SUMMARY
Nod2 has been extensively characterized as a bacterial sensor that induces an antimicrobial and inflammatory gene expression program. Therefore, it is unclear why Nod2 mutations that disrupt bacterial recognition are paradoxically among the highest risk factors for Crohn’s disease, which involves an exaggerated immune response directed at intestinal bacteria. Here, we identified several abnormalities in the small intestinal epithelium of Nod2−/− mice including inflammatory gene expression and goblet cell dysfunction, which were associated with excess interferon-γ production by intraepithelial lymphocytes and Myd88 activity. Remarkably, these abnormalities were dependent on the expansion of a common member of the intestinal microbiota, Bacteroides vulgatus, which also mediated exacerbated inflammation in Nod2−/− mice upon small intestinal injury. These results indicate that Nod2 prevents inflammatory pathologies by controlling the microbiota, and support a multi-hit disease model involving specific gene-microbe interactions.
INTRODUCTION
The surfaces of the body, particularly the gastrointestinal tract, are exposed to a large number of diverse microbes that are collectively referred to as the microbiota. Individual members of the microbiota are generally referred to as commensals since their presence in most individuals is innocuous or beneficial. However, a breakdown in tolerance towards these microbes is considered central to the origin of Crohn’s disease (CD), a major type of inflammatory bowel disease (IBD) characterized by excessive inflammatory cytokine production by T cells and damage to the intestinal epithelium (Abraham and Cho, 2009). IBD patients typically display imbalances in the composition of the microbiota, and reducing exposure to intestinal bacteria has been shown to ameliorate recurrent inflammation (Packey and Sartor, 2009). Additionally, genetic susceptibility factors that have been identified are involved in host-microbe interactions, as exemplified by mutations in NOD2 (Jostins et al., 2012). Nod2 is a cytosolic bacterial sensor that induces cytokine and antimicrobial gene expression in response to the peptidoglycan constituent muramyl dipeptide (Girardin et al., 2003; Inohara et al., 2003). Therefore, Nod2 function may be key to understanding how equilibrium between the immune system and commensal bacteria is maintained.
The three major mutations in the gene encoding NOD2 that are associated with CD have been extensively demonstrated to reduce or ablate the ability to sense bacterial products in cell culture assays (Billmann-Born et al., 2011; Chamaillard et al., 2003), primary cells from humans (Cooney et al., 2010; Netea et al., 2005; Travassos et al., 2010), and mouse models in vivo (Kim et al., 2011; Watanabe et al., 2008). Hence, Nod2−/− mice have been widely used to investigate the basic immune function of this gene as well as its association with CD. Despite progress in understanding Nod2 function during infectious disease settings, the effect of Nod2 deletion on the immune response to the intestinal microbiota remains unclear. Nod2−/− mice on a mixed genetic background were initially demonstrated to display reduced expression of antimicrobial defensins by Paneth cells, an epithelial cell type in the small intestinal crypt that generates antimicrobial granules (Kobayashi et al., 2005). However, a subsequent study did not observe decreases in defensin expression in Nod2−/− mice backcrossed to the C57BL/6 (B6) background (Shanahan et al., 2013). In other studies examining the composition of the intestinal microbiota, both Nod2−/− mice and CD patients with NOD2 mutation were shown to have an overrepresentation of the Bacteroides genus (Couturier-Maillard et al., 2013; Mondot et al., 2012; Petnicki-Ocwieja et al., 2009; Rehman et al., 2011). In spite of this concordance between mice and humans, cohousing Nod2−/− and control mice eliminated differences in the composition of commensal bacteria, suggesting that the expansion of Bacteroides is non-specific (Robertson et al., 2013). Because mutation of NOD2 remains one of the strongest CD risk factors and associates with small intestinal involvement, we sought to resolve these issues by examining the small intestinal abnormalities present in Nod2−/− mice.
Here, we find that Nod2−/− mice display abnormalities in the small intestinal epithelium including defects in mucus production by goblet cells. These epithelial abnormalities were associated with an increase in interferon-γ (IFNγ) expressing intraepithelial lymphocytes (IELs), and were dependent on the innate immune signaling molecule Myd88. Remarkably, these intestinal abnormalities were dependent on the expansion of the commensal bacterium Bacteroides vulgatus. Finally, we demonstrate that B. vulgatus mediated exacerbated small intestinal inflammation in Nod2−/− mice following non-steroidal anti-inflammatory drug (NSAID) treatment. Taken together, we have identified a role for Nod2 in preventing inflammation by restricting the pathological expansion of a common member of the intestinal microbiota.
RESULTS
Nod2−/− mice display inflammatory gene expression in the small intestinal epithelium
The following precautions have been taken because previous studies have revealed that differences in background and housing conditions are confounding variables for investigating Nod2 function: (1) all mutant mice have been backcrossed to the B6 background; (2) mice were maintained in a barrier facility where they were routinely checked for the absence of specific infectious agents including murine norovirus (MNV), Helicobacter, and segmented filamentous bacteria (SFB); (3) mutant mice were compared to wild-type (WT) B6 control mice bred onsite in the same facility; (4) mutant mice were maintained since birth in separate cages from WT mice in the initial set of experiments, and cohousing experiments were performed later in the study where indicated.
We previously reported that mice deficient in another CD susceptibility gene, Atg16L1, develop Paneth cell granule abnormalities in a manner dependent on murine norovirus (MNV) infection (Cadwell et al., 2010). Because the role of Nod2 in Paneth cells is still uncertain, we examined Paneth cell morphology in Nod2−/− mice by light microscopy and lysozyme immunofluorescence (IF) in the presence and absence of MNV. In contrast to MNV-infected Atg16L1 mutant mice, Nod2−/− and WT mice did not display aberrant Paneth cell morphology by light microscopy even after MNV infection (Figure S1A–D). Similarly, only mild abnormalities in lysozyme distribution were detected in the Nod2−/− samples, independently of MNV (Figure S1A–D). These results are consistent with observations made in CD patients, where individuals with the ATG16L1 risk allele display a more severe alteration in lysozyme distribution compared to patients with NOD2 risk alleles (Cadwell et al., 2008; Vandussen et al., 2013).
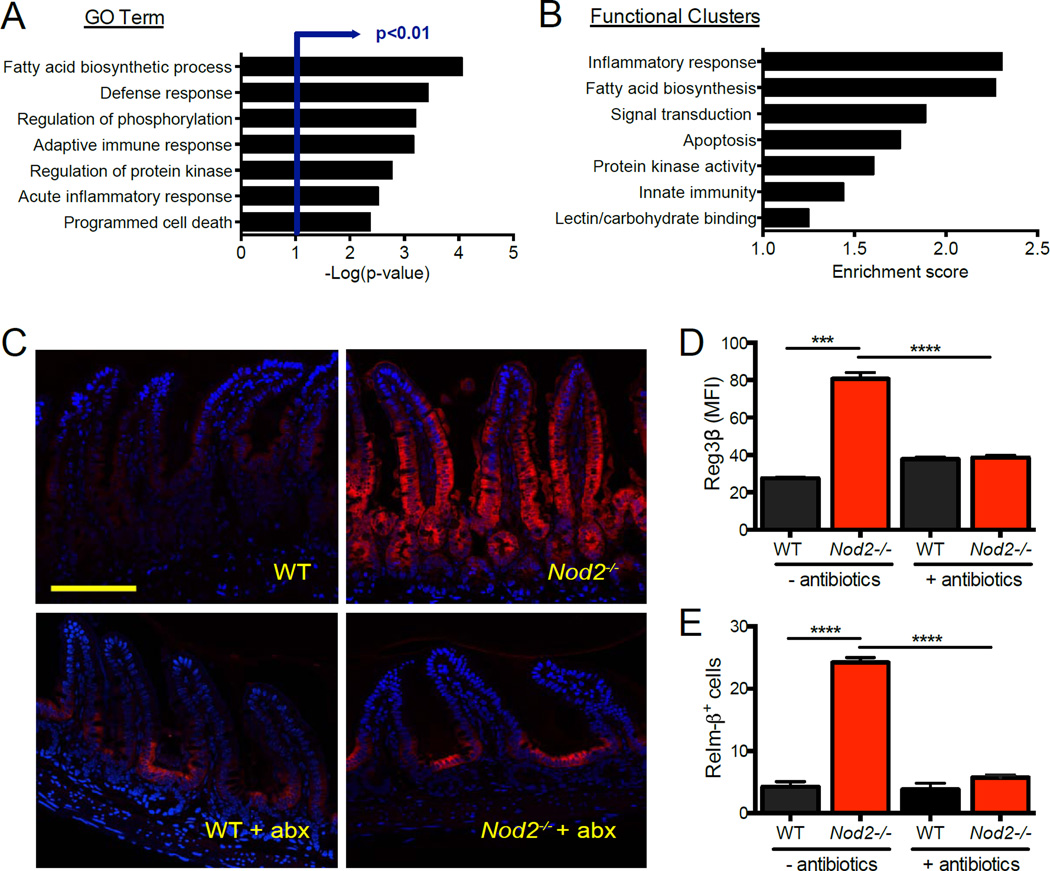
Because Nod2 was suggested to mediate antimicrobial gene expression by Paneth cells, we performed a microarray analysis on laser-captured epithelial cells from the crypt base where Paneth cells are located. Unexpectedly, pathway analysis indicated that Nod2−/− samples were enriched, rather than deficient, in transcripts associated with inflammation, fatty acid biosynthesis (eicosanoid pathway), and host defense (Figure 1A, B, S1E, Table S1). Gene set enrichment analysis (GSEA) confirmed that Nod2−/− mice displayed a pro-inflammatory gene expression signature in the epithelium. Many of the genes with the highest differential expression encode antimicrobial molecules such as Reg3β, Relm-β, Pla2, and Defb1 (Figure S1F). We validated these results by staining small intestinal sections for Reg3β since a human homolog has been shown to display increased expression in IBD patients (Ogawa et al., 2003). Compared to the minimal staining observed in WT mice, high levels of Reg3β were detected in the Nod2−/− crypt epithelium that extended partway up the length of the villi (Figure 1C, D, S1G, H). Relm-β, which has been shown to mediate inflammation (McVay et al., 2006), was also highly expressed in Nod2−/− small intestinal sections compared to WT (Figure 1D, S1I, J). Overproduction of Reg3β and Relm-β is most likely due to a response to bacteria because treatment with an antibiotic cocktail reduced expression to WT levels (Figure 1C, D, E). Increased Reg3β and Relm-β staining was not observed in the colon, and unlike the small intestine, colonic Relm-β levels increased with antibiotics as previously described (Hill et al., 2010) (Figure S1I, K). Thus, Nod2-deficiency leads to a pro-inflammatory gene expression signature specifically in the small intestinal epithelium.
Figure 1. Nod2−/− mice display inflammatory gene expression in the small intestinal epithelium.
(A) Microarray analysis of transcripts displaying >1.25 fold enrichment in crypt-base epithelial cells from Nod2−/− compared to WT mice. Gene ontology (GO) terms with the highest significance are shown. (B) Functional clusters displaying greatest enrichment from (a). n = 3 mice per genotype. (C) Immunofluorescence (IF) staining of Reg3β in WT and Nod2−/− small intestinal sections with and without treatment with antibiotics (metronidazole, ampicillin, vancomycin, neomycin). Scale bar = 100µm. (D) Quantification of the mean fluorescence intensity (MFI) of Reg3β in small intestinal sections as in (C). n ≥ 5 mice per genotype. (E) Quantification of MFI of Relm-β in WT and Nod2−/− small intestinal sections with and without antibiotics treatment. n ≥ 3 mice per genotype. ****p<0.0001 by ANOVA with Holm-Sidak multiple comparisons test for (D) and (E). Data are represented as mean ± SEM from at least two independent experiments in (D) and (E). See also figure S1.
Nod2 is required to maintain small intestinal goblet cell function
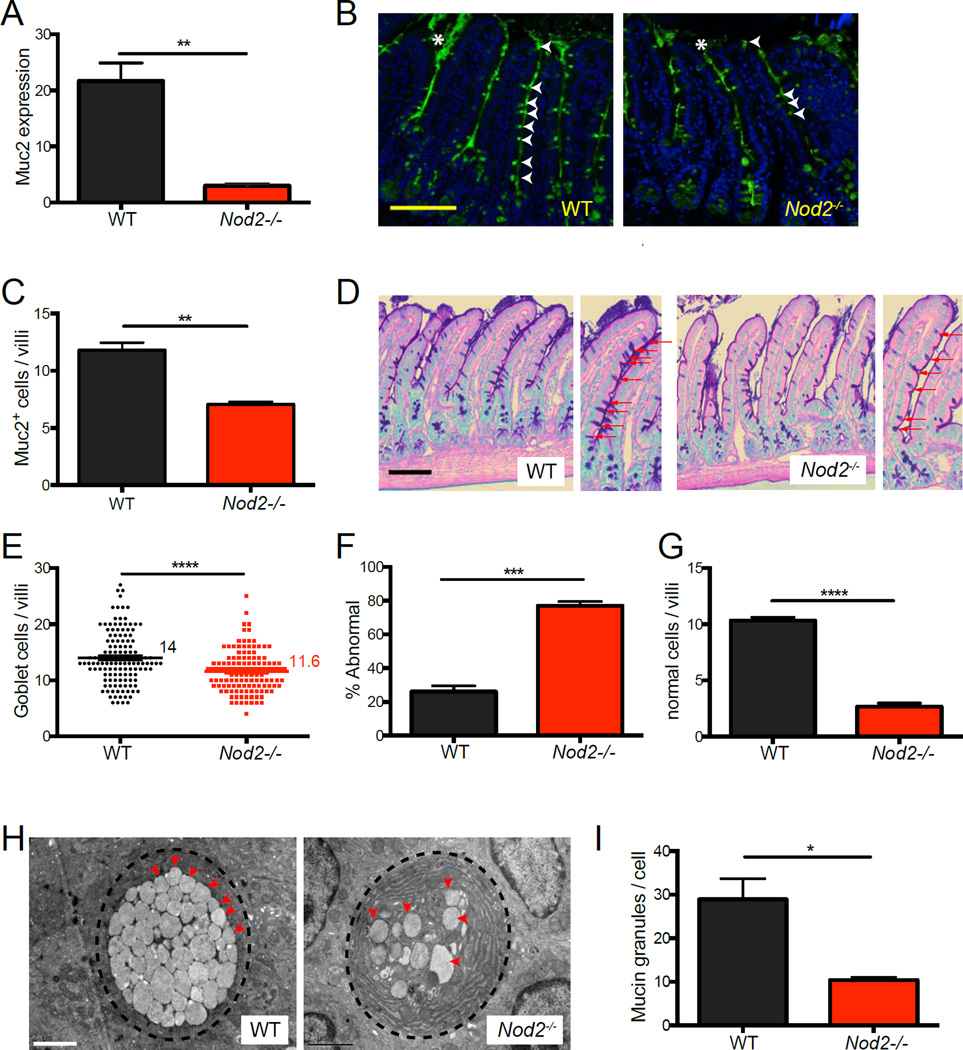
We next investigated the possibility that Nod2−/− mice develop other abnormalities in the small intestinal epithelium. Mucus secreted by goblet cells forms a physical barrier that separates intestinal bacteria from the epithelium. Goblet cells in IBD patients have decreased Muc2 expression, and mice deficient in this dominant intestinal mucin display increased expression of antimicrobial genes including Reg3β (Burger-van Paassen et al., 2012; Sheng et al., 2012). Consistent with these observations, we observed decreased Muc2 expression in the small intestine of Nod2−/− mice compared to WT mice (Figure 2A). IF analysis also indicated that there was a decrease in the number of cells in the villi that stained positive for Muc2, and a corresponding decrease in the amount of Muc2 in the lumen between villi (Figure 2B and C). Additionally, light microscopy analysis of PAS-Alcian blue-stained sections confirmed the reduction and staining intensity of goblet cells in the villi of Nod2−/− mice (Figure 2D–G). Transmission electron microscopy (TEM) revealed that Nod2−/− mice had much fewer mucin granules per goblet cell, and many of these granules had an abnormal fused appearance that was rarely detected in WT samples (Figure 2H, I, S2A–B). Thus, Nod2 has an unappreciated function in maintaining the goblet cell compartment.
Figure 2. Nod2−/− mice display goblet cell abnormalities in the small intestine.
(A) qPCR analysis of Muc2 expression normalized to Gapdh in small intestinal tissue harvested from WT and Nod2−/− mice. (B) IF staining of Muc2 in small intestinal sections. Muc2 positive cells and extracellular staining are denoted by white arrow heads and asterisks respectively. (C) Number of Muc2+ cells per villi from (B). (D) PAS-Alcian blue staining of small intestinal sections from WT and Nod2−/− mice. Red arrows denote goblet cells. (E–G) Quantification of the number of goblet cells per villi (E), the proportion displaying translucent staining or atypical morphology (F), and number of goblet cells displaying normal morphology per villi (G) from the PAS-Alcian blue-stained sections in (B). (H) Transmission electron microscopy analysis of goblet cell morphology in WT and Nod2−/− small intestine. Red arrowheads denote individual granules. (I) Quantification of the number of mucin granules per goblet cell in (H), performed by manually counting individual granules per cell in at least 25 cells per mouse. n ≥ 3 mice per genotype. **p<0.01, ***p<0.001 and ****p<0.0001 by unpaired two-tailed t test in (A), (C), (E), (F), (G), and (I). Scale bar = 100µm in (B) and (D), and 2µm in (H). All bar graphs display mean ± SEM from at least two independent experiments. Bars and numbers represent the mean in (E). See also figure S2.
Goblet cell abnormalities are associated with an increased proportion of IFN-y expressing lymphocytes
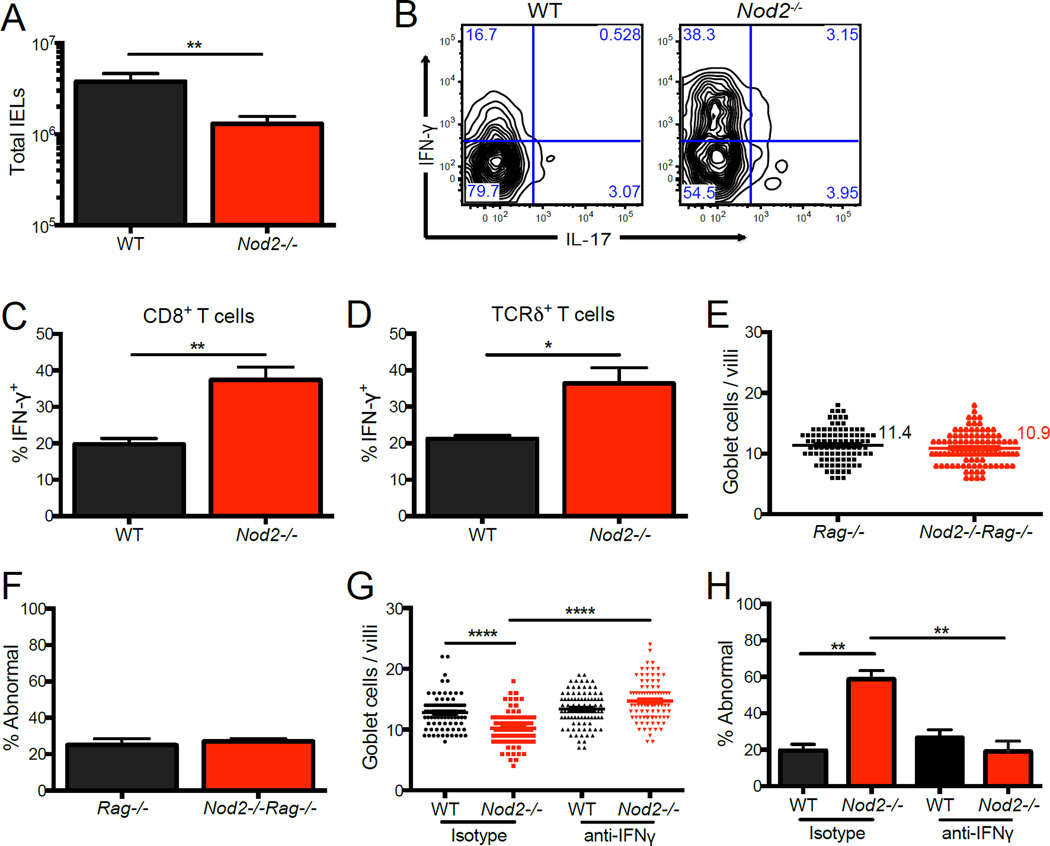
IFN-γ production by lymphocytes has been suggested to contribute to the development of intestinal pathologies observed in CD patients (Abraham and Cho, 2009). A previous observation indicating that Nod2−/− mice have an increase in IFN-γ+ lymphocytes in Peyer’s patches and mesenteric lymph nodes (MLNs) (Barreau et al., 2010) has been shown to be downstream of Helicobacter infection (Biswas et al., 2010). Although our mice are maintained in a Helicobacter-negative environment, we considered the possibility that cytokine production by intraepithelial lymphocytes (IELs) would be preferentially affected in Nod2−/− mice due to the proximity of these cells to the intestinal epithelium and lumenal bacteria. Nod2−/− mice were recently shown to have a reduction in the number of IELs (Jiang et al., 2013), an observation that we confirmed (Figure 3A). However, we found that a higher proportion of the IELs from Nod2−/− mice expressed IFN-γ compared to control mice, which included both CD8+ and TCRγδ+ T cell subsets (Figure 3B–D). In contrast, we did not detect differences in the amount of IFN-γ+ lymphocytes in the small intestinal and colonic lamina propria (LP), MLNs, or Peyer’s patches when comparing WT and Nod2−/− mice (Figure S3A, B).
Figure 3. Nod2−/− mice have an increased proportion of IFN-y expressing intraepithelial lymphocytes.
(A) Total number of intraepithelial lymphocytes (IELs) harvested from WT and Nod2−/− mice. n ≥ 16 mice per genotype. (B) Representative flow cytometry plot of CD8+ IELs from (A) stained for intracellular IFN-γ and IL-17 expression following stimulation with PMA and ionomycin gated on CD3+ live cells. (C–D) Quantification of the proportion of CD8+ (C) and TCRδ+ (D) IELs expressing IFN-γ from (B). n ≥ 6 mice per genotype. (E–F) Quantification of the number of goblet cells per villi (E) and the proportion displaying abnormal morphology (F) from PAS-Alcian blue-stained small intestinal sections in Rag−/− and Nod2−/−Rag−/− mice. n ≥ 3 mice per genotype. (G–H) Quantification of the number of goblet cells per villi (G) and the proportion displaying abnormal morphology (H) from PAS-Alcian blue-stained small intestinal sections in WT and Nod2−/− mice treated with antibody to IFN-γ or isotype control. n ≥ 3 mice per genotype. *p<0.05, **p<0.01, and ****p<0.0001 by unpaired two-tailed t test in (A) (C) and (D), and ANOVA with Holm-Sidak multiple comparisons test for (G) and (H). All bar graphs display mean ± SEM from at least two independent experiments. Bars and numbers represent the mean in (E) and (G). See also figure S3.
To assess the role of lymphocytes in the goblet cell defects, we crossed Nod2−/− mice with lymphocyte-deficient Rag1−/− mice. We found that double mutant Nod2−/−Rag−/− mice had goblet cells with normal morphology and a similar number of goblet cells per villi compared to single mutant Rag−/− mice (Figure 3E, F). In contrast, Reg3β levels remained high in Nod2−/−Rag−/−mice (Figure S3C). Thus, defects in goblet cells, but not necessarily antimicrobial gene expression, are dependent on the presence of lymphocytes. Additionally, treatment of Nod2−/−mice with an anti-IFN-γ depleting antibody, but not isotype control, led to full restoration of goblet cell morphology and numbers (Figure 3G, H). Thus, goblet cell abnormalities in Nod2−/−mice are dependent on lymphocytes and IFN-γ.
Small intestinal abnormalities are downstream of Myd88 signaling
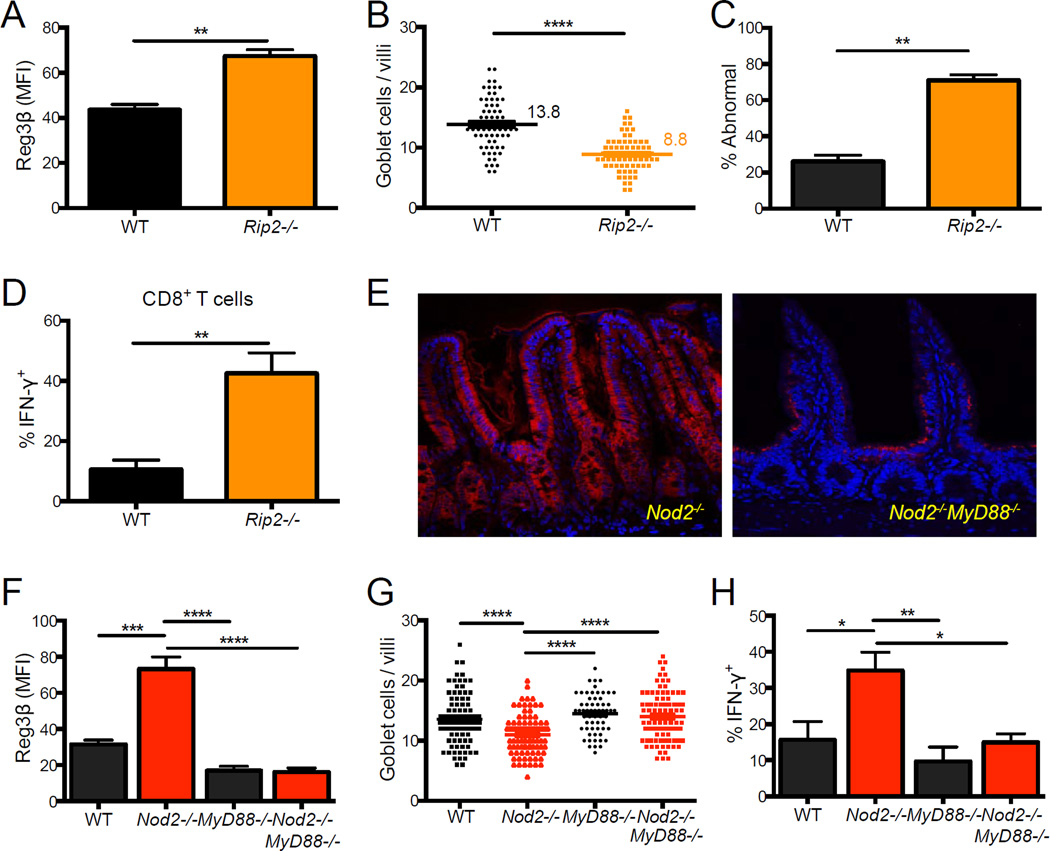
Nod2-mediated gene expression is dependent on the adaptor and kinase Rip2, although Rip2-independent functions of Nod2 in inflammasome and autophagy activation have been described (Hsu et al., 2008; Travassos et al., 2010). We examined Rip2−/− mice for intestinal abnormalities to determine if the above observations reflect a Rip2-dependent function of Nod2. Rip2−/− mice displayed a similar increase in Reg3β expression in small intestinal sections as Nod2−/− mice (Figure 4A). Also, Rip2−/− mice had a substantial decrease in the number of goblet cells per villi, and the cells displayed aberrant morphology (Figure 4B, C). In contrast, Atg16L1 mutants did not display these abnormalities (Figure S4B–F). Consistent with a relationship between goblet cell abnormalities and IFN-γ production by lymphocytes, Rip2-deficiency also led to an increase in the proportion of IFN-γ+ IELs (Figure 4D). Thus, Rip2−/− and Nod2−/− mice display similar small intestinal abnormalities.
Figure 4. Intestinal abnormalities in Nod2−/− mice are observed in Rip2−/− mice and dependent on Myd88.
(A–D) Quantification of Reg3β IF staining (A), number of goblet cells per villi (B) and proportion displaying abnormal morphology by light microscopy (C), and the proportion of IFN-γ+ IELs by flow cytometry (D) in the small intestine of WT and Rip2−/− mice. (E) Representative IF staining of Reg3β in Nod2−/− and Nod2−/−MyD88−/− small intestinal sections. Scale bar = 100 µm. (F) Quantification of Reg3β IF in WT, Nod2−/−, MyD88−/− and Nod2−/−MyD88−/−small intestine. (G) Quantification of the number of goblet cells per villi in WT, Nod2−/−, MyD88−/−and Nod2−/−MyD88−/− small intestine. (H) Flow cytometry analysis of the proportion of IFN-γ+ IELs in WT, Nod2−/−, MyD88−/− and Nod2−/−MyD88−/− mice. n ≥ 3 mice per genotype, **p<0.01 and ****p<0.0001 by unpaired two-tailed t test in (A), (B), (C), and (D), and ANOVA with a Holm-Sidak multiple comparisons test for (F), (G) and (H). All bar graphs display mean ± SEM from at least two independent experiments. Bars and numbers represent the mean in (B) and (G). See also figure S4
The toll-like receptor (TLR) adaptor molecule Myd88 functions in the small intestinal epithelium to regulate the expression of antimicrobial molecules including those that we identified as increased in Nod2−/− mice such as Reg3β and Relm-β (Vaishnava et al., 2008). Also, Nod2-deficiency has been suggested to lead to unrestricted TLR signaling (Watanabe et al., 2008; Watanabe et al., 2004). Although Myd88 is typically necessary for maintaining the epithelial barrier, Myd88-deletion protects against epithelial damage in certain models of intestinal inflammation (Asquith et al., 2010; Hoshi et al., 2012; Raetz et al., 2012). To examine the role of Myd88 in the generation of the above small intestinal abnormalities, we generated double mutant Nod2−/−Myd88−/− mice and found that the abnormalities were eliminated. Nod2−/−Myd88−/− mice did not display increases in Reg3β or aberrant goblet cell morphology, and the number of goblet cells were restored (Figure 4E–G). The proportion of IFNγ+ IELs was also reduced to WT levels (Figure 4H). These results indicate that the absence of Nod2 function induces small intestinal abnormalities in a manner dependent on MyD88.
Nod2 prevents expansion of Bacteroides vulgatus
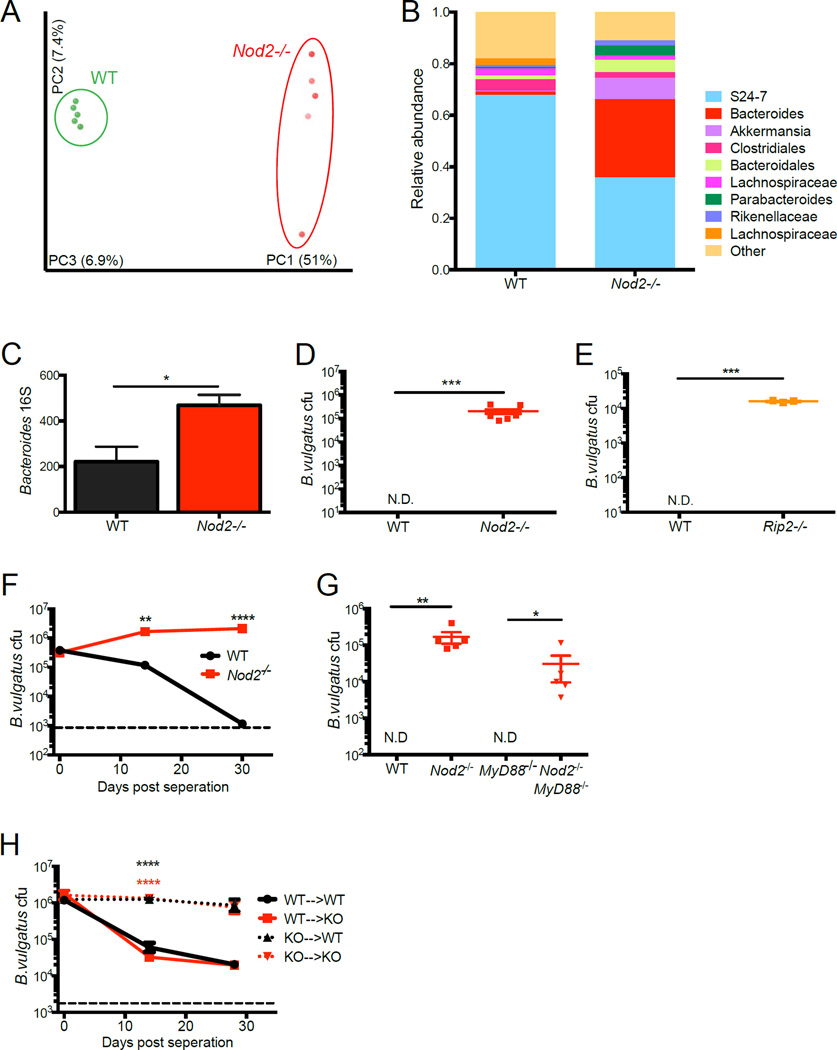
Our finding that the treatment of Nod2−/− mice with a combination of antibiotics reduces the expression of Reg3β and Relm-β (Figure 1C, D) implicates commensal bacteria in the generation of intestinal abnormalities. To compare the composition of the intestinal microbiota, we performed deep sequencing of 16S rRNA gene amplicons from stool harvested from WT and Nod2−/− mice maintained in separate cages as in previous experiments. Principle coordinate analysis (PCoA) revealed that bacterial community structures were significantly different between WT and Nod2−/− mice (Figure 5A). Similar to previous studies in which Nod2−/− and control mice were maintained in separate cages, we found a striking overrepresentation of the Bacteroides genus in Nod2−/− mice, which we further confirmed by qPCR (Figure 5B, C, S5B). When we plated stool on agar selective for Bacteroides under anaerobic conditions, we observed that colonies of a uniform morphology consistently grew out from Nod2−/− samples but not from WT (Figure 5D, S5C). Sequencing of the entire 16S rRNA gene revealed a 100% match with Bacteroides vulgatus. Additional sequencing analysis of 15 colonies (3 each from 5 different Nod2−/− mice) confirmed that they were all B. vulgatus, and we did not detect any other species on these plates. Examination by fluorescent in situ hybridization (FISH) also indicated that B. vulgatus was present throughout the gastrointestinal tract of Nod2−/− mice and undetectable in controls (Figure S5A). In contrast, we occasionally detected other Bacteroides species in WT mice that were not detected in Nod2−/− mice (Figure S5C).
Figure 5. Nod2 prevents the expansion of Bacteroides vulgatus.
(A) PCoA plot generated from unweighted UniFrac distance matrix displaying the distinct clustering pattern of intestinal microbial communities in WT and Nod2−/− mice. (B) Relative abundances of taxonomic groups averaged across WT and Nod2−/− mice. (C) Quantification of Bacteroides genus-specific 16S rRNA gene detected by qPCR normalized to total bacterial 16S in stool of WT and Nod2−/− mice. (D) Quantification of colony forming units (cfu) of anaerobic bacteria identified as Bacteroides vulgatus from stool of WT and Nod2−/− mice plated on BBE agar. n = 10 mice per genotype. (E) Quantification of B. vulgatus in stool of WT and Rip2−/− mice. n = 5 mice per genotype. (F) Quantification of B. vulgatus in stool of WT and Nod2−/− mice that were co-housed and separated by genotype on day 0. n = 5 mice per genotype. (G) Quantification of B. vulgatus in stool of WT, Nod2−/−, MyD88−/− and Nod2−/−MyD88−/− mice. n ≥ 3 mice per genotype. (H) Quantification of B. vulgatus in stool harvested from bone marrow chimeras generated from WT and Nod2−/− mice that were co-housed and then separated by genotype. Black asterisks refer to WT->WT compared to KO->WT and red asterisks refer to WT->WT compared to KO->KO. N.D. = not detected, dotted line refers to limit of detection. n ≥ 5 mice per genotype. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001 by unpaired two-tailed t test in (C), Wilcoxon-Mann-Whitney test in (D), (E) and (G), and Kruskal-Wallis with Dunn’s multiple comparisons test for (F) and (H). Data are represented as mean ± SEM in (C) and (G). Bars represent the mean in (D) and (E). See also figure S5.
It is possible that only Nod2−/− mice were exposed to B. vulgatus in a previous generation and remained colonized through vertical transmission, thus leading to a ‘legacy effect’. It is unlikely that vertical transmission alone accounts for colonization of Nod2−/− mice since Rip2−/−mice also displayed high amounts of B. vulgatus in the stool, and this bacterium was not recovered from other mouse lines such as Atg16L1 mutants (Figure 5E, Figure S4A). To further rule out a legacy effect, we intentionally exposed WT mice to the microbiota of Nod2−/− mice. As with the previous study in which mice were cohoused (Robertson et al., 2013), we found that WT mice placed in the same cage with Nod2−/− mice became readily colonized with B. vulgatus. However, upon re-separation, B. vulgatus decreased to an undetectable level over time in WT mice, but remained stable in Nod2−/− mice (Figure 5F). These results indicate that WT mice require continual exposure to remain colonized, while B. vulgatus levels are stable in Nod2−/−mice and can be transmitted to the next generation. In addition, mice heterozygous for Nod2 deletion displayed levels of B. vulgatus colonization and intestinal abnormalities that were in-between WT and Nod2−/− mice (Figure S4G–K). This gene dosage effect strongly argues that our observations are due to Nod2-deficiency rather than an artifact of housing conditions. Finally, we did not detect B. vulgatus colonization in Myd88−/− mice, but Nod2−/−Myd88−/− mice remained highly colonized (Figure 5G). These results highlight a specific relationship between deficiency in the Nod2/Rip2 pathway and Bacteroides overgrowth.
Nod2 is highly expressed in myeloid and dendritic cell lineages, but it can also be detected in epithelial cell types (Kobayashi et al., 2005). To differentiate between the function of Nod2 in these compartments, we generated bone marrow chimeras using WT and Nod2−/− mice as both donors and recipients (Figure S5F). Chimeric mice of different origins were cohoused with each other for 4 weeks to allow equal colonization by B. vulgatus (day 0 in Figure 5H), and bacterial burden was then measured after separating mice by genotype. We found that B. vulgatus levels steadily decreased over time in WT and Nod2−/− mice that received WT bone marrow (Figure 5H). In contrast, WT and Nod2−/− mice that received Nod2−/− bone marrow continued to shed high amounts of B. vulgatus (Figure 5H). Thus, Nod2-deficiency in the hematopoietic compartment confers susceptibility to B. vulgatus. Importantly, this experiment definitively rules out a legacy effect since the genotype of the donor rather than the recipient mouse determined B. vulgatus colonization.
B. vulgatus mediates intestinal abnormalities and inflammation in Nod2−/− mice
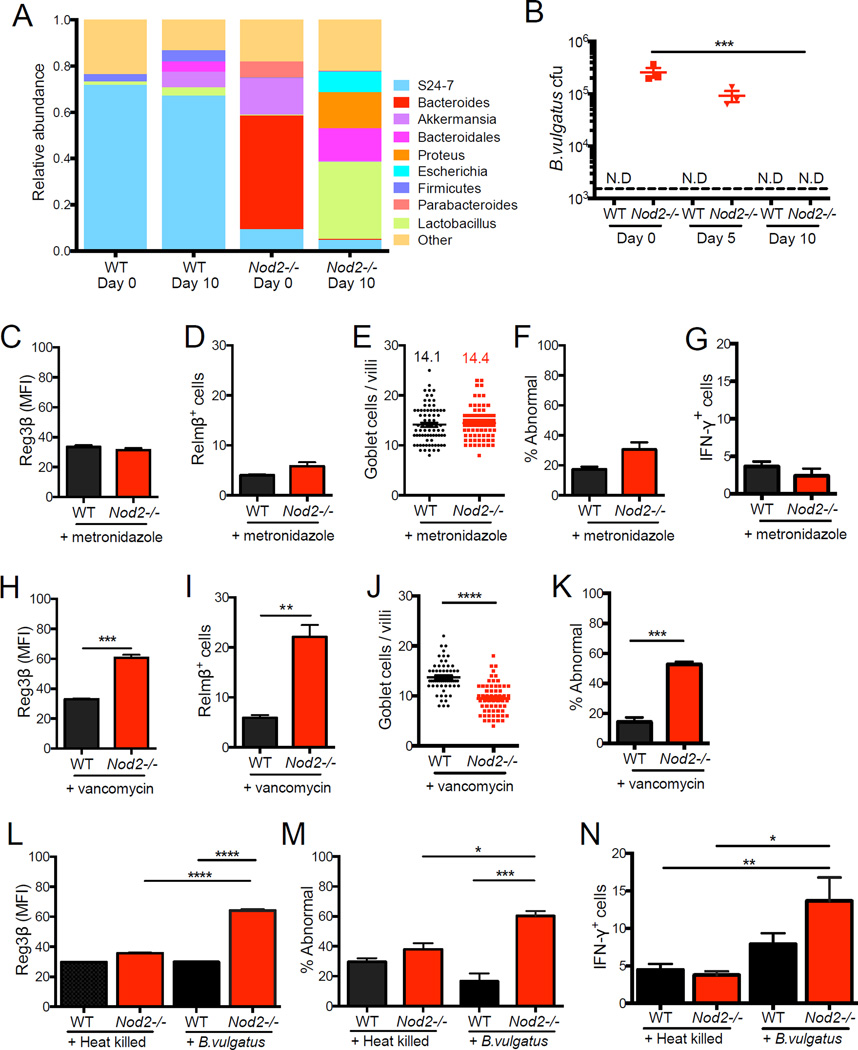
In the above chimeric mice, we found that WT mice reconstituted with Nod2−/− bone marrow developed goblet cell abnormalities, while Nod2−/− mice that received WT bone marrow were ‘cured’ of this defect (Figure S5C–E). Thus, epithelial abnormalities appear to be linked to B. vulgatus levels. To further assess the functional consequence of B. vulgatus colonization in Nod2−/− mice, we treated mice with metronidazole, an antibiotic that targets anaerobic bacteria. 16S deep sequencing and plating on selective media confirmed that metronidazole treatment eliminated the Bacteroides overgrowth observed in Nod2−/− mice (Figure 6A, B, S6A). Nod2−/−mice that received metronidazole displayed decreased epithelial Reg3p levels, restoration of normal goblet cell numbers and morphology, and reduced frequency of IFN-γ+ IELs (Figure 6C–G). In contrast, vancomycin treatment, which targets gram-positive bacteria, did not deplete B. vulgatus and did not reverse intestinal abnormalities (Figure 6H–K, S6D).
Figure 6. B. vulgatus mediates small intestinal abnormalities in Nod2−/− mice.
(A) Relative abundances of intestinal microbial communities in WT and Nod2−/− mice before and after metronidazole treatment. (B) Quantification of B. vulgatus in stool of WT and Nod2−/− mice after 0, 5, and 10 days of metronidazole treatment. (C–G) Quantification of IF staining of Reg3β (C) and Relm-β (D), number of goblet cells per villi (E) and the proportion displaying abnormal morphology (F) by light microscopy, and IFN-γ+ IELs by flow cytometry in WT and Nod2−/− mice treated with metronidazole. (H–K) Quantification of IF staining of Reg3β (H) and Relm-β (I), and number of goblet cells per villi (J) and the proportion displaying abnormal morphology (K) by light microscopy in WT and Nod2−/− mice treated with vancomycin. (L–N) Quantification of IF staining of Reg3β (L), proportion of goblet cells displaying abnormal morphology by light microscopy (M), and IFN-γ+ IELs by flow cytometry in WT and Nod2−/− mice treated with metronidazole and gavaged with 108 cfu B. vulgatus or equivalent amount of heat-killed bacteria. Dotted line denotes limit of detection. n ≥ 3 mice per genotype in (A)-(K), n = 9–10 mice per genotype in (L)-(N). *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001 by Kruskal-Wallis with Dunn’s multiple comparisons test for (B), unpaired two tailed t-test in (C)-(K) and ANOVA with a Holm-Sidak multiple comparisons test for (L), (M) and (N). Data are represented as mean ± SEM from at least two independent experiments in (B)-(D) and (F)-(N). Bars and numbers represent the mean in (E) and (J). See also figure S6.
Next, we re-introduced B. vulgatus by orally gavaging metronidazole-treated WT and Nod2−/− mice with 1×108 colony forming units (CFU) of the cultured bacteria or an equivalent amount of heat-killed bacteria. Only Nod2−/− mice that received live B. vulgatus displayed abnormalities in Reg3p expression, goblet cells, and IFN-γ+ IELs (Figure 6L–N). Interestingly, metronidazole-pretreatment allowed WT mice to be transiently colonized by B. vulgatus during the two-week timeframe of this experiment, indicating that B. vulgatus cannot induce intestinal abnormalities in a WT host even when exposure is forced (Figure S6B). The intestinal abnormalities may require the ability of this bacterium to access regions proximal to the epithelium since a substantially higher amount of tissue-associated B. vulgatus was recovered from Nod2−/− mice under these same conditions (Figure S6C). To test if these observations are specific to B. vulgatus, we inoculated metronidazole-treated WT and Nod2−/− mice with Bacteroides thetaiotaomicron VPI-5482, a commensal strain that is frequently used to investigate host responses in mouse models (Goodman et al., 2009; Hooper et al., 2001; Peterson et al., 2007). Neither WT nor Nod2−/− mice developed small intestinal abnormalities in response to B. thetaiotaomicron on day 3 or 12 post-inoculation despite detection of a high amount of bacteria in the stool (Figure S6E–H, I–L). Another genus member, Bacteroides uniformis, was able to stably colonize metronidazole-treated WT mice but not Nod2−/− mice (Figure S6M). Also, we were able to detect Bacteroides acidophilus, Bacteroides eggerthii, and Parabacteroides goldstenii in WT mice but not in Nod2−/− mice (Figure S5C). These observations suggest that efficiency or location of colonization may have an important role in determining if a given Bacteroides species can induce intestinal abnormalities in Nod2−/− mice.
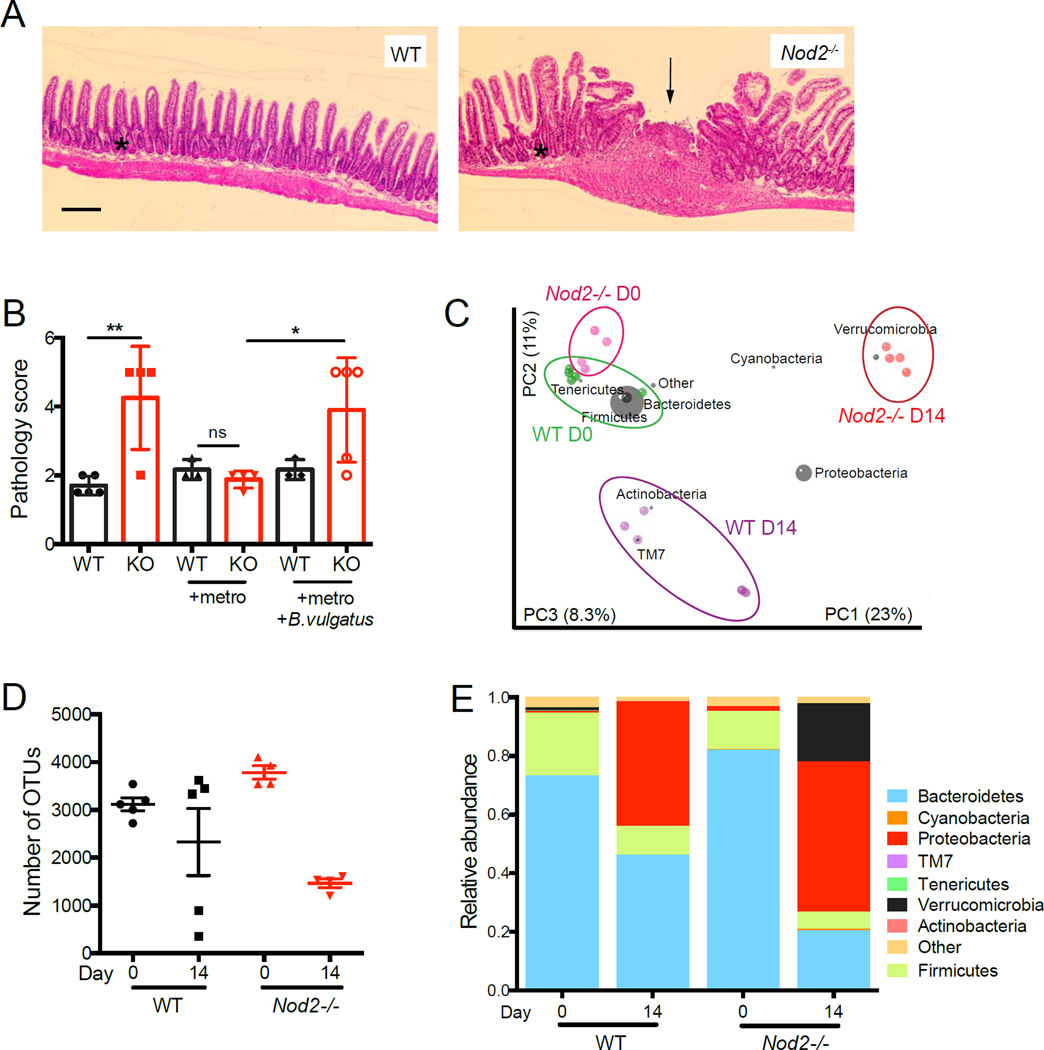
To relate these findings to IBD, we analyzed the role of B. vulgatus in Nod2−/− mice treated with piroxicam, a NSAID that can induce small intestinal injury (Han et al., 2009). Compared to similarly treated WT mice, Nod2−/− mice receiving piroxicam displayed increases in focal ulceration, epithelial hyperplasia, and peri-cryptal and submucosal infiltrates (Figure 7A, B). Moreover, we found that only Nod2−/− mice displayed blood in the small intestinal lumen and attachment of the wall of the ileal-cecal junction to the surrounding tissue of the peritoneal cavity (Figure S7A, B). Piroxicam did not induce these pathologies in Nod2−/− mice that received metronidazole (Figure 7B, S7B). In contrast, inoculation of metronidazole-treated Nod2−/− mice with B. vulgatus prior to piroxicam treatment restored the appearance of the pathologies described above (Figure 7B, S7B). Consistent with a relationship between intestinal bacteria and exacerbated disease, 16S deep sequencing revealed that the difference in the microbiota between WT and Nod2−/− mice was magnified following piroxicam treatment (Figure 7C). The composition of the intestinal microbiota in Nod2−/− mice shifted away from the initial B. vulgatus dominated flora towards a typical ‘dysbiotic’ flora with decreased diversity and phylum-level changes (Figure 7D, E). In general, the effect of piroxicam on the microbiota was exaggerated in Nod2−/− mice compared to WT mice, and included increases in Proteobacteria and decreases in Bacteroidetes and Clostridiales (Figure 7C, E, S7C). These changes are similar to those recently described in early-onset CD patients (Gevers et al., 2014). Thus, B. vulgatus is essential for at least initiating intestinal inflammation in Nod2−/− mice following piroxicam treatment, which induces additional large-scale changes in the composition of the microbiota that potentially contributes to the ensuing pathologies.
Figure 7. Nod2−/− mice are susceptible to piroxicam induced small intestinal inflammation in the presence of B. vulgatus.
(A) Representative H&E-stained small intestinal sections from piroxicam-treated WT and Nod2−/− mice. Black arrow and asterisk point to a focal ulcer and epithelial hyperplasia respectively. Scale bar = 200µm. (B) Quantification of small intestinal pathology in WT and Nod2−/− (KO) mice treated with piroxicam. (C) Graph represents a PCoA plot generated from unweighted UniFrac distance matrices of WT and Nod2−/− mice before and after piroxicam treatment, overlaid with the distribution of different bacterial taxa based on phylogenetic information. (D) Quantification of the number of observed taxonomic units (OTUs) in WT and Nod2−/− mice before and after piroxicam treatment. (E) Relative abundances of intestinal microbial communities in WT and Nod2−/− mice before and after piroxicam treatment. n = 4–5 mice per group. *p<0.05, **p<0.01 and ns = not significant by ANOVA with Holm-Sidak multiple comparisons test for (B). Data are represented as mean ± SEM in (B) and (D).
DISCUSSION
Despite the discovery of NOD2 as a susceptibility gene more than a decade ago, it is still unclear why mutations that disrupt the ability of Nod2 to mediate an immune response to bacteria would increase the risk of developing CD, which is a disease characterized by an excessive immune response to intestinal bacteria. Addressing this question has been hindered by the lack of spontaneous inflammatory abnormalities reported in Nod2−/− mice. This problem has been compounded by inconsistent observations by individual laboratories due to differences in the background, housing conditions, and health status of the mice. In this study, we identified several inflammatory abnormalities in the small intestine of Nod2−/− mice - antimicrobial gene expression, goblet cell defects, and an increase in the proportion of IFN-γ expressing IELs. All three of these abnormalities, as well as increased inflammation induced by piroxicam, were associated with the susceptibility of Nod2−/− mice to B. vulgatus expansion and stable colonization. Thus, our results indicate that Nod2 prevents harmful immune responses by controlling the expansion of a pro-inflammatory member of the microbiota.
Recent studies indicate that defective cytokine production by myeloid or dendritic cells in response to bacteria leads to impaired immunity in Nod2−/− mice (Davis et al., 2011; Jiang et al., 2013). Our finding that Nod2-deficiency in the hematopoietic compartment is primarily responsible for uncontrolled B. vulgatus colonization could reflect such a role for Nod2. For instance, the inability of Nod2−/− dendritic cells to produce cytokines such as IL-15 that support lymphoid cells (Jiang et al., 2013) provides a potential explanation for the initial susceptibility to B. vulgatus. Although the exact antimicrobial response necessary for resistance to B. vulgatus requires further elucidation, our results are consistent with a model in which unrestricted B. vulgatus expansion evokes a sub-clinical inflammatory response including IFN-γ production that drives goblet cell abnormalities and is further exacerbated by piroxicam-induced inflammation (Figure S7D).
The intestinal abnormalities in Nod2−/− mice we describe are likely relevant to CD. The increased expression of Reg3 family members and defective Muc2 production by goblet cells has been documented in patients (Ogawa et al., 2003; Sheng et al., 2012). The alterations in goblet cell number and morphology in Nod2−/− mice are particularly striking, and defects in this cell type are known to have devastating effects on barrier function (Sheng et al., 2012). Also, our results demonstrating that these goblet cell abnormalities are dependent on lymphocytes and IFN-γ are consistent with related findings in patients suggesting that an excessive T cell response causes damage to the epithelium (Abraham and Cho, 2009). These observations in a mouse model are also consistent with the preferential association of NOD2 mutation with small intestinal involvement in humans (Abraham and Cho, 2009). For this reason, we chose to demonstrate the adverse effect of B. vulgatus colonization by examining the response of Nod2−/−mice to piroxicam, a small intestinal injury model that is frequently used to investigate the relationship between NSAID and recurrent IBD (Han et al., 2009). Notably, the overgrowth of Bacteroides in Nod2−/− mice under homeostatic condition is masked by large-scale phylum level changes following piroxicam challenge. While such shifts in microbial communities are consistent with observations made in CD patients (Gevers et al., 2014), our results raise the possibility that expansion of a bacterial species that initiated disease would no longer be easily detectable once disease symptoms become apparent. Hence, the major shifts in composition that occur in an inflammatory environment could make it difficult to identify specific proinflammatory bacteria in patients.
It will be critical in future studies to identify microbial factors that regulate virulence in addition to investigating host pathways. Bacteroides species are generally referred to as commensals because they are ubiquitous members of the gut microbiota and innocuous in most settings (Wexler, 2007). Although the susceptibility of Nod2−/− mice appears to be specific to a single Bacteroides genus member, a limitation of our study is that we examined only a subset of genus members. For instance, the B. thetaiotomicron strain we used in this study has been shown to colonize and evoke immune responses in mice, but is originally a human isolate (Goodman et al., 2009; Hooper et al., 2001; Peterson et al., 2007). Thus, the ability of a given Bacteroides strain to induce abnormalities could require host species-specific adaptations. However, such microbial factors should not be investigated in isolation. Pioneering studies have demonstrated that B. vulgatus induces colitis in germfree HLA-B27 transgenic rats but not in non-transgenic germfree controls (Rath et al., 1999). A more recent study showed that Bacteroides species mediate disease in mice severely deficient in immune-regulatory pathways (Il10r2−/−Tgfbr2DN mice) but not cohoused controls (Bloom et al., 2011). Similarly, we found that inoculation of metronidazole-treated WT mice with B. vulgatus did not reproduce intestinal abnormalities observed in Nod2−/− mice despite a similarly high degree of colonization during the timeframe of the experiment. This feature of B. vulgatus distinguishes it from other members of the microbiota that can expand in a susceptible background and transfer disease to WT counterparts through cohousing (Elinav et al., 2011; Garrett et al., 2007). Interestingly, Nod2−/−mice maintained at Jackson Laboratories are not colonized by B. vulgatus (data not shown). Therefore, housing conditions and animal facilities can clearly influence experimental results between institutions. The complex epidemiology of IBD in humans may reflect such variability in microbial exposure. When taken together, our experiments support a multi-hit model in which neither NOD2 mutation nor members of the microbiota are sufficient to cause intestinal disease in isolation.
Finally, the susceptibility of Nod2−/− mice to B. vulgatus shares certain similarities with our previous findings with Atg16L1 mutant mice and a specific strain of MNV (Cadwell et al., 2010). In both cases, a specific gene-microbe interaction leads to epithelial abnormalities in the intestine, and additional inflammatory pathologies develop in response to chemical injury. Thus, although the disease manifestation may be similar, the precise combination of genetic and environmental factors that initiate disease could vary considerably between individuals. Future treatment options targeting early events in disease progression may need to be personalized for patients to complement strategies that target downstream manifestations of inflammation such as TNFα production.
EXPERIMENTAL PROCEDURES
Mice
Nod2−/− mice backcrossed to the B6 background for at least 12 generations were previously described (Kim et al., 2008). MyD88−/− and Rag−/− B6 mice were from Jackson Laboratory and crossed to Nod2−/− mice to generate double mutants. Rip2−/− and Atg16L1HM B6 mice were previously described (Marchiando et al., 2013). Unless stated otherwise, animals for experiments were produced by breeding male and female mice homozygous for the mutant allele in a specific pathogen free (SPF) facility at NYU School of Medicine. WT B6 mice were purchased from Jackson Laboratory and bred onsite to generate controls for experiments. For cohousing experiments, 2–3 mice of one genotype were placed in the same cage with 2–3 mice of the other genotype for at least 2 weeks prior to separation, at which point only mice of the same genotype were placed in the same cage. Bone marrow chimeras were generated by lethally irradiating 8-week old female recipient mice (1100 CGy in two divided doses) followed by i.v. injection of 5×106 T cell-depleted bone marrow cells from donor female mice. For anti-IFN-γ treatment, mice were injected IP with 0.25mg anti-IFNγ (clone H22) or isotype control (polyclonal hamster IgG) (BioXCell) every 3 days for 9 days and animals were sacrificed on day 10. All animal studies were performed according to protocols approved by the NYU School of Medicine Institutional Animal Care and Use Committee (IACUC).
Microarray
The Leica LMD6000 Laser Microdissection system was used to capture crypt base epithelial cells from methacarn-fixed tissue as previously described (Cadwell et al., 2008) and Arcturus PicoPure RNA isolation kit was used to extract RNA. Enrichment of Paneth cells was confirmed by qPCR analysis of α-defensin1 normalized to Gapdh relative to whole tissue scraped from the paraffin embedded section. The Ovation Pico WTA System V2 from NuGEN was used to prepare and amplify cDNA, which was hybridized to a GeneChip Mouse Gene ST array (Affymetrix). Data has been deposited in NCBI’s Gene Omnibus and are accessible through the GEO accession number GSE58948. Pathway analysis of transcripts displaying >1.25 fold enrichment in Nod2−/− samples was performed by Database of Annotation, Visualization and Integrated Discovery (DAVID). Gene Set Enrichment Analysis (GSEA) was used to compare results to the Molecular Signatures Database (MSigDB).
Microscopy
Quantification of all microscopy data was performed blind. Intestinal sections were prepared and stained with PAS-Alcian blue or H&E as previously described (Cadwell et al., 2008; Cadwell et al., 2010). Goblet cells were quantified by counting the total number per villi and graphed as individual values. IF analyses was performed by staining formalin or methacarn fixed paraffin-embedded small sections with anti-Reg3β (R&D Systems), anti-Relm-β (Peprotech), or anti-Muc2 (Santa Cruz) followed by the appropriate secondary antibody. Sections were imaged using the Zeiss 710 confocal microscope.
Isolation of IELs and LP lymphocytes
Small intestine (Peyer’s patches removed) or colon was cut longitudinally and rinsed in HBSS. The tissue was washed in HBSS containing HEPES, sodium pyruvate, 5mM EDTA, and 1mM DTT for 15 min to obtain the IEL fraction. The intestines were further washed in HBSS containing HEPES, sodium pyruvate, and EDTA, and digested for 20 min using Collagenase VIII (Sigma) to obtain the LP fraction. Both IEL and LP fractions were filtered and fractionated on a Percoll gradient (40% and 80%). The cells at the interphase of the gradient were collected and washed twice with complete RPMI.
Bacterial culture and inoculation
Fecal bacteria were quantified by dilution plating on selective BBE agar (BD) in an anaerobic chamber (AS-580, Anaerobe Systems) at 37°C. Colonies of a single color and morphology grew from Nod2−/− samples within 24–36 hours but not WT samples. 20 colonies from 3 Nod2−/− samples were chosen at random to identify the bacterium by sequencing of the 16S rRNA gene (Fwd primer 1 5’-AGAGTTTGATCCTGGCTCAGGAT-3’, Rev primer 1 5’-TCCTTTGAGTTTCACCGTTGCC-3’, Fwd primer 2 5’-CAGGATTAGATACCCTGGTAGTCCA-3’, Rev primer 2 5’-GACTTAGCCCCAGTCACCAGT-3’). 100% of the sequenced colonies were identified as B. vulgatus. For inoculation into mice, B. vulgatus was anaerobically cultured in PYG broth at 37°C for 48 hrs and metronidazole-treated mice were gavaged with 1×108 bacteria. Metronidazole was lifted 24 hrs prior to inoculation. Heat-killed B. vulgatus was prepared by heating the culture at 80°C in aerobic conditions for 1 hr. B. thetaiotaomicron (VPI-5482) and B. uniformis (Eggerth and Gagnon) were kindly provided by Eric Martens and were cultured and administered to mice using a similar procedure.
Administration of antibiotics and piroxicam
Mice were given 1g/L metronidazole alone, 500mg/L vancomycin alone, or in combination with 1g/L ampicillin and 1g/L neomycin sulfate with 1% sucrose (Sigma Aldrich) in their drinking water for 10 days. Stool from treated mice was plated on blood agar and BBE agar to confirm reduction in total bacteria or B. vulgatus respectively. WT and Nod2−/− mice were treated with 60mg/kg piroxicam and 80mg/kg piroxicam in powdered mouse chow for 7 days each. For piroxicam experiments, mice received metronidazole for 10 days and after reduction of B. vulgatus was confirmed, metronidazole was removed and mice were treated with the 2 doses of piroxicam. A separate group of mice treated with metronidazole were gavaged with 108 cfu B. vulgatus and treated with piroxicam concurrently. H&E stained small intestinal sections of mice treated with piroxicam were used for histopathological scoring in a blinded fashion. Each mouse was given an individual cumulative score (max = 5) based on the following criteria: number of focal ulcers (0 = none, 0.5 = 1–2, 1 = 3 or more), the extent of epithelial hyperplasia (0 = none, 0.5 = elongated villi and crypts, 1 = severe hyperplasia where the crypt villus axis is 2 times higher than the crypt villus axis in untreated mice), the presence of immune infiltrates (0 = none, 0.5 = pericryptal infiltrates, 1 = submucosal infiltrates), the presence of intestinal bleeding (1), and attachment of the intestine to the peritoneal cavity (1).
Statistical analysis
Analyses except for microarray data used GraphPad Prism v6. An unpaired two-tailed t test was used to evaluate differences between two groups. An ANOVA with the Holm-Sidak multiple comparisons test was used to evaluate experiments involving multiple groups. For experiments requiring non-parametric analyses, the Wilcoxon-Mann-Whitney test or Kruskal-Wallis with Dunn’s multiple comparisons test were used.
Supplementary Material
Highlights.
Nod2−/− mice develop inflammatory gene expression and goblet cell defects
Intestinal abnormalities are associated with an excessive immune response
Nod2−/− mice are susceptible to expansion and colonization by B. vulgatus
B. vulgatus mediates intestinal abnormalities and inflammation in Nod2−/− mice
ACKNOWLEDGEMENTS
We would like to thank Dan Littman and Susan Schwab for providing reagents, Yan Deng for assistance with confocal microscopy (NCRR S10 RR023704-01A1), Jiri Zavadil for assistance with microarray analysis, NYULMC OCS microscopy core for their assistance with transmission electron microscopy, and genome technology center, flow cytometry, and histopathology cores (Cancer Center Support Grant, P30CA016087) for assistance with sample preparation and analyses. This research was supported by NIH grants R01 DK093668 (DR and KC) and AI093811 (PL); The Broad Medical Research Program (PL); Kevin and Marsha Keating Family Foundation (PL); NIH/NCATS UL1 TR000038 (KC and PL); Crohn’s & Colitis Foundation of America Genetics Pilot Award (KC); and New York University Whitehead Fellowship (KC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
D.R., P.L., and K.C. formulated the original hypothesis, designed the study, and analyzed results. D.R. performed the experiments. M.S.T., and R.B. assisted with sample preparation for bacterial 16S deep sequencing, and M.S.T. performed the computational analysis. D.R. and K.C. wrote the manuscript, and all authors commented on the manuscript, data, and conclusions before submission.
REFERENCES
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith MJ, Boulard O, Powrie F, Maloy KJ. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. 2010;139:519–529. doi: 10.1053/j.gastro.2010.04.045. 529 e511–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau F, Madre C, Meinzer U, Berrebi D, Dussaillant M, Merlin F, Eckmann L, Karin M, Sterkers G, Bonacorsi S, et al. Nod2 regulates the host response towards microflora by modulating T cell function and epithelial permeability in mouse Peyer's patches. Gut. 2010;59:207–217. doi: 10.1136/gut.2008.171546. [DOI] [PubMed] [Google Scholar]
- Billmann-Born S, Till A, Arlt A, Lipinski S, Sina C, Latiano A, Annese V, Hasler R, Kerick M, Manke T, et al. Genome-wide expression profiling identifies an impairment of negative feedback signals in the Crohn's disease-associated NOD2 variant L1007fsinsC. Journal of immunology. 2011;186:4027–4038. doi: 10.4049/jimmunol.1000085. [DOI] [PubMed] [Google Scholar]
- Biswas A, Liu YJ, Hao L, Mizoguchi A, Salzman NH, Bevins CL, Kobayashi KS. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14739–14744. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell host & microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M, Lu P, Van Goudoever JB, Wells JM, Dekker J, et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3beta, Reg3gamma and angiogenin-4. PLoS One. 2012;7:e38798. doi: 10.1371/journal.pone.0038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Philpott D, Girardin SE, Zouali H, Lesage S, Chareyre F, Bui TH, Giovannini M, Zaehringer U, Penard-Lacronique V, et al. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3455–3460. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. The Journal of clinical investigation. 2013 doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. The Journal of clinical investigation. 2011;121:3666–3676. doi: 10.1172/JCI57761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Hugot JP, Sansonetti PJ. Lessons from Nod2 studies: towards a link between Crohn’s disease and bacterial sensing. Trends in immunology. 2003;24:652–658. doi: 10.1016/j.it.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Uchida K, Jurickova I, Koch D, Willson T, Samson C, Bonkowski E, Trauernicht A, Kim MO, Tomer G, et al. Granulocyte-macrophage colony-stimulating factor autoantibodies in murine ileitis and progressive ileal Crohn's disease. Gastroenterology. 2009;136:1261–1271. doi: 10.1053/j.gastro.2008.12.046. e1261–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal immunology. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Hoshi N, Schenten D, Nish SA, Walther Z, Gagliani N, Flavell RA, Reizis B, Shen Z, Fox JG, Iwasaki A, et al. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nature communications. 2012;3:1120. doi: 10.1038/ncomms2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LC, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. The Journal of biological chemistry. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang X, Zeng B, Liu L, Tardivel A, Wei H, Han J, Macdonald HR, Tschopp J, Tian Z, et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. The Journal of experimental medicine. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Park JH, Shaw MH, Franchi L, Inohara N, Nunez G. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–257. doi: 10.1016/j.immuni.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kim YG, Shaw MH, Warner N, Park JH, Chen F, Ogura Y, Nunez G. Cutting edge: Crohn’s disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. Journal of immunology. 2011;187:2849–2852. doi: 10.4049/jimmunol.1001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, Wang C, Ziel JW, van Rooijen N, Nunez G, et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell host & microbe. 2013;14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVay LD, Keilbaugh SA, Wong TM, Kierstein S, Shin ME, Lehrke M, Lefterova MI, Shifflett DE, Barnes SL, Cominelli F, et al. Absence of bacterially induced RELMbeta reduces injury in the dextran sodium sulfate model of colitis. The Journal of clinical investigation. 2006;116:2914–2923. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Barreau F, Al Nabhani Z, Dussaillant M, Le Roux K, Dore J, Leclerc M, Hugot JP, Lepage P. Altered gut microbiota composition in immune-impaired Nod2(−/−) mice. Gut. 2012;61:634–635. doi: 10.1136/gutjnl-2011-300478. [DOI] [PubMed] [Google Scholar]
- Netea MG, Ferwerda G, de Jong DJ, Werts C, Boneca IG, Jehanno M, Van Der Meer JW, Mengin-Lecreulx D, Sansonetti PJ, Philpott DJ, et al. The frameshift mutation in Nod2 results in unresponsiveness not only to Nod2- but also Nod1-activating peptidoglycan agonists. J Biol Chem. 2005;280:35859–35867. doi: 10.1074/jbc.M504924200. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, Kitayama T, Matsuno S, Ohtani H, Takasawa S, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Current opinion in infectious diseases. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz M, Hwang SH, Wilhelm CL, Kirkland D, Benson A, Sturge CR, Mirpuri J, Vaishnava S, Hou B, Defranco AL, et al. Parasite-induced T(H)1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nature immunology. 2012 doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath HC, Wilson KH, Sartor RB. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infection and immunity. 1999;67:2969–2974. doi: 10.1128/iai.67.6.2969-2974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, Philpott DJ. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan MT, Carroll IM, Grossniklaus E, White A, von Furstenberg RJ, Barner R, Fodor AA, Henning SJ, Sartor RB, Gulati AS. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut. 2013 doi: 10.1136/gutjnl-2012-304190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng YH, Hasnain SZ, Florin TH, McGuckin MA. Mucins in inflammatory bowel diseases and colorectal cancer. Journal of gastroenterology and hepatology. 2012;27:28–38. doi: 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature immunology. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandussen KL, Liu TC, Li D, Towfic F, Modiano N, Winter R, Haritunians T, Taylor KD, Dhall D, Targan SR, et al. Genetic Variants Synthesize to Produce Paneth Cell Phenotypes That Define Subtypes of Crohn's Disease. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJ, Strober W. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nature immunology. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.