subset AUC was slightly increased for MLR, similar for NBC and slightly lower for ANN as compared to the full data set (table 5). Variables in this subset included birth weight, Apgar at 5 minutes, maternal betamethasone, lag time, rate ratio, FiO2, PIP, inspiratory time, tidal volume, pH, PaCO2, PAO2, SaO2, pulse, blood pressure, minute volume, surfactant, caffeine. As a result of the consistently low performance across all algorithms no decision-support tool using the most accurate prediction methods was developed.
Table 5.
Performance of algorithms as measured by the area under the curve (AUC) for dataset including only statistically significant variables (median AUC from 100 resampling steps).
| Algorithm | Training | Validation |
|---|---|---|
| ANN | 0.921 | 0.682 |
| BDT | 1.000 | 0.507 |
| MLR | 0.853 | 0.776 |
| NBC | 0.769 | 0.607 |
| SVM | - | 0.519 |
Discussion
Development of machine learning models for prediction has recently moved towards use of homogeneous and heterogeneous sets of algorithms to capitalize on better performance and generalizability of the combined results. However, better performance of a set of methods can only be achieved if accuracy among the individual members of the set is high, i.e., the predictions are better than guessing, results are diverse, and the methods produce errors that are different from those of other methods for a given set of input variables. In our results, MLR achieved the highest median performance (AUC=0.78) using the data set including only variables that showed a statistically significant relationship with extubation in prior descriptive analyses. This AUC value can be loosely interpreted as having at best 78% of the extubations among this group of premature infants predicted correctly. In a previous study, the predictive performance of clinicians was directly compared to the performance of two algorithms, ANN and MLR [6,14]. On average, clinicians were 70% accurate in the validation set with a range from 51–79% when limited to the same information (variables) provided to the machine learning algorithms. This wide spread reflected the level of experience among the clinicians (i.e., years as neonatologist working in the NICU) as well as differing preferences such as extubating an infant rather “too early” than “too late”. In contrast, the current data set contained only 12% of extubation failures, indicating that clinicians predicted extubation success with 88% accuracy. However, none of the algorithms used in this study achieved sufficiently high accuracy to be included in a tool intended to provide decision-support for clinicians.
Inferring from these results when variable selection is used as underlying method in algorithms processing these types of data it is likely to fail due to batch effects found in such datasets. The term “batch effect” was initially used in micro-array experiments where differences were found between different batches of experiments when trying to combine data sets. This phenomenon has since been found in other areas of research such as prediction of outcomes using machine learning. If batch effects are present in the data validation using resampling procedures, for example using a subset of a given dataset, will not do well since it is likely that data points from the same batch that are very similar to each other exist in both data sets, training and validation, which will cause selection of variables relevant to one batch but not others. In our study, MLR resulted in better performance than ANN supporting the above hypothesis since MLR can be considered special cases of ANN models. Three methods, MLR, ANN and NBC, performed best in the full data set and the set including only variables showing a statistically significant relationship with extubation outcome with AUCs ranging from 0.63 to 0.78. In contrast, two methods, SVM and BDT, methods tended to over-fit the training data resulting in poor performance (AUC ~0.5) in all data sets using validation data.
Therefore, we hypothesize that an additional pre-processing step is needed prior to model development in which the dimensionality of the dataset is reduced. This step would decrease the number of variables that would be considered for inclusion during model development and may improve performance of the individual methods sufficiently to be included individually or in combination in a decision-support tool. However, this requires that the additional step for variable reduction can deal with potential batch effects present in the data and could, for example, be configured to rank variables as to how much batch effects they exhibit. As discussed in Leek et al. [15], handling of batch effects is an active area of research where current solutions still depends mostly on multivariate exploratory analysis rather than on development and inclusion of a reliable preprocessing step for variable selection prior to model development.
Limitations
A large data set was obtained retrospectively from a period of several years. During this time, NICU procedures may have changed such as implementation of a weaning protocol starting in summer of 2006 (see Appendix I.). This change may not have been fully captured by inclusion of a variable whether or not a given infant was treated using this weaning protocol. In addition, NICU personnel may have changed during the study period. Further, only variables that were available from the medical record and routinely obtained during the care for a premature infant with ventilator-assisted breathing could be included. Lastly, the outcome variable was severely unbalanced in this data set with only 12% of infants included in this data set failing extubation. This imbalance reduced the ability of the prediction methods to learn from these data.
Conclusions
To date clinicians still outperform machine-learning prediction models and the medical field remains a challenge for artificial intelligence methodologies such as those used in this study. All of these methods use the available data to make predictions. Logically, these methods are disadvantaged compared to clinicians when decisions are based on experience that reflects implicit awareness of covariates resulting from information gained from long term exposure and experience, i.e., hours spent in the NICU. To our knowledge, such information has not been reliably captured to provide machine-learning or statistical methods with data comparable to those processed in the brains of clinical experts. However, since the skill of making accurate predictions is based on many years spent at the bedside, we feel that a tool providing reasonable decision-support to inexperienced clinicians would be valuable in clinical practice. To this date, development of a tool that reliably achieves prediction accuracy comparable to those of expert clinicians has not been accomplished and especially in the population of premature infants a “pretty good” prediction is simply not good enough.
Our results suggest that a critical component in the development of prediction algorithms is still missing when dealing with complex medical data that likely contain batch effects. This conclusion is consistent with a trend towards approaches using relatively undirected large data that rely on the “unreasonable effectiveness of data” as described by Halevy, Norvig & Pereira [16]. In other words, the results reported here support the view that maximizing data capture describing the context of complex biomedical processes offers more promise for predictive modeling than trimming the parameters recorded to the few that can be systematically acquired and orderly fed to conventional machine learning tools.
Supplementary Material
Acknowledgments
This work was supported by NIH/NHLBI grant #5R21HL090598-02.
We would like to thank Kathy Ray, M.S.N. for her dedication to this work and her many suggestions during data collection.
Abbreviations
- ANN
artificial neural network
- AUC
area under the curve
- BDT
boosted decision tree
- CPAP
continuous positive airway pressure
- HFJV
high frequency jet ventilation
- HFOV
High frequency oscillatory ventilation
- IRB
institutional review board
- MLR
multivariable logistic regression
- NBC
naïve Bayesian classifier
- NICU
neonatal intensive care unit
- RDS
respiratory distress syndrome
- ROC
operating receiver characteristic
- PINS
Perinatal Information System
- PIP
peak inspiratory pressure
- SIMV
synchronized intermittent mandatory ventilation.
- SVM
support vector machine
Appendix I
Neonatal Ventilator Management Order for Conventional Pressure-Limited Ventilation (“Weaning Protocol.pdf”)
References
- 1.Halliday HL. What interventions facilitate weaning from the ventilator? A review of the evidence from systematic reviews. Paediatr Respir Rev. 2004;5(Suppl A):S347–S352. doi: 10.1016/s1526-0542(04)90060-7. [DOI] [PubMed] [Google Scholar]
- 2.Bancalari E, Claure N. Weaning Infants from Mechanical Ventilation. Neonatology. 2008;94:197–202. doi: 10.1159/000143722. doi: 10.1159/000143722. [DOI] [PubMed] [Google Scholar]
- 3.Greenough A, Prendergast M. Difficult extubation in low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 2008;93:F242–F245. doi: 10.1136/adc.2007.121335. doi:10.1136/adc.2007.121335. [DOI] [PubMed] [Google Scholar]
- 4.Barrington KJ. Extubation failure in the very preterm infant. J Pediatr (Rio J) 2009;85(5):375–377. doi: 10.2223/JPED.1937. doi:10.2223/JPED.1937. [DOI] [PubMed] [Google Scholar]
- 5.Gupta S, Sinha SK, Tin W, Donn SM. A Randomized Controlled Trial of Post-extubation Bubble Continuous Positive Airway Pressure Versus Infant Flow Driver Continuous Positive Airway Pressure in Preterm Infants with Respiratory Distress Syndrome. J Pediatr. 2009;154:645–50. doi: 10.1016/j.jpeds.2008.12.034. Doi:10.1016/j.jpeds.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Mueller M, Wagner CL, Annibale DJ, Hulsey TC, Knapp RG, Almeida JS. Predicting extubation outcome in preterm newborns: a comparison of neural networks with clinical expertise and statistical modeling. Pediatr Res. 2004;56(1):11–8. doi: 10.1203/01.PDR.0000129658.55746.3C. doi: 10.1203/01.PDR.0000129658.55746.3C. [DOI] [PubMed] [Google Scholar]
- 7.Aksela M. Comparison of Classifier Selection Methods for Improving Committee Performance. In: Windeatt T, Roli F, editors. MCS 2003. Springer-Verlag; Berlin Heidelberg: 2003. pp. 84–93. LNCS 2709. [Google Scholar]
- 8.Cohen P, Cohen J, West AG, Aiken LS. Applied Multiple Regression/Correlation Analysis for the Behavioral Sciences. Third edition Routledge Academic; 2003. pp. 486–496. [Google Scholar]
- 9.Bishop CM. Neural Networks for Pattern Recognition. Oxford University Press; 1995. pp. 332–364. [Google Scholar]
- 10.Burges JC. A Tutorial on Support Vector Machines for Pattern Recognition. Data Min Knowl Discov. 1998;2:121–167. [Google Scholar]
- 11.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Mach Learn. 1997;29:131–163. [Google Scholar]
- 12.Dietterich TG. An experimental comparison of three methods for constructing ensembles of decision trees: Bagging, boosting and randomization. Mach Learn. 2000;40(2):1–19. [Google Scholar]
- 13.Mueller M, Wagner CL, Stanislaus R, Almeida JS. IEEE IJCNN. 2013. Machine learning to predict extubation outcome in premature infants. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller M, Wagner CL, Annibale DJ, Knapp RG, Hulsey TC, Almeida JS. Parameter selection for and implementation of a web-based decision-support tool to predict extubation outcome in premature infants. BMC Med Inform Decis. 2006;6:11. doi: 10.1186/1472-6947-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leek JT, Scharpf R, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halevy A, Norvig P, Pereira F. The Unreasonable Effectiveness of Data. IEEE Intell Syst. 2009;24(2):8–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.