Abstract
We have utilized 3D printing technology to create an inexpensive spectroelectrochemical cell insert that fits inside a standard cuvette and can be used with any transmission spectrometer. The cell positions the working, counter, and reference electrodes and has an interior volume of approximately 200 microliters while simultaneously providing a full 1 cm path length for spectroscopic measurements. This method reduces the time required to perform a potentiometric titration on a molecule compared with standard chemical titration methods and achieves complete electrolysis of protein samples within minutes. The device thus combines the best aspects of thin-layer cells and standard potentiometry.
Keywords: safranine, spectroelectrochemistry, reduction potential, 3D printing
Knowledge of the reduction potentials of the cofactors involved in electron transfer in respiration and metabolism is a central concern of the fields of bioenergetics and enzymology [1]. The most common method used to determine these is potentiometry, in which anaerobic solutions of the protein of interest are oxidized and reduced with chemical reagents in a custom-made piece of glassware known as the Dutton cuvette [2]. However, these experiments require a large volume of relatively concentrated protein and take hours to complete.
Spectroelectrochemistry is another powerful hybrid characterization technique [3]. In this method, a custom-built cell is created in which the solution potential is fixed electrodically. The ratio of the total solution volume to the electrodes surface area is kept as low as possible in order to speed the approach to solution equilibrium. Thus this type of cell is typically called an Optically Transparent Thin Layer Electrochemical, or OTTLE, cell. Spectral measurements are taken through the working electrode which can be a wire mesh or a transparent glass coating such as indium-tin oxide. Small molecule mediator compounds are used to relay electrons between the protein and the electrode. This technique has many obvious benefits, but the necessity of constructing a custom experimental setup in order to perform reliable measurements has limited its use. Furthermore, electrochemistry and spectroscopy place competing constraints on cell design. Thin-layer conditions at the electrode surface results in excessively short optical path lengths, requiring unreasonably high protein concentrations.
In this work we demonstrate that 3D printing technology can be utilized to make a microchanneled plastic insert which fits inside the standard 1 cm square cuvettes used in conventional transmission spectrometers. A printable 3D design allows for reduction potential determination of protein or small molecule analytes without the need for an optically transparent electrode or an extremely thin optical path length. While not as fast as a custom thin-layer setup, the cell reaches equilibrium in tens of seconds with a thirty-fold reduction in solution volume. Titrations are completed in roughly half the time previously required. This device combines the best aspects of thin-layer cells and standard reduction potential measurements, is inexpensive and rapidly made, and uses standard cuvettes and spectrophotometers.
Materials
Fullcure 720 was purchased from Stratasys. Safranine O was purchased from Sigma-Aldrich and purified as described previously [4]. Equine heart Cytochrome C (Sigma-Aldrich) was used without further purification. The artificial hemoprotein HH was expressed and purified as described previously [5]. All other solvents and reagents were purchased from VWR. Gold foil working electrodes and platinum wire counter electrodes were purchased from Sigma-Aldrich. The Ag/AgCl reference electrode is a Microelectrodes Inc. MI-402 flexible microelectrode with a 2 mm diameter. Before each use, gold foil electrodes are sonicated in ethanol for 20 minutes at room temperature, polished using the Aldrich electrode polishing kit. Potentials were set using a BAS, Inc. PWR-3 power module.
Cell design and assembly
The cell body (Figure 1) was printed on an Objet Eden 333 3D printer (Stratasys) using Fullcure 720 acrylic polymer. It is a rectangular box 9.8 × 9.8 × 30 mm. An STL file containing the coordinates for printing this cell from the majority of commercial or homemade 3D printers is in the supplementary material and may also be downloaded at: www.thingiverse.com/thing:40429. Alternatively, the piece may be ordered from any of several online 3D print services for less than $20.
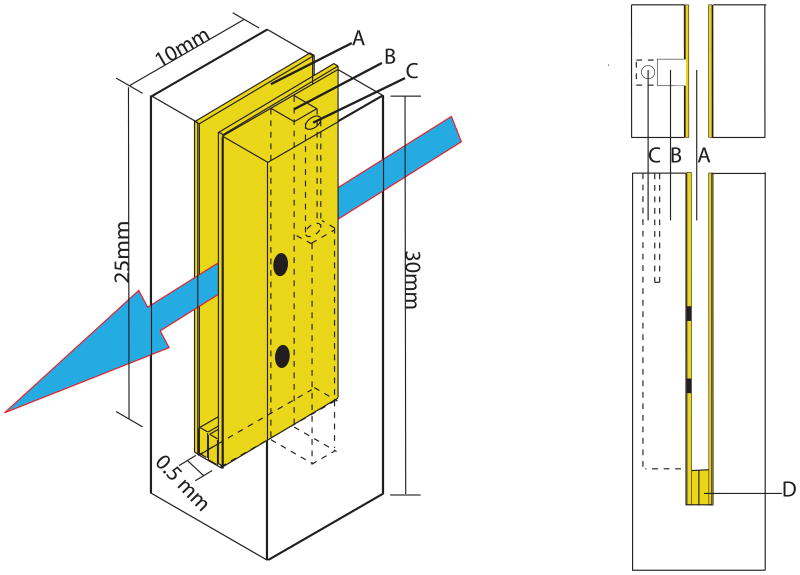
Figure 1.
Schematic diagram of the 3D printed electrode holder/spacer. A) gold working electrode, B) 2mm × 2mm reference electrode channel, C) 1mm diameter channel for platinum counter electrode, D) gold foil spacer. Side view shows continuous fluid connection of working, reference and counter electrode compartments out lined in (- -). Black spots are holes punched in the working electrode to enable fluidic connections between compartments.
The working electrode is composed of two gold foils 25 mm tall, 10 mm wide and 0.25 mm thick. A small piece of additional gold foil is folded in half and placed between the bottoms of the two foils so as to separate them by 0.5 mm while maintaining them in electrical contact. The first step in cell assembly is to slide the two gold foils and spacer that comprise the working electrode into the 1.2 mm thick, central slit of the cell body. The cell with gold foils is then placed into a standard cuvette with a 10 mm2 base. Once the 0.5 mm diameter gold wire which connects the working electrode to the power module is placed between the top of the two foils, all of the working electrode components are in electrical contact. This arrangement leaves a light path which is 15 mm tall by 0.5 mm wide by 10 mm deep and thus requires 75 microliters of sample to fill while still providing a 1 cm path length for the light, allowing clear spectra of dilute samples.
The gold foils have 1 mm diameter holes drilled into them at the indicated positions to allow flow from the working electrode to the reference and counter electrode chambers via the internal microchannels in the cell body. The reference electrode chamber is a 2 mm square cavity that runs 15 mm deep into the cell body and is placed directly between the working electrode slit and the counter electrode chamber, at the position of highest current density. The counter electrode is a 1 mm diameter platinum wire which slides into the 1mm diameter, 15 mm deep chamber immediately adjacent to the reference electrode chamber. The reference and counter electrode chambers are separate for the top 5 mm of their length to prevent them from coming into contact, but open to one another for the bottom 10 mm of their length to allow for the entire sample to be in a continuous, single conducting phase. This arrangement, with internal microchannels, is conveniently fabricated with the use of a 3D printer. The reference, counter, and gold wire leading to the gold foil working electrodes are all permanently affixed into the Teflon cuvette cap, so that all three slide into their respective chambers as the lid is placed onto the cuvette and sealed to make gas tight. The last step is to place the cuvette into the spectrometer stage and insert the N2 gas inlet and outlet needles.
Cell kinetics
Electron transfer kinetics of the cell were determined by monitoring the reduction and oxidation of a 1 mM potassium ferricyanide, 1M sodium nitrate solution purged under continuous de-oxygenated N2 gas flow. Samples were equilibrated at fully oxidized or reduced conditions for 30 seconds, after which the polarization of applied potential is reversed and the absorbance at 425 nm was monitored first for oxidizing and then for reducing potential settings (-600 mV and +600 mV vs Ag AgCl) respectively.
Spectroelectrochemistry
Analytes were dissolved in 0.1 M KCl, 0.125 M KOH, 0.25 M Borate pH 9.0. Samples are degassed under vacuum for 30 minutes prior to insertion into the cell while the cell is also purged with nitrogen gas for 30 minutes prior to sample injection. For protein titrations, the gold working electrode was coated with a self-assembled monolayer by soaking the slides in a 1-proponal solution containing 1% 1-mercaptohexanol (v/v) for 20 minutes [6]. Protein titrations were performed with a small molecule mediator mixture [7] which mediates electron transfer in the -500 to +300 millivolt range. The Ag/AgCl reference electrode was calibrated using a quinhydrone standard (saturated quinhydrone, phosphate buffer pH 7.0 = +255 mV vs. the Normal Hydrogen Electrode (NHE)).
A visible spectrum was taken of each sample before applying external voltage to the cell. The initial potential was then set to a fully reducing value while taking time-course spectra to monitor the reduction. Once the absorbance reached equilibrium (typically 1-2 minutes after setting the potential), another full -spectra of the sample is taken. The potential was stepped back towards an oxidizing value at 20 millivolt increments, with time course spectra (typically complete in less than 1 minute for small potential steps) accompanying the approach to equilibrium at the given potential. Full visible spectra were then taken between each potential step, so as to build up both a kinetic profile and Nernst-curve data in a single titration.
Cell design and assembly
This cell grew out of our unsatisfactory attempts to create an inexpensive OTTLE cell capable of supporting the analysis of sol-gel thin films. A 3D printed OTTLE cell holder has recently been reported [8], but the small path length of the resultant cell precludes the analysis of samples with limited solubility such as the majority of proteins. Instead, a cell insert maintaining a 1 cm light path while minimizing the solution volume of a standard cuvette was created. 3D printing enables the creation of an insert which fixes the position of each electrode at the optimal positions and orientations to enable rapid equilibration of the bulk solution with the electrode, which is one of the primary advantages of the OTTLE configuration.
The cell design depicted in Figure 1 requires a 3D printer capable of a 250 μm or smaller feature size. This allows conductive fluidic connections between the various electrode-containing compartments while minimizing the total fluid volume of the system. The placement depicted minimizes heating while taking advantage of the fact that the reference electrode is encased in an insulating sleeve which precludes contact between the reference and auxiliary electrodes.
Cell Kinetics
Ferricyanide reduction is a common metric for the reduction kinetics of different OTTLE cell designs [9]. The kinetics of oxidation/reduction transformations in the cell are largely set by the ratio of the working electrode surface area to the solution volume. The cell design maximizes this ratio while avoiding construction of a labor-intensive custom apparatus. The cell slit width of 0.5 millimeters is too thick to satisfy “thin-layer” electrochemical. Nevertheless, the cell's surface area to volume ratio allows for complete electrolysis of the solution contents to be achieved with a half-life of 30 s (not shown).
Small molecule reduction potential determination
Figure 2A depicts the potentiometric titration of safranine O, chosen as a test for small molecule reduction potential determination because of our interest in safranine analogues [4]. Safranin reduction and oxidation were tracked at its single visible absorption peak located at 522 nm (inset, Figure 2A) and the data fit with the Nernst equation with n=2.0 electrons. Reduction kinetics (not shown) were identical to that observed for ferricyanide, and the observed two-electron reduction midpoint potential, -350 mV vs NHE, is identical to that determined earlier potentiometrically [4].
Figure 2.
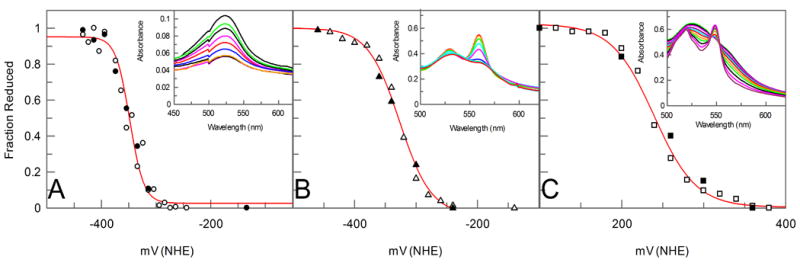
Potentiometric Nernst plots (• reduction points, ◦ oxidation points) fit to indicated parameters (A) Safranine O (n=2, Em=-350 mV), (B) de novo protein HH (n=1, Em=-327 mV), and (C) horse heart Cytochrome-C (n=1, Em =+240 mV). All midpoints are vs. NHE. Inserts show spectral changes monitored.
Protein reduction potential determination
Equine heart cytochrome c and the artificial diheme helical bundle HH were utilized to evaluate cell performance on cofactor-containing proteins. Figure 2B depicts the titration of a solution of 60 μM HH and 10 μM hemin (to ensure complete binding in the reduced state [5]) and Figure 2C depicts the titration of a 10 μM solution of cytochrome C. Reduction was reversible for both of the molecules analyzed with kinetics 4-fold slower than observed for the small molecule samples, presumably due to the increased minimum possible distance between the protein-bound cofactor and the SAM-covered electrode surface. Data fit with the Nernst equation with n=1.0 electron. Both reduction midpoint potentials are in agreement with reported literature values [10].
In conclusion, we have described the design of a new 3D printed cell insert which fits into a standard cuvette and allows rapid spectroelectrochemical titrations to be performed while retaining the full path length of the cuvette. The performance of the cell has been demonstrated using both small molecule and protein samples. This inexpensive insert combines the best features of standard Dutton cells and OTTLE devices in a form factor which allows the use of standard optical glass cuvettes and laboratory spectrophotometers.
Supplementary Material
References
- 1.Moser CC, Page CC, Dutton PL. Darwin at the molecular scale: selection and variance in electron tunnelling proteins including cytochrome c oxidase. Philosophical Transactions Of The Royal Society B-Biological Sciences. 2006;361:1295–1305. doi: 10.1098/rstb.2006.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods in Enzymology. 1978;54:411–35. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 3.Heineman WR, Anderson CW, Halsall HB, Hurst MM, Johnson JM, Kreishman GP, Norris BJ, Simone MJ, Su CH. Studies of biological redox systems by thin-layer electrochemical techniques. Advances in Chemistry Series. 1982:1–21. [Google Scholar]
- 4.Raju G, Capo J, Lichtenstein BR, Cerda JF, Koder RL. Manipulating reduction potentials in an artificial safranin cofactor. Tetrahedron Letters. 2012;53:1201–1203. doi: 10.1016/j.tetlet.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Anderson JLR, Ahmed I, Norman JA, Negron C, Mutter AC, Dutton PL, Koder RL. Manipulating Cofactor Binding Thermodynamics in an Artificial Oxygen Transport Protein. Biochemistry. 2011;50:10254–10261. doi: 10.1021/bi201242a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtenstein BR, Cerda JF, Koder RL, Dutton PL. Reversible proton coupled electron transfer in a peptide-incorporated naphthoquinone amino acid. Chemical Communications. 2009:168–170. doi: 10.1039/b815915g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shifman JM, Moser CC, Kalsbeck WA, Bocian DF, Dutton PL. Functionalized de novo designed proteins: Mechanism of proton coupling to oxidation/reduction in heme protein maquettes. Biochemistry. 1998;37:16815–16827. doi: 10.1021/bi9816857. [DOI] [PubMed] [Google Scholar]
- 8.Wilson RA, Pinyayev TS, Membreno N, Heineman WR. Rapid Prototyped Optically Transparent Thin-Layer Electrode Holder for Spectroelectrochemistry in Bench-Top Spectrophotometers. Electroanalysis. 2010;22:2162–2166. [Google Scholar]
- 9.Heineman WR, Hawkridge FM, Blount HN. Spectroelectrochemistry at optically transparent electrodes .2. Electrodes under thin-layer and semi-infinite diffusion conditions and indirect coulometric titrations. Electroanalytical Chemistry. 1984;13:1–113. [Google Scholar]
- 10.Myer YP, Saturno AF, Verma BC, Pande A. Horse heart Cytochrome-C - Oxidation-reduction potential and protein structures. Journal of Biological Chemistry. 1979;254:1202–1207. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.