Abstract
Introduction
Antiretroviral resistance leads to treatment failure and resistance transmission. Resistance data in western Kenya are limited. Collection of non-plasma analytes may provide additional resistance information.
Methods
We assessed HIV diversity using the REGA tool, transmitted resistance by the WHO mutation list and acquired resistance upon first-line failure by the IAS–USA mutation list, at the Academic Model Providing Access to Healthcare (AMPATH), a major treatment programme in western Kenya. Plasma and four non-plasma analytes, dried blood-spots (DBS), dried plasma-spots (DPS), ViveSTTM-plasma (STP) and ViveST-blood (STB), were compared to identify diversity and evaluate sequence concordance.
Results
Among 122 patients, 62 were treatment-naïve and 60 treatment-experienced; 61% were female, median age 35 years, median CD4 182 cells/µL, median viral-load 4.6 log10 copies/mL. One hundred and ninety-six sequences were available for 107/122 (88%) patients, 58/62 (94%) treatment-naïve and 49/60 (82%) treated; 100/122 (82%) plasma, 37/78 (47%) attempted DBS, 16/45 (36%) attempted DPS, 14/44 (32%) attempted STP from fresh plasma and 23/34 (68%) from frozen plasma, and 5/42 (12%) attempted STB. Plasma and DBS genotyping success increased at higher VL and shorter shipment-to-genotyping time. Main subtypes were A (62%), D (15%) and C (6%). Transmitted resistance was found in 1.8% of plasma sequences, and 7% combining analytes. Plasma resistance mutations were identified in 91% of treated patients, 76% NRTI, 91% NNRTI; 76% dual-class; 60% with intermediate-high predicted resistance to future treatment options; with novel mutation co-occurrence patterns. Nearly 88% of plasma mutations were identified in DBS, 89% in DPS and 94% in STP. Of 23 discordant mutations, 92% in plasma and 60% in non-plasma analytes were mixtures. Mean whole-sequence discordance from frozen plasma reference was 1.1% for plasma-DBS, 1.2% plasma-DPS, 2.0% plasma-STP and 2.3% plasma-STB. Of 23 plasma-STP discordances, one mutation was identified in plasma and 22 in STP (p<0.05). Discordance was inversely significantly related to VL for DBS.
Conclusions
In a large treatment programme in western Kenya, we report high HIV-1 subtype diversity; low plasma transmitted resistance, increasing when multiple analytes were combined; and high-acquired resistance with unique mutation patterns. Resistance surveillance may be augmented by using non-plasma analytes for lower-cost genotyping in resource-limited settings.
Keywords: HIV, drug resistance, subtype, diversity, Kenya, analyte, AMPATH
Introduction
Antiretroviral therapy (ART) resistance is a cause and a consequence of treatment failure [1, 2]. Optimizing treatment in resource-limited settings (RLS) is problematic due to limited availability and high cost of new ART regimens and treatment monitoring [3–9]. Surveillance studies to evaluate HIV-transmitted drug resistance (TDR) before ART [10] and to characterize acquired resistance upon treatment failure [11, 12] rely on technology not easily accessible for patient management. Less costly, simpler analytes, including dried blood spots (DBS), dried plasma spots (DPS) and ViveSTs (ST; formerly Sample Tankers®), could facilitate ART management [13–18].
Adult HIV prevalence in Kenya (5.6–6.1% in 2012) is the 12th highest worldwide, representing a high health-burden on the country [19–21]. HIV-1 infection is highly diverse with co-circulation of subtypes A (50–80%), D (10–20%) and C (5–15%), and multiple recombinants (10–20%) [22–34]. ART access has significantly increased in Kenya since 2001, with positive patient outcomes [35, 36]. First-line ART has included zidovudine/stavudine, lamivudine and nevirapine/efavirenz, with a recent, still on-going, substitution of tenofovir for stavudine, in keeping with recent World Health Organization (WHO) guidelines [37]. Data on drug resistance (DR) in the country are limited. TDR from mother to child was reported in 29–67% [38, 39], and in adults among 1.1–7.5% in the coast and Nairobi [22, 40–44]. Resistance upon ART failure has only been reported in three studies, one describing 14% resistance in 58 injecting drug users in Mombasa with no available treatment histories [45]; the second in 15% of 100 patients from two clinics in Mombasa and Nairobi as part of a multi-site African study [46]; and the third from our work reporting 94% resistance in 28 patients from Burnt Forest, a rural Academic Model Providing Access to Healthcare (AMPATH) clinic [34].
To gain preliminary insight on the magnitude of resistance in diverse circulating subtypes and its potential impact, we examined HIV diversity, TDR in newly diagnosed patients and acquired resistance in treatment-experienced patients failing first-line ART at AMPATH, a large HIV treatment programme in western Kenya [47]. Genotyping from plasma and non-plasma analytes as alternate options for resistance testing were compared, to investigate alternative monitoring strategies in western Kenya and other RLS.
Methods
Study setting
As of March 2013, AMPATH [47–50] provided comprehensive clinical services to 138,736 HIV-positive patients. Of those, 78,064 are in care, and 58,841 receive ART. The Moi Teaching and Referral Hospital (MTRH) clinic, AMPATH's largest, enrolled 26,791 adults, 42% on ART. Patients are managed with an electronic medical record [51] according to protocols based on WHO and AMPATH guidelines. At the time of this study, first-line ART included zidovudine/stavudine, lamivudine and efavirenz/nevirapine.
Patient enrolment
Between May 2006 and November 2007, two groups of HIV-positive adults attending MTRH were offered enrolment. Inclusion criteria for treatment-naïve patients included: 1) no prior ART and 2) most recent CD4 count>350 or<200 cells/mL, in an attempt to identify recent versus chronic infections. Inclusion criteria for treatment-experienced patients included: 1) on WHO-recommended first-line ART >6 months; 2) ART adherence >50% based on patient-report; and 3) suspected of failing therapy based on a CD4 count drop >25% over six months prior to enrolment, or no increase in CD4 after 12 months of ART.
Consenting patients were enrolled sequentially until the desired number of samples was obtained – about 60 treatment-naïve and 60 experienced, 20% higher than the desired enrolment of 100, to account for sample deterioration. Upon enrolment, patients were interviewed, charts were reviewed for demographic, clinical and laboratory characteristics, and blood was obtained. The study was approved by Lifespan and Moi University ethics review boards.
Laboratory methods
CD4 (FACSCalibur system; Becton Dickenson, San Jose, CA, USA) and viral load (VL) (Amplicor 1.5; Roche Molecular, Pleasanton, CA, USA) testing were done at the AMPATH reference laboratory, which participates in the United Kingdom Quality Assessment Service and National Institutes of Health Department of AIDS Viral Quality Assurance Programs. Virologic failure and detectable VL were defined as VL>400 copies/mL.
Each sample was prepared as: 1) plasma; 2) DBS; 3) DPS; 4) STB; and 5) STP. Plasmas were shipped on dry-ice to the US, and frozen at −80°C. DBS and DPS were prepared with 100 µL/spot on Whatman 903 Protein-Saver Cards. First-generation STs, a novel dried transportation matrix device [13, 16], were prepared with 1 mL plasma (STP) or blood (STB) per ST. Filter and ST analytes were hood-dried overnight upon initial sample preparation. To increase the number of STP sequences, a second STP batch was prepared in the US from thawed frozen-plasma and dried on driDOC, a prototype drying device, according to manufacturer's instructions. Analyses included sequences from both batches. DBS and DPS were stored with desiccant at room temperature, hand-carried to the US and frozen at −80°C. STs were shipped at room temperature to Research Think Tank (Alpharetta, GA, USA) [52] and stored at ambient room temperature for 10–90 days before evaluating the built-in colour indicator capsule and testing.
For DBS and DPS, spots were placed directly into 9 mL of Nuclisens lysis buffer and agitated for 2 hours at room temperature and RNA extraction completed according to the Nuclisens minMag protocol (Biomerieux, Durham, NC, USA). Genotyping was performed as previously described [53]. Briefly, RNA was reverse transcribed via Life Technologies (Carlsbad, CA, USA) SuperScript III One-Step RT-PCR kit and a second-round PCR was performed using Life Technologies Platinum Taq. PCR products were Sanger sequenced. Sequence assembly was with Sequencher (Gene Codes Corporation, Ann Arbor, MI, USA). Amplification was attempted for all plasma, STP and STB and a portion of DBS (64%) and DPS (37%). For success rate calculations, only the portion of STP (37%) and STB (43%) that had “blue” colour indicators were counted. Such indicators are built into the STs pointing to adequate (blue) or inadequate (pink) drying [54].
ST genotyping was performed as previously described [54], and accomplished by recovering ST-dried samples with version 1 kit supplied eluent buffer. RNA extraction was with QIAmp Viral RNA mini Kit (QIAGEN, USA). Sequences were obtained with TRUGENE® HIV-1 Genotyping System (Siemens Healthcare Diagnostics, Tarrytown, NY, USA).
Data analysis
Patient-level data included age, gender, previous ART and previous CD4. Sample covariates included time spans between sample acquisition and shipment, and between arrival in the US and genotyping.
Sequence quality control was with SQUAT [55]. Hypermutation and susceptibility analyses were with Stanford Database tools, accessed 1 August 2012 [56]. TDR among treatment-naïve patients was interpreted with the WHO mutation list [57] and compared to the IAS–USA list [58]. Acquired resistance among treatment-experienced patients was with the IAS–USA list. Mixtures were considered mutant. Subtype was derived with REGA version 2.0 [59], with manual bootscan assessment.
For plasma and DBS, multivariable logistic regression analyses were used to examine the relationships between genotyping success, as the dependent variable, and log10 VL, the duration between sample acquisition and shipping, the duration between shipping and genotyping, and patient stratum (naïve/experienced). For DPS, STP and STB, the logistic regression analyses included only log10 VL and patient stratum as independent variables. The linearity assumption was evaluated in all models as an exploratory step using generalized additive models.
For resistance detection concordance between analytes, resistance-associated amino acids were counted as distinct mutations. Mutations between analyte pairs were compared using McNemar's test.
The number of nucleic acid (NA) differences was calculated for each non-plasma/plasma sequence pair. Both complete and partial NA differences were counted and summarized by analyte type. For each non-plasma analyte, discordance rates between treated and naïve and among subtypes were compared using Poisson regression, fit using generalized estimating equations, an unstructured correlation structure between analyte types and an offset corresponding to the log-length of the sequence overlap in the pair. Outcome was number of discordances and dependent variables were analyte type, treatment status and subtype. Robust standard errors were used to calculate 95% confidence intervals and p-values.
Results
Patients and genotypes
A total of 122 patients, 62 ART-naïve and 60 ART-experienced, were enrolled. The treatment-naïve group was sequentially enrolled from May to August 2006, after screening 436 patients. Of those screened for the treatment naïve group and not enrolled, 102 were not new to clinic, 114 were treated, 40 did not fit CD4 criteria, 111 missed their appointment, three were missed by a research assistant and four refused.
The ART-experienced group was enrolled from January to November 2007 using clinical and immunological WHO criteria. As previously reported as a result of this study, misclassification of virologic failure using these criteria was high [7]. Of 209 patients who met CD4 enrolment criteria and had VL testing, 60 (29%) had detectable VL, and were included. Specific breakdown of patients screened for the treatment experienced group is not available.
Table 1 shows demographic, clinical and genotype characteristics according to patient stratum. Median age of participants was 35 years, 61% were female, median enrolment CD4 was 182 cells/µL (14%) and median VL 4.6 log10 copies/mL. Treated patients were on ART a median 2.4 years, most (88%) with stavudine+lamivudine+nevirapine/efavirenz.
Table 1.
Demographic, clinical and genotypic characteristics of study cohorta
| Variable | Naïve: CD4<200 | Naïve: CD4>350 | Treated and failing | Total |
|---|---|---|---|---|
| Number | 28 | 34 | 60 | 122 |
| Age | 34 (22, 55) | 34 (19, 68) | 37 (20, 64) | 35 (19, 68) |
| Male | 12/28, 43% | 11/34, 32% | 24/60, 40% | 47/122, 39% |
| CD4 count | 99 (8, 195) | 507 (367, 1984) | 153 (8, 719) | 182 (8, 1984) |
| CD4% | 8 (1, 30) | 25.5 (10, 60) | 11 (1, 30) | 14 (1, 60) |
| Viral load (log10 copies/mL) | 5.1 (3.2, 6.3) | 4.3 (2.6, 5.3) | 4.4 (3.8, 6.0) | 4.6 (2.6, 6.3) |
| Antiretroviral regimen | ||||
| 3TC, D4T, NVP | 40/59, 68% | |||
| 3TC, AZT, NVP | 5/59, 8% | |||
| 3TC, D4T, EFV | 12/59, 20% | |||
| 3TC, AZT, EFV | 2/59, 3% | |||
| Years on antiretroviral therapy | 2.4 (0.92, 5.3) | |||
| Subtype | ||||
| A | 16/28, 57% | 21/30, 70% | 29/49, 59% | 66/107, 62% |
| D | 5/28, 18% | 3/30, 10% | 8/49, 16% | 16/107, 15% |
| AD | 2/28, 7% | 4/30, 13% | 6/49, 12% | 12/107, 11% |
| C | 4/28, 14% | 1/30, 3% | 1/49, 2% | 6/107, 6% |
| AC | 0/28, 0% | 1/30, 3% | 2/49, 4% | 3/107, 3% |
| Otherb | 1/28, 4% | 0/30, 0% | 3/49, 6% | 4/107, 4% |
Values are presented as median (range) for continuous variables and n/N, % for categorical variables;
AG (1/107, 1%); CD (1/107, 1%); undetermined (2/107, 2%). 3TC, lamivudine; D4T, stavudine; AZT, zidovudine; NVP, nevirapine; EFV, efavirenz.
Genotyping success
A total of 196 sequences were available for 107/122 (88%) patients, 58/62 (94%) naïve and 49/60 (82%) treated. Genotyping success rates were 100/122 (82%) for plasma; 37 of the 78 attempted DBS (47%); 16 of the 45 attempted DPS (36%); 14 of the 44 attempted field-plasma STP with blue indicators (32%) and 23 of the 34 frozen-plasma STP (68%) (one additional STP sequence was subsequently obtained); and 5 of the 42 attempted STB (12%). Differences in STP success rates may be attributable to sample (fresh/frozen), sample handling or drying methods. No participant had sequences from all five analytes; seven had sequences from four analytes; 16 from three; 36 from two; 48 from one (43/48 plasma); and 15 from none (12%). All intra-patient sequences clustered phylogenetically with high bootstrap values. No hyper-mutation was identified.
For plasma, odds of successful genotyping were higher at higher VL but in a non-linear manner suggesting a threshold at approximately 3.5 log10 VL. Odds of successful genotyping with VL>3.5 log copies/mL were 9.5 times the odds for VL≤3.5 log copies/mL (95% CI=1.3–69.6, p=0.03). Longer shipment-to-genotyping time (mean 6.0 months, range 4.2–8.9) was also predictive of decreased success, with an odds ratio (OR) of 0.66 per one month longer duration (95% CI 0.46–0.96, p=0.03), but sampling-to-shipping time was not predictive of genotyping success. For DBS, higher log10 VL (OR=3.4, CI=1.3–9.4, p=0.01) was associated with higher success but, unlike plasma, the relationship between log odds of success and log10 VL was linear and therefore, a threshold was not examined. Longer sampling-to-shipping time (median 2.9 months, range 0.5–16.1) was also associated with lower DBS success, with an OR of 0.5 per one month longer duration (CI=0.3–0.8, p=0.003). Higher VL was also associated with STP genotyping success (OR=3.5 per 1 log10 higher VL; CI=1.4–8.8; p=0.01), but not with DPS. The pink colour indicator within the ST, which identifies inadequate drying, was highly predictive of genotyping success. The association between genotyping success and VL was not examined for STB because of the small sample size (five sequences).
HIV diversity
Common subtypes were A (62%; 60% A1 and 2% A2), D (15%) and C (6%), followed by various unique recombinants, including AD/DA/DAD/ADA (11%), AC/CA (3%), CD in (1%), AG (1%) and 2% undetermined (Table 1).
DR in treatment-naïve patients
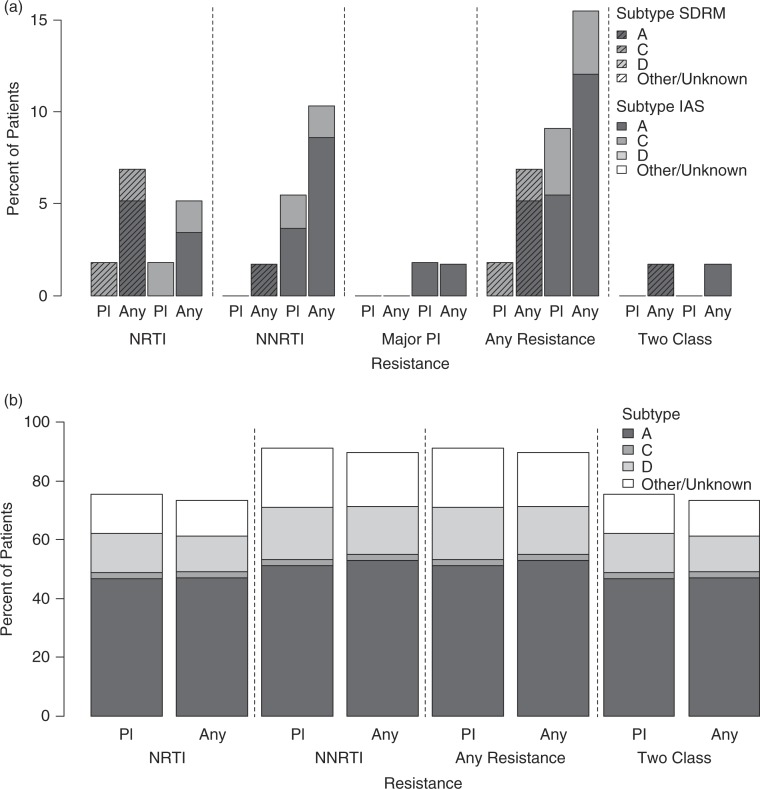
Plasma sequences were available for 55/62 (89%), and from any analyte from 58/62 (94%) naïve patients (Figure 1a). Based on the WHO list 1/55 (1.8%, 95% binomial CI 0.04–9.7%) had resistance in plasma (RT L210W; subtype C). However, combining analytes, TDR was observed in 4/58 (7%, CI 1.9–16.7%), including RT T215ST, Y181C (STP; subtype A); D67DN, K219KQ (DPS/STP; subtype A); F77FL (STP; subtype A) and L210LW (plasma/DBS/STP; subtype C). There were no differences in TDR between the two CD4 groups, or among different subtypes.
Figure 1.
HIV drug resistance in ART naïve (a) and treated (b) patients according to HIV-1 subtype. Bars show percent of patients with transmitted (a) or acquired (b) nucleoside reverse transcriptase inhibitor (NRTI), non-nucleoside reverse transcriptase inhibitor (NNRTI), major protease inhibitor (PI; in (a) only), any and two-class drug resistance. Results for each resistance category are shown for plasma and for any analyte (combining plasma and non-plasma analytes), according to HIV-1 subtype. Results for treatment naïve patients (a) are shown for the World Health Organization surveillance drug resistance mutation (SDRM) list (hashed bars) and for the International Antiviral Society–USA (IAS–USA) list (solid bars). “Pl,” plasma.
Using the IAS–USA list, four additional ART-naïve participants, eight in total (14%), had RT resistance; 0/4 in plasma only, 2/4 in non-plasma analytes only and 2/4 combined. One patient had a major PI resistance mutation (Q58E; subtype A) and all had minor mutations, most commonly K20R, M36I, H69K and L89M for subtype A; M36I, H69K and I93L for subtype C; and M36I, I64V and L63P for subtype D. Two-class TDR was seen in 1/58 (1.7%) patients according to either mutation list and 2/58 (3.4%) combining lists.
DR in treatment experienced patients
Plasma sequences were available for 45/60 (75%) drug-experienced patients and from any analyte for 49/60 (82%) patients (Figure 1b). In plasma, 91% (41/45, CI 78.8–97.5%) had RT-associated mutations, 76% NRTI and 91% NNRTI. No major PI mutations were observed. Fifteen percent had one-class (all NNRTI, CI 6–29%) and 76% dual-class resistance (CI 60–87%). In plasma, the median number of mutations per patient was 4 (range 0–10), 2 (range 0–7) NRTIs and 2 (range 0–5) NNRTIs; 60% had ≥4, and 47% had ≥5 mutations (Table 2). Common NRTI mutations were M184V (76%); T215F/Y (42%); D67N (27%); and M41L (22%). K65R was not observed. Common NNRTI mutations were K103N/S (40%); G190A/S (31%); Y181C (22%); and K101E/H (18%). The number of NRTI, NNRTI or total mutations did not differ by subtype.
Table 2.
Drug resistance mutation patterns in treated patients failing first-line ART, according to subtype and number of resistance mutations
| ID | Subtype | P/NP | NRTI | NNRTI |
|---|---|---|---|---|
| 1 | A | 1/1 | 41LMa, 67N, 70KR, 75IMV, 184V, 215F, 219KQ | 101H, 106M, 190A |
| 2 | A | 1/0 | 67N, 70R, 184V, 215F, 219Q | 101E, 190A |
| 3 | A | 1/0 | 41L, 62V, 184V, 215Y | 108I, 181C, 221Y |
| 4 | A | 1/3 | 67N, 70R, 184V, 215F, 219E | 103NSb, 181C |
| 5 | A | 1/2 | 67N, 70R, 184V, 219Qc | 101E, 190A |
| 6 | A | 1/0 | 70EK, 116Y, M184V | 101E, 181CY, 190A |
| 7 | A | 1/0 | 184V, 215FIST | 108IV, 181C, 190A, 221Y |
| 8 | A | 0/2 | 41Ld, 184Vd, 210Wd, 215Yd | 103Nd, 138Gd |
| 9 | A | 1/1 | 75IM, 184V, 215F | 103N, 190AGa |
| 10 | A | 1/0 | 184V, 215F | 103N, 138Q, 179L |
| 11 | A | 1/1 | 41La, 184V, 215F | 103N, 138Q |
| 12 | A | 1/0 | 184V, 215F | 103N, 106I, 230L |
| 13 | A | 1/0 | 41L, 184V, 215Y | 103N, 138Q |
| 14 | A | 1/1 | 75I, 184V | 90I, 181C, 221Y |
| 15 | A | 1/0 | 41L, 70R, 184V | 188L |
| 16 | A | 1/0 | 67N, 184V | 101E, 190A |
| 17 | A | 1/0 | 67N, 70R, 184V | 181C |
| 18 | A | 1/0 | 184V, 215Y | 101E, 190A |
| 19 | A | 1/1 | 184V | 103N |
| 20 | A | 1/0 | A62V, M184V | 103N |
| 21 | A | 1/0 | 184V | 181C, 221Y |
| 22 | A | 0/2 | 184Vd | 103Nd |
| 23 | A | 1/2 | 184V | 190S |
| 24 | A | 0/1 | Noned | 103Nd |
| 25 | A | 1/3 | None | 188L |
| 26 | A | 1/0 | None | 103N |
| 27 | A | 0/1 | Noned | Noned |
| 28 | A | 1/0 | None | None |
| 29 | A | 1/0 | None | None |
| 30 | AC | 1/0 | 41L, 67N, 184V, 210W, 215F | 181CY, 190A |
| 31 | AC | 1/1 | 41L, 184V, 215Y | 98AGa, 106IVa, 188L |
| 32 | AD | 1/1 | 41La, 184V, 215F | 103N, 106I, 108Ia, 221Ya, 230L |
| 33 | AD | 1/2 | 67N, 70R, 184V, 215F, 219Q | 101E, 190A |
| 34 | AD | 1/0 | 184V, 215Y | 90I, 103N, 138A |
| 35 | AD | 1/0 | 67N, 184V | 101E, 190A |
| 36 | AD | 1/0 | None | 138A |
| 37 | AD | 1/1 | None | None |
| 38 | AG | 1/2 | None | 103N |
| 39 | C | 1/1 | 41LMa, 67N, 70R, 184V, 215F, 219EQb | 106M, 179D, 230LM |
| 40 | CD | 1/1 | None | None |
| 41 | D | 1/1 | 67N, 184V, 210W, 215Y | 108I, 181C, 221Yc |
| 42 | D | 1/0 | 41L, 67N, 184V, 210W, 215Y | 103N |
| 43 | D | 1/1 | 184V | 103N, 108I, 225H |
| 44 | D | 1/1 | 184V | 103N, 138GQR |
| 45 | D | 1/1 | 184V | 103S, 190A |
| 46 | D | 1/0 | 184V | 190A |
| 47 | D | 1/1 | None | 103KNa |
| 48 | D | 1/0 | None | 103N |
| 49 | ? | 1/0 | None | 181CY |
The table is sorted by overall number of mutations in descending order within subtype. P/NP is whether there is a plasma sequence (yes=1, no=0)/the number of non-plasma sequences;
Non-plasma analyte available, but mutation found only in plasma;
219EQ found in non-plasma analyte and 219E in plasma; 103NS found in non-plasma analyte and 103N in plasma;
plasma sequence available, but mutation found only in non-plasma analytes (DBS and STP for 219Q; DBS for 219EQ and 221Y);
plasma sequence not available.
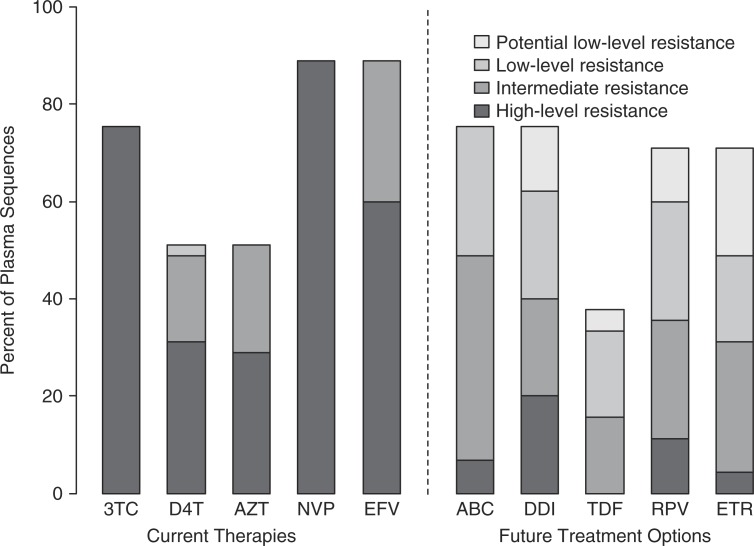
In plasma sequences, 89% of patients had intermediate-to-high predicted resistance to first-line regimens, 13% for one, 27% for two and 49% for all three drugs; 40/45 (89%) for nevirapine and efavirenz, (34/45) 76% for lamivudine, (22/45) 49% for stavudine, and (23/45) 51% for zidovudine (Figure 2). Sixty percent had intermediate or high-level resistance to subsequent second-line RT inhibitors treatment options, including 11% to all five medications.
Figure 2.
Predicted drug resistance to current ART and future treatment options in treated patients. “Current therapies” to the left of the dashed line, refers to medications taken by participants at the time of the study; “Future therapies” to the right of the dashed line refers to potentially available subsequent second-line RT inhibitor options at the time of the study. Bars show percent of plasma sequences with predicted resistance according to four categories, based on resistance scores available at the Stanford HIV Sequence Database [56]. 3TC, lamivudine; D4T, stavudine; AZT, zidovudine; NVP, nevirapine; EFV, efavirenz; ABC, abacavir; DDI, didanosine; TDF, tenofovir; RPV, rilpivirine; ETR, etravirine.
Examination of co-occurrence of resistance mutations in this non-B subtype cohort, checked against sequences with at least two resistance mutations in the Stanford Database [56], revealed unique patterns in 11/21 (52%) subtype A sequences, that were not found among 295 subtype A sequences from patients on NRTIs+NNRTI; and 5/21 (24%) that were not found among 10,767 subtype B sequences with the same ART. For subtype D, 4/6 (67%) demonstrated unique patterns compared to 188 subtype D Stanford sequences, one of which (17%) was seen in only one subtype B sequence. The one subtype C sequence had a pattern that was not seen in 1636 subtype C or the subtype B sequences. Additionally, V106M, a subtype-C specific NNRTI mutation [60] was observed in our dataset in one subtype A isolate, observed previously only in one subtype A sequence from South Africa [61].
Combining analytes, 90% had RT-associated mutations, 73% NRTI and 90% NNRTI; 16% had one-class resistance, and 73% dual-class. Five patients did not have evidence for any DR despite treatment failure. This is most likely due to poor medication adherence, though all five did report full adherence.
Analyte concordance for resistance mutations
A total of 97 mutations (50 NRTI; 47 NNRTI, at 45 positions) appeared in plasma sequences that had non-plasma analyte pairs. Of those, 65/74 (88%) were identified in DBS (39/43, 91% NRTI; 26/31, 84% NNRTI); 17/19 (89%) in DPS (8/8, 100% NRTI; 9/11, 82% NNRTI); and 34/36 (94%) in STP (19/19, 100% NRTI; 15/17, 88% NNRTI). Mutation detection was not significantly different by analyte type.
Additional mutations, not identified in plasma, included three in DBS (2 NRTI; 1 NNRTI); two in DPS (1 NRTI; 1 NNRTI); and six in STP (3 NRTI; 3 NNRTI). Combining non-plasma analytes, of 97 plasma mutations, 84 (87%) were also identified by non-plasma analytes (46/50, 79% NRTI; 38/47, 81% NNRTI); and 10 additional mutations, not identified in plasma, were detected in non-plasma analytes (5 NRTI; 5 NNRTI). Of 23 discordant mutations, 12/13 (92%) in plasma only and 6/10 (60%) in non-plasma analytes only were mixtures (p=0.13).
Table 3 provides details on specific mutations discordance among analytes for the seven patients with sequences from four analytes. Of 12 mutations (7 NRTI; 5 NNRTI), 4 (33%) were discordant between plasma and non-plasma analytes, two appearing in DPS only and two in STP only.
Table 3.
Discordant resistance and non-resistance mutations among patients with sequences from plasma and three non-plasma analytes
| Discordant resistance mutations | Discordant non-resistance mutationsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| IDb | Subtype | Stratum | NRTI (totalc) | NNRTI (totalc) | Totalf | Plasma | DBS | DPS | STP |
| 25 | A | Treated | None (0) | None (1) | 17 | D123N | D123DGNS | D123DN, K154KT, N175DN | D123N, G141EG |
| 4 | A | Treated | None (5) | K103Sd (3) | 16 | P140Q | T165R, Q242HQ | ||
| 50 | DA | Naïve | None (0) | None (0) | 23 | D121H, K122E, D123S, S162HNR, S I244IV | D121H, K122E, D123S, G141EG, S162R | D121H, K122E, D123S, A158AP, S162HNRS, I244IV | T39IT, P52HP, D110N, D113E, A114S, V118D, L120FL, D121X, K122X, D123X, T131NT, N137IN, S162HN, I244V |
| 51 | A | Naïve | D67DNd K219KQe (2) | E138Ge (1) | 17 | K173AS, R211KR | E169DE, K173AS, R211KR | E169D, K173S, F214FL, L234IL, H235HP | I142N, R143K, M164MV, E169D, F171FL, K173S, I202IM, R211KR, G231D |
| 52 | A | Naïve | None (0) | None (0) | 15 | S68RS, E79DEV, N81KN, R83RS, D86AD | |||
| 53 | A | Naïve | None (0) | None (0) | 13 | ||||
| 54 | AD | Naïve | None (0) | None (0) | 9 | R211KR | R211KR | R211KR | |
Table is sorted by treatment status then by number of total discordant mutations;
amino acids are shown for each analyte at positions where mutations were found to be discordant in at least one analyte;
ID<50 match those in Table 2; values>49 were given to naïve patients as to not overlap with Table 2.
total number of mutations occurring in sequences from all analytes are shown in parenthesis;
DPS only;
STP only;
total number of non-resistance-associated mutations occurring in sequences from all analytes, compared to HIV-1 subtype B. DBS, dried blood spots; DPS, dried plasma spots; STP, ViveST plasma.
Analyte concordance for whole sequences
In 59 patients with sequences from ≥2 analytes, mean plasma-DBS NA discordance was 1.1% (n=33; range 0.4–2.3%; 1.2% for naïves and 1.1% for treated, Table 4). Similar values for plasma-DPS discordance were 1.2% (n=15; range 0.3–2.2%; 1.4% naïves and 0.9% treated); 2.0% for plasma-STP discordance (n=34; range 0.5–7.1%; 2.2% naïves and 1.2% treated); and 2.3% for plasma-STB discordance (n=5; range 0.8–5.3%; 2.3% naïves; no data for treated).
Table 4.
Nucleic acid discordance between plasma and non-plasma sequence pairs by treatment status and subtype
| DBS | DPS | STP | STB | ||
|---|---|---|---|---|---|
| Total | |||||
| N | 33 | 15 | 34 | 5 | |
| % Discordance; Range | 1.1; 0.4–2.3 | 1.2; 0.3–2.2 | 2.0; 0.5–7.1 | 2.3; 0.8–5.3 | |
| Patient stratum | |||||
| Treated | |||||
| N | 17 | 5 | 7a | 0 | |
| % Discordance; Range | 1.1; 0.4–2.2 | 0.9; 0.3–1.3 | 1.2; 0.7–2.0 | – | |
| Naïve | |||||
| N | 16b | 10b | 27 | 5 | |
| % Discordance; Range | 1.2; 0.5–2.3 | 1.4; 0.4–2.2 | 2.2; 0.5–7.1 | 2.3; 0.8–5.3 | |
| Treated vs. Naïve | RR (95% CI), p |
0.9 (0.7, 1.3) p=0.70 |
0.7 (0.5, 1.1) p=0.09 |
0.6 (0.4, 0.8) p=0.002 |
– |
| Subtype | |||||
| A | |||||
| N | 17 | 10 | 18b | 2 | |
| % Discordance; Range | 1.1; 0.4–2.3 | 1.3; 0.7–2.0 | 2.0; 0.5–4.7 | 2.0; 1.4–2.6 | |
| D | |||||
| N | 6 | 2 | 5 | 0 | |
| % Discordance; Range | 1.0; 0.4–2.2 | 1.5; 0.9–2.2 | 1.0; 0.7–1.6 | – | |
| D vs. A | RR (95% CI), p |
0.9 (0.5, 1.6) p=0.64 | 1.1 (0.8, 1.6) p=0.50 |
0.5 (0.4, 0.7) p<0.001 |
– |
| Viral load | |||||
| Per 1-log unit higher | RR (95% CI) p |
0.7 (0.6, 0.9) p=0.001 |
0.9 (0.7, 1.4) p=0.31 |
0.9 (0.6, 1.3) p=0.59 |
0.8 (0.5, 1.3) p=0.32 |
Numbers represent mean (range) percent discordance. RR, rate ratios (95% confidence interval) of discordant nucleic acids comparing patient treatment status and subtype by analyte type. “–” means that there were no STB/plasma sequence pairs;
two missing protease sequences;
one missing protease sequence. DBS, dried blood spots. DPS, dried plasma spots. STP, ViveST plasma. STB, ViveST blood.
In the seven patients with sequences from four analytes (Table 3), of 110 non-resistance mutations, only one (P140Q) was found in plasma only and 31 in a non-plasma analyte. Of the five discordant plasma-DBS mutations, three were in plasma and two in DBS; of 11 plasma-DPS discordances, two were in plasma and nine in DPS; of 23 plasma-STP discordances, one was in plasma and 22 in STP.
The comparison of plasma-to-non-plasma discordance rates demonstrated that naïve and treated patients had similar rates in DBS, and treated patients had lower rates in DPS (RR=0.7, CI=0.5–1.1, p=0.09, Table 4) and STP (RR=0.6, CI 0.4–0.8, p=0.002), compared to naïve patients. We found similar discordance rates among subtypes A and D in both DBS and DPS, but subtype D had lower discordance rates in STP compared to subtype A (RR=0.5, CI=0.4–0.7, p<0.001). For all analytes, discordance was inversely related to VL (i.e. lower VL had higher discordance), but significantly so only for DBS (RR=0.7 per 1-log higher VL, 95% CI=0.6–0.9, p=0.001).
Discussion
In AMPATH, HIV-1 infected patients demonstrated high subtype diversity, infrequent TDR and high acquired DR with unique mutation patterns upon first-line failure. Virologic failure was identified among ART-experienced patients using WHO guidelines in a setting where VL testing is limited [7]. The enrolment of both ART-naïve and experienced patients and the use of multiple analytes for resistance testing enabled capacity building to conduct resistance studies. In this setting higher VL and a shorter shipment-to-genotyping time were associated with genotyping success.
The HIV-1 subtype distribution at AMPATH in western Kenya is mostly consistent with prior reports from the wider region, where subtypes A, D and C predominate [25, 29, 30, 33, 62]. We identified unique pol recombinants in 12%, similar to one Uganda report [63], and higher than prior reports. Identification of inter- and intra-subtype recombination depends on the genomic region and subtyping methods [64–66].
TDR was seen in 1.8% of plasma sequences, a “low” WHO TDR threshold [67]. This is reasonable, given ART introduction to routine clinical care at AMPATH only in 2001 [10]. These low (<5%) TDR estimates contrast with more recent reports of higher-level (6% and 7.4%) TDR from Nairobi, the Kenyan coast and neighbouring Uganda [42, 68]. The TDR increase to 7% combining multiple analytes deserves further attention. Using multiple analytes may be advantageous, and analogous to accumulating sequences over time when estimating resistance, rather than a cross sectional assessment [69]. Whether non-plasma analytes are more sensitive in detecting TDR deserves further study. The WHO SDRM list was designed specifically to account for subtype diversity [57] and the high (14%) TDR based on the IAS–USA mutation list emphasizes the importance of using a reference adapted to global subtypes. Despite recent reports [70], we found no difference in TDR between higher and lower CD4 groups or between subtypes, although these conclusions are limited due to small sample-sizes.
Resistance mutations were found in 91% of patients failing a first-line regimen, at the high end of reports from other regional settings (60–80%) [4, 46, 71]. Similar high resistance rates were reported in Burnt-Forest, a rural AMPATH clinic [34]. These high rates may be related to adherence, lack of virologic monitoring and treatment failure misclassification by immunologic criteria, each of which contributes to resistance accumulation [7, 72].
Our results are relevant even with samples collected in 2006–2007 and WHO guidelines to phase out stavudine and phase in tenofovir [37]. Implementation of such guidelines is slow and resistance can be transmitted and relevant for tenofovir-based regimens. Similarly, there are significant implications of high resistance levels; 76% 2-class resistance, 47% ≥5 mutations and 60% intermediate–high predicted resistance to future ART. These findings provide a strong rationale for the use of a boosted PI in second-line ART with an active NRTI to limit transmission and continued accumulation of resistance. Continued monitoring of resistance as well as routine VL testing are important, to enable early failure identification and limit resistance accumulation [73]. No differences in resistance development were observed among subtypes, although larger studies suggest such differences [71, 74, 75], justifying further research. Unique resistance patterns in diverse subtypes identified here may support subtype-specific and host-specific resistance pathways, though other viral, host and environmental factors should be considered.
Non-plasma analytes have been explored for resistance testing in RLS, mostly for their cost and simplicity [14, 17, 76]. This is the first report of comparison of four different non-plasma analytes to plasma. Though these pilot results demonstrated lower genotyping yield in non-plasma analytes, we demonstrate their feasibility in these “real life” laboratory settings. The relatively low levels of successful genotyping may be ascribed to potential sample mishandling, freeze thawing, prolonged exposure to higher temperature and duration between collection and assay. Reduced rates of reverse transcription and amplification of HIV RNA were also documented in a study from Asian and African sites which compared storage temperatures and duration [77]. Lower-yield results with field-plasma on ST resemble a recent report [54], however the higher success rates of genotyping using frozen-plasma (highest among non-plasma analytes), under controlled drying conditions, more closely resemble previously published success rates for VL from ST [16]. These findings highlight the importance of complete sample drying with any ST devices. Furthermore, the ST colour indicator provides an additional cost-benefit in preventing unnecessary reagent loss. With higher ART and VL monitoring access and use of non-plasma analytes [78, 79], the need to increase sensitivity, yield and local laboratory capacity to use such analytes will rise [80].
High (93–99%) full sequence concordance was demonstrated between plasma and non-plasma analytes, confirming prior results with DBS [17, 76] and one study with ST [54], and providing support for non-plasma analytes for resistance testing. Our results confirm recent findings of an inverse relationship between VL and sequence discordance [81]. Most of the resistance mutations that were not identified in plasma were seen in DPS and STP rather than DBS. Such findings, as well as resistance mutations observed in plasma but not non-plasma analytes in seven patients, are relevant to global DBS resistance surveillance recommended by WHO, and additional research is needed to better understand this phenomenon and its implications. Resistance mutation concordance between plasma and non-plasma analytes was slightly lower (88–94%), with most discordances being mixtures. Unique data from seven patients with sequences from four analytes even demonstrated 33% discordant resistance mutations. Whether these findings are related to different sensitivities of filter analytes to hold stable DNA and/or RNA, the different sequencing methodology used for ST, or variables such as sampling, extraction or amplification methods or VL, remains to be determined.
Our findings from AMPATH in western Kenya are relevant to HIV care and resistance monitoring in other RLS, where the rising treatment roll-out can lead to increasing selection and transmission of resistance. The provision of prevention, treatment and clinical care may benefit from on-going examination of resistance and sequence diversity, to avoid evolution of extensive resistance, particularly in settings with limited ART regimens. As treatment programmes expand to meet WHO guidelines for increased VL monitoring [82], the need for resistance testing will increase as well, and better low-cost methods are needed. Challenges to the currently recommended plasma genotyping include cost and complexity of on-site phlebotomy, centrifugation and separation, and maintenance of a transport cold-chain. These challenges may be overcome by the use of analytes such as DBS, and ST. This evaluation of multiple analytes for genotyping in a RLS with subtype diversity and with barriers like minimal infrastructure, demanding transportation requirements, and high temperatures and humidity, demonstrates feasibility and further optimization needs. This study also underscores the potential advantage of using multiple analytes in resistance determination. Although the demonstrated sequence concordance among analytes is encouraging, their utility in research and clinical care will require larger scale evaluation of feasibility and effectiveness.
It is important to recognize that this was a pilot study, reflecting on its small sample size, limiting our ability to robustly examine resistance, subtype effects and plasma-non-plasma analyte concordance, or draw inference to other settings. Yet it is the first examination of HIV resistance and diversity in a large clinic in western Kenya. Other limitations include use of early DBS and ST versions, reducing genotyping yield and usage of different genotyping methods for ST versus other analytes.
Conclusions
High levels of HIV diversity and resistance in multiple subtypes were observed in western Kenya. Although new antiretroviral agents and classes are in development, resistance remains a major challenge to treatment, particularly in RLS with a high burden of infection, diverse HIV variants, increasing treatment access, low treatment monitoring capacity and limited medications. Lower cost, robust analytes and assays to monitor resistance are important and useful in maintaining the benefit of ART.
Acknowledgements
This research was funded by a developmental grant from the Lifespan/Tufts/Brown Center for AIDS Research, an NIH funded programme (P30AI42853); NIH grant R01AI066922, the Rhode Island Foundation and the Friendship Foundation.
Competing interests
The authors have no competing interests to declare.
Authors' contributions
RK conceived the study and was responsible for all its aspects. RK and LD designed the study protocol. MB, LS, RML, FM and SM conducted laboratory assays. LK, WI, DK, NB and LD supervised study procedures. RK, LS, MB, ADL and JH conducted sequence and statistical analysis. RK wrote the first draft of the manuscript and worked with co-authors to finalize the writing. RK, ADL, JH, RML, MB, LD and DK critically revised the manuscript. All authors read and approved the final manuscript.
References
- 1.Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs. 2012;72(9):e1–25. doi: 10.2165/11633630-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350(10):1023–35. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Antiretroviral therapy for HIV infection in adults and adolescents: Recommendation for a public health approach 2010 revision. Geneva, Switzerland [Internet]. [cited 2014 Aug 4]. Available from: http://whqlibdoc.who.int/publications/2010/9789241599764_eng.pdf. [PubMed]
- 4.Hosseinipour MC, Gupta RK, Van Zyl G, Eron JJ, Nachega JB. Emergence of HIV drug resistance during first- and second-line antiretroviral therapy in resource-limited settings. J Infect Dis. 2013;207(Suppl 2):S49–56. doi: 10.1093/infdis/jit107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UNAIDS. MSF. Speed Up Scale-Up. Strategies, tools and policies to get the best HIV treatment to more people, sooner [Internet] [cited 2014 Aug 4]. Available from: http://aids2012.msf.org/wp-content/uploads/2012/07/MSF_Speed_up_Scale-up_report-webres.pdf.
- 6.Schooley RT. Viral load testing in resource-limited settings. Clin Infect Dis. 2007;44(1):139–40. doi: 10.1086/510090. [DOI] [PubMed] [Google Scholar]
- 7.Kantor R, Diero L, Delong A, Kamle L, Muyonga S, Mambo F, et al. Misclassification of first-line antiretroviral treatment failure based on immunological monitoring of HIV infection in resource-limited settings. Clin Infect Dis. 2009;49(3):454–62. doi: 10.1086/600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigaloff KC, Hamers RL, Wallis CL, Kityo C, Siwale M, Ive P, et al. Unnecessary antiretroviral treatment switches and accumulation of HIV resistance mutations; two arguments for viral load monitoring in Africa. J Acquir Immune Defic Syndr. 2011;58(1):23–31. doi: 10.1097/QAI.0b013e318227fc34. [DOI] [PubMed] [Google Scholar]
- 9.Hosseinipour MC, van Oosterhout JJ, Weigel R, Phiri S, Kamwendo D, Parkin N, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009;23(9):1127–34. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012;380(9849):1250–8. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Cajas JL, Pai NP, Klein MB, Wainberg MA. Differences in resistance mutations among HIV-1 non-subtype B infections: a systematic review of evidence (1996–2008) J Int AIDS Soc. 2009;12:11. doi: 10.1186/1758-2652-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamers RL, Kityo C, Lange JM, de Wit TF, Mugyenyi P. Global threat from drug resistant HIV in sub-Saharan Africa. BMJ. 2012;344:e4159. doi: 10.1136/bmj.e4159. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd RM, Jr., Burns DA, Huong JT, Mathis RL, Winters MA, Tanner M, et al. Dried-plasma transport using a novel matrix and collection system for human immunodeficiency virus and hepatitis C virus virologic testing. J Clin Microbiol. 2009;47(5):1491–6. doi: 10.1128/JCM.02354-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckton AJ. New methods for the surveillance of HIV drug resistance in the resource poor world. Curr Opin Infect Dis. 2008;21(6):653–8. doi: 10.1097/QCO.0b013e3283186d1a. [DOI] [PubMed] [Google Scholar]
- 15.Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review. Antivir Ther. 2009;14(5):619–29. [PubMed] [Google Scholar]
- 16.Zanoni M, Cortes R, Diaz RS, Sucupira MC, Ferreira D, Inocencio LA, et al. Comparative effectiveness of dried plasma HIV-1 viral load testing in Brazil using ViveST for sample collection. J Clin Virol. 2010;49(4):245–8. doi: 10.1016/j.jcv.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Bertagnolio S, Soto-Ramirez L, Pilon R, Rodriguez R, Viveros M, Fuentes L, et al. HIV-1 drug resistance surveillance using dried whole blood spots. Antivir Ther. 2007;12(1):107–13. [PubMed] [Google Scholar]
- 18.Plantier JC, Dachraoui R, Lemee V, Gueudin M, Borsa-Lebas F, Caron F, et al. HIV-1 resistance genotyping on dried serum spots. AIDS. 2005;19(4):391–7. doi: 10.1097/01.aids.0000161768.98534.e7. [DOI] [PubMed] [Google Scholar]
- 19.NASCOP. Kenya AIDS Indicator Survey 2012 [Internet] [cited 2014 Aug 4]. Available from: http://nascop.or.ke/library/3d/Preliminary Report for Kenya AIDS indicator survey 2012.pdf.
- 20.Maina WK, Kim AA, Rutherford GW, Harper M, K'Oyugi BO, Sharif S, et al. Kenya AIDS Indicator Surveys 2007 and 2012: implications for public health policies for HIV prevention and treatment. J Acquir Immune Defic Syndr. 2014;66(Suppl 1):S130–7. doi: 10.1097/QAI.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CIA. The World Factbook [Internet] [cited 2014 Aug 4]. Available from: https://www.cia.gov/library/publications/the-world-factbook/rankorder/2155rank.html.
- 22.Lihana RW, Khamadi SA, Lubano K, Lwembe R, Kiptoo MK, Lagat N, et al. HIV type 1 subtype diversity and drug resistance among HIV type 1-infected Kenyan patients initiating antiretroviral therapy. AIDS Res Hum Retroviruses. 2009;25(12):1211–17. doi: 10.1089/aid.2009.0007. [DOI] [PubMed] [Google Scholar]
- 23.Nyagaka B, Kiptoo MK, Lihana RW, Khamadi SA, Makokha EP, Kinyua JG, et al. HIV type 1 gag genetic diversity among antenatal clinic attendees in North Rift Valley, Kenya. AIDS Res Hum Retroviruses. 2012;28(5):523–6. doi: 10.1089/AID.2011.0223. [DOI] [PubMed] [Google Scholar]
- 24.Oyaro M, Mbithi J, Oyugi F, Laten A, Anzala O, Engelbrecht S. Molecular characterization of HIV type 1 among HIV-infected respondents in a cohort being prepared for HIV Phase III vaccine clinical trials, Western Kenya. AIDS Res Hum Retroviruses. 2011;27(3):257–64. doi: 10.1089/aid.2010.0061. [DOI] [PubMed] [Google Scholar]
- 25.Yang C, Li M, Shi YP, Winter J, van Eijk AM, Ayisi J, et al. Genetic diversity and high proportion of intersubtype recombinants among HIV type 1-infected pregnant women in Kisumu, western Kenya. AIDS Res Hum Retroviruses. 2004;20(5):565–74. doi: 10.1089/088922204323087822. [DOI] [PubMed] [Google Scholar]
- 26.Robbins KE, Kostrikis LG, Brown TM, Anzala O, Shin S, Plummer FA, et al. Genetic analysis of human immunodeficiency virus type 1 strains in Kenya: a comparison using phylogenetic analysis and a combinatorial melting assay. AIDS Res Hum Retroviruses. 1999;15(4):329–35. doi: 10.1089/088922299311295. [DOI] [PubMed] [Google Scholar]
- 27.Neilson JR, John GC, Carr JK, Lewis P, Kreiss JK, Jackson S, et al. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 1999;73(5):4393–403. doi: 10.1128/jvi.73.5.4393-4403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lihana RW, Khamadi SA, Lwembe RM, Kinyua JG, Muriuki JK, Lagat NJ, et al. HIV-1 subtype and viral tropism determination for evaluating antiretroviral therapy options: an analysis of archived Kenyan blood samples. BMC Infect Dis. 2009;9:215. doi: 10.1186/1471-2334-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khamadi SA, Lihana RW, Osman S, Mwangi J, Muriuki J, Lagat N, et al. Genetic diversity of HIV type 1 along the coastal strip of Kenya. AIDS Res Hum Retroviruses. 2009;25(9):919–23. doi: 10.1089/aid.2009.0005. [DOI] [PubMed] [Google Scholar]
- 30.Kageha S, Lihana RW, Okoth V, Mwau M, Okoth FA, Songok EM, et al. HIV type 1 subtype surveillance in central Kenya. AIDS Res Hum Retroviruses. 2012;28(2):228–31. doi: 10.1089/aid.2011.0089. [DOI] [PubMed] [Google Scholar]
- 31.Arroyo MA, Sateren WB, Foglia G, Kibaya R, Langat L, Wasunna M, et al. Short communication: HIV type 1 genetic diversity among tea plantation workers in Kericho, Kenya. AIDS Res Hum Retroviruses. 2009;25(11):1061–4. doi: 10.1089/aid.2009.0092. [DOI] [PubMed] [Google Scholar]
- 32.Dowling WE, Kim B, Mason C, Wasunna KM, Alam U, Elson L, et al. Forty-one near full-length HIV-1 sequences from Kenya reveal an epidemic of subtype A and A-containing recombinants. AIDS. 2002;16(13):1809–20. doi: 10.1097/00002030-200209060-00015. [DOI] [PubMed] [Google Scholar]
- 33.Hue S, Hassan AS, Nabwera H, Sanders EJ, Pillay D, Berkley JA, et al. HIV type 1 in a rural coastal town in Kenya shows multiple introductions with many subtypes and much recombination. AIDS Res Hum Retroviruses. 2012;28(2):220–4. doi: 10.1089/aid.2011.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann M, Diero L, Kemboi E, Mambo F, Rono M, Injera W, et al. Antiretroviral treatment interruptions induced by the Kenyan postelection crisis are associated with virological failure. J Acquir Immune Defic Syndr. 2013;64(2):220–4. doi: 10.1097/QAI.0b013e31829ec485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wools-Kaloustian K, Kimaiyo S, Diero L, Siika A, Sidle J, Yiannoutsos CT, et al. Viability and effectiveness of large-scale HIV treatment initiatives in sub-Saharan Africa: experience from western Kenya. AIDS. 2006;20(1):41–8. doi: 10.1097/01.aids.0000196177.65551.ea. [DOI] [PubMed] [Google Scholar]
- 36.UNAIDS. HIV and AIDS Estimates. [cited 2014 Aug 4]. Available from: http://www.unaids.org.
- 37.WHO. Rapid advice. Antiretroviral therapy for HIV infection in adults and adolescents [Internet] 2009 Nov [cited 2014 Aug 4]. Available from: http://www.who.int/hiv/pub/arv/rapid_advice_art.pdf?ua=1.
- 38.Kiptoo M, Ichimura H, Wembe RL, Ng'Ang'a Z, Mueke J, Kinyua J, et al. Prevalence of nevirapine-associated resistance mutations after single dose prophylactic treatment among antenatal clinic attendees in north rift Kenya. AIDS Res Hum Retroviruses. 2008;24(12):1555–9. doi: 10.1089/aid.2008.0018. [DOI] [PubMed] [Google Scholar]
- 39.Zeh C, Weidle PJ, Nafisa L, Lwamba HM, Okonji J, Anyango E, et al. HIV-1 drug resistance emergence among breastfeeding infants born to HIV-infected mothers during a single-arm trial of triple-antiretroviral prophylaxis for prevention of mother-to-child transmission: a secondary analysis. PLoS Med. 2011;8(3):e1000430. doi: 10.1371/journal.pmed.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamers RL, Wallis CL, Kityo C, Siwale M, Mandaliya K, Conradie F, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis. 2011;11(10):750–9. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 41.Hassan AS, Mwaringa SM, Obonyo CA, Nabwera HM, Sanders EJ, Rinke de Wit TF, et al. Low prevalence of transmitted HIV type 1 drug resistance among antiretroviral-naive adults in a rural HIV clinic in Kenya. AIDS Res Hum Retroviruses. 2013;29(1):129–35. doi: 10.1089/aid.2012.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigaloff KC, Mandaliya K, Hamers RL, Otieno F, Jao IM, Lyagoba F, et al. Short communication: high prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2012;28(9):1033–7. doi: 10.1089/AID.2011.0348. [DOI] [PubMed] [Google Scholar]
- 43.Hassan AS, Nabwera HM, Mwaringa SM, Obonyo CA, Sanders EJ, Rinke de Wit TF, et al. HIV-1 virologic failure and acquired drug resistance among first-line antiretroviral experienced adults at a rural HIV clinic in coastal Kenya: a cross-sectional study. AIDS Res Ther. 2014;11(1):9. doi: 10.1186/1742-6405-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wamalwa DC, Lehman DA, Benki-Nugent S, Gasper MA, Gichohi R, Maleche-Obimbo E, et al. Long-term virologic response and genotypic resistance mutations in HIV-1 infected Kenyan children on combination antiretroviral therapy. J Acquir Immune Defic Syndr. 2013;62(3):267–74. doi: 10.1097/QAI.0b013e31827b4ac8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman S, Lihana RW, Kibaya RM, Ishizaki A, Bi X, Okoth FA, et al. Diversity of HIV type 1 and drug resistance mutations among injecting drug users in Kenya. AIDS Res Hum Retroviruses. 2013;29(1):187–90. doi: 10.1089/AID.2012.0182. [DOI] [PubMed] [Google Scholar]
- 46.Hamers RL, Sigaloff KC, Wensing AM, Wallis CL, Kityo C, Siwale M, et al. Patterns of HIV-1 drug resistance after first-line antiretroviral therapy (ART) failure in 6 sub-Saharan African countries: implications for second-line ART strategies. Clin Infect Dis. 2012;54(11):1660–9. doi: 10.1093/cid/cis254. [DOI] [PubMed] [Google Scholar]
- 47.AMPATH. Academic Model Providing Access to Healthcare [Internet] [cited 2014 May 5]. Available from: http://www.who.int/hiv/pub/prev_care/ampath/en/
- 48.Einterz RM, Kelley CR, Mamlin JJ, Van Reken DE. Partnerships in international health. The Indiana University-Moi University experience. Infect Dis Clin North Am. 1995;9(2):453–5. [PubMed] [Google Scholar]
- 49.Einterz RM, Kimaiyo S, Mengech HN, Khwa-Otsyula BO, Esamai F, Quigley F, et al. Responding to the HIV pandemic: the power of an academic medical partnership. Acad Med. 2007;82(8):812–18. doi: 10.1097/ACM.0b013e3180cc29f1. [DOI] [PubMed] [Google Scholar]
- 50.Inui TS, Nyandiko WM, Kimaiyo SN, Frankel RM, Muriuki T, Mamlin JJ, et al. AMPATH: living proof that no one has to die from HIV. J Gen Intern Med. 2007;22(12):1745–50. doi: 10.1007/s11606-007-0437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tierney WM, Rotich JK, Hannan TJ, Siika AM, Biondich PG, Mamlin BW, et al. The AMPATH medical record system: creating, implementing, and sustaining an electronic medical record system to support HIV/AIDS care in western Kenya. Stud Health Technol Inform. 2007;129(Pt 1):372–6. [PubMed] [Google Scholar]
- 52.Research Think Tank [Internet] [cited 2014 May 5]. Available from: http://www.researchthinktank.com/
- 53.Winters MA, Coolley KL, Girard YA, Levee DJ, Hamdan H, Shafer RW, et al. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–75. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diallo K, Lehotzky E, Zhang J, Zhou Z, de Rivera IL, Murillo WE, et al. Evaluation of a dried blood and plasma collection device, sampletanker, for HIV type 1 drug resistance genotyping in patients receiving antiretroviral therapy. AIDS Res Hum Retroviruses. 2014;30(1):67–73. doi: 10.1089/aid.2013.0127. [DOI] [PubMed] [Google Scholar]
- 55.Delong AK, Wu M, Bennett D, Parkin N, Wu Z, Hogan JW, et al. Sequence quality analysis tool for HIV type 1 protease and reverse transcriptase. AIDS Res Hum Retroviruses. 2012;28(8):894–901. doi: 10.1089/aid.2011.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shafer R. Stanford HIV Sequence Database. [cited 2012 Aug 1]. Available from: http://hivdb.stanford.edu.
- 57.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer R, et al. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2011;19(4):156–64. [PMC free article] [PubMed] [Google Scholar]
- 59.de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21(19):3797–800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 60.Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2003;17(1):F1–5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmann CJ, Charalambous S, Sim J, Ledwaba J, Schwikkard G, Chaisson RE, et al. Viremia, resuppression, and time to resistance in human immunodeficiency virus (HIV) subtype C during first-line antiretroviral therapy in South Africa. Clin Infect Dis. 2009;49(12):1928–35. doi: 10.1086/648444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nyamache AK, Muigai AW, Khamadi SA. Circulating trends of non-B HIV type 1 subtypes among Kenyan individuals. AIDS Res Hum Retroviruses. 2013;29(2):400–3. doi: 10.1089/AID.2012.0213. [DOI] [PubMed] [Google Scholar]
- 63.Eshleman SH, Gonzales MJ, Becker-Pergola G, Cunningham SC, Guay LA, Jackson JB, et al. Identification of Ugandan HIV type 1 variants with unique patterns of recombination in pol involving subtypes A and D. AIDS Res Hum Retroviruses. 2002;18(7):507–11. doi: 10.1089/088922202317406655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ntemgwa M, Gill MJ, Brenner BG, Moisi D, Wainberg MA. Discrepancies in assignment of subtype/recombinant forms by genotyping programs for HIV type 1 drug resistance testing may falsely predict superinfection. AIDS Res Hum Retroviruses. 2008;24(7):995–1002. doi: 10.1089/aid.2008.0064. [DOI] [PubMed] [Google Scholar]
- 65.Gifford R, de Oliveira T, Rambaut A, Myers RE, Gale CV, Dunn D, et al. Assessment of automated genotyping protocols as tools for surveillance of HIV-1 genetic diversity. AIDS. 2006;20(11):1521–9. doi: 10.1097/01.aids.0000237368.64488.ae. [DOI] [PubMed] [Google Scholar]
- 66.Yebra G, de Mulder M, Martin L, Perez-Cachafeiro S, Rodriguez C, Labarga P, et al. Sensitivity of seven HIV subtyping tools differs among subtypes/recombinants in the Spanish cohort of naive HIV-infected patients (CoRIS) Antiviral Res. 2011;89(1):19–25. doi: 10.1016/j.antiviral.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Jordan MR, Bennett DE, Wainberg MA, Havlir D, Hammer S, Yang C, et al. Update on World Health Organization HIV drug resistance prevention and assessment strategy: 2004–2011. Clin Infect Dis. 2012;54(Suppl 4):S245–9. doi: 10.1093/cid/cis206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nazziwa J, Njai HF, Ndembi N, Birungi J, Lyagoba F, Gershim A, et al. Short communication: HIV type 1 transmitted drug resistance and evidence of transmission clusters among recently infected antiretroviral-naive individuals from Ugandan fishing communities of Lake Victoria. AIDS Res Hum Retroviruses. 2013;29(5):788–95. doi: 10.1089/aid.2012.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Punyacam P, Iemwimangsa N, Chantratita W, Sukasem C, Sungkanuparph S. HIV drug resistance interpreted by cumulative versus last genotypes in HIV-infected patients with multiple treatment failures. Curr HIV Res. 2012;10(3):271–4. doi: 10.2174/157016212800618129. [DOI] [PubMed] [Google Scholar]
- 70.Yanik EL, Napravnik S, Hurt CB, Dennis A, Quinlivan EB, Sebastian J, et al. Prevalence of transmitted antiretroviral drug resistance differs between acutely and chronically HIV-infected patients. J Acquir Immune Defic Syndr. 2012;61(2):258–62. doi: 10.1097/QAI.0b013e3182618f05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kyeyune F, Nankya I, Metha S, Akao J, Ndashimye E, Tebit DM, et al. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. AIDS. 2013;27(12):1899–909. doi: 10.1097/QAD.0b013e3283610ec7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sigaloff KC, Ramatsebe T, Viana R, Wit TF, Wallis CL, Stevens WS. Accumulation of HIV drug resistance mutations in patients failing first-line antiretroviral treatment in South Africa. AIDS Res Hum Retroviruses. 2012;28(2):171–5. doi: 10.1089/aid.2011.0136. [DOI] [PubMed] [Google Scholar]
- 73.Smith DM, Schooley RT. Running with scissors: using antiretroviral therapy without monitoring viral load. Clin Infect Dis. 2008;46(10):1598–600. doi: 10.1086/587110. [DOI] [PubMed] [Google Scholar]
- 74.Kantor R, Katzenstein D, Efron B, Carvalho AP, Wynhoven B, Soares M, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotypic evolution: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wainberg MA, Brenner BG. Role of HIV subtype diversity in the development of resistance to antiviral drugs. Viruses. 2010;2(11):2493–508. doi: 10.3390/v2112493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bertagnolio S, Parkin NT, Jordan M, Brooks J, Garcia-Lerma JG. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev. 2010;12(4):195–208. [PubMed] [Google Scholar]
- 77.Wallis C, Aga E, Ribaudo H, Chevallier C, Klingman K, Stevens W, et al. Refining approaches to viral load monitoring using dried blood spots; 18th Conference on Retroviruses and Opportunistic Infections; GA: Atlanta; 2013. Mar 3–5, [Google Scholar]
- 78.Aitken SC, Kliphuis A, Bronze M, Wallis CL, Kityo C, Balinda S, et al. Development and evaluation of an affordable real-time qualitative assay for determining HIV-1 virological failure in plasma and dried blood spots. J Clin Microbiol. 2013;51(6):1899–905. doi: 10.1128/JCM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inzaule S, Yang C, Kasembeli A, Nafisa L, Okonji J, Oyaro B, et al. Field evaluation of a broadly sensitive HIV-1 in-house genotyping assay for use with both plasma and dried blood spot specimens in a resource-limited country. J Clin Microbiol. 2013;51(2):529–39. doi: 10.1128/JCM.02347-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Land S, Zhou J, Cunningham P, Sohn AH, Singtoroj T, Katzenstein D, et al. Capacity building and predictors of success for HIV-1 drug resistance testing in the Asia-Pacific region and Africa. J Int AIDS Soc. 2013;16(1):18580. doi: 10.7448/IAS.16.1.18580. doi: http://dx.doi.org/10.7448/IAS.16.1.18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji H, Li Y, Liang B, Pilon R, MacPherson P, Bergeron M, et al. Pyrosequencing dried blood spots reveals differences in HIV drug resistance between treatment naive and experienced patients. PLoS One. 2013;8(2):e56170. doi: 10.1371/journal.pone.0056170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.WHO. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach 2013 [Internet] [cited 2014 Aug 4]. Available from: http://www.who.int/hiv/pub/guidelines/arv2013/download/en/ [PubMed]