Abstract
Wheat (Triticum aestivum L.) is one of the most important crops in the world. Squamosa-promoter binding protein (SBP)-box genes play a critical role in regulating flower and fruit development. In this study, 10 novel SBP-box genes (TaSPL genes) were isolated from wheat ((Triticum aestivum L.) cultivar Yanzhan 4110). Phylogenetic analysis classified the TaSPL genes into five groups (G1–G5). The motif combinations and expression patterns of the TaSPL genes varied among the five groups with each having own distinctive characteristics: TaSPL20/21 in G1 and TaSPL17 in G2 mainly expressed in the shoot apical meristem and the young ear, and their expression levels responded to development of the ear; TaSPL6/15 belonging to G3 were upregulated and TaSPL1/23 in G4 were downregulated during grain development; the gene in G5 (TaSPL3) expressed constitutively. Thus, the consistency of the phylogenetic analysis, motif compositions, and expression patterns of the TaSPL genes revealed specific gene structures and functions. On the other hand, the diverse gene structures and different expression patterns suggested that wheat SBP-box genes have a wide range of functions. The results also suggest a potential role for wheat SBP-box genes in ear development. This study provides a significant beginning of functional analysis of SBP-box genes in wheat.
Keywords: Expression profile, grain yield, squamosa-promoter binding protein-box genes, Triticum aestivum
INTRODUCTION
Flowering plants undergo a series of developmental stages from seed germination, vegetative growth, reproductive phase, and senescence. With development of new organs, each phase displays unique morphological and physiological characteristics (Chuck et al. 2011). The correct timing of the transition from vegetative growth to flowering is of utmost importance to ensure reproductive success (Mathieu et al. 2009; Huijser and Schmid 2011; Wang et al. 2011). Floral transition also has a large influence on agronomic traits; for example, a small delay in flowering time leads to increases in biomass yield, and premature flowering results in reduced biomass and seed set (Demura and Ye 2010; Huijser and Schmid 2011). Wheat (Triticum aestivum L.) is one of the most important crops in the world. Increased grain yield is always a major objective for breeders. From the jointing to booting stages, wheat goes through a transition from vegetative to reproductive growth. The shoot apical meristem changes from production of new leaves to production of a young spike. All developmental periods of the spike, from spikelet formation and floret development until flowering, will influence the final grain yield.
Squamosa-promoter binding protein (SBP)-box genes encode a plant-specific family of transcription factors, which play an important role in regulation of phase transitions (Cardon et al. 1999; Chen et al. 2010). Squamosa-promoter binding protein-box genes were first identified in Antirrhinum majus. They were named SBP1 and SBP2, and function as transcriptional activators of floral meristem identity gene SQUAMOSA (Klein et al. 1996). Based on sequence analysis, 17 non-redundant SBP-box genes in Arabidopsis and 19 in rice were predicted (Cardon et al. 1999; Xie et al. 2006; Yang et al. 2008). The SBP domain contains a highly conserved 76 amino acid residue region consisting of two, non-interleaved Zn-finger-like structures and a bipartite nuclear localization signal (NLS). They recognize and bind specifically to the promoters of target genes by the core motif (TNCGTACAA, where N represents any base), which is conserved from single cell algae to higher plants (Yamasaki et al. 2004; Birkenbihl et al. 2005).
Recent studies in various species suggested that SBP-box genes affect a broad range of developmental processes, especially promotion of the transition from juvenile to the adult and flowering stages (Wu and Poethig 2006; Schwarz et al. 2008). The gene functions of this family members are well characterized in Arabidopsis. Squamosa-promoter binding protein-box genes, known as SBP-like (SPL) genes in Arabidopsis, show extensive functional redundancy. AtSPL3, AtSPL4, and AtSPL5 (AtSPL3/4/5) have similar functions in promoting vegetative phase changes and flowering (Cardon et al. 1997; Wu and Poethig 2006; Gandikota et al. 2007). Single and double mutant phenotype analysis of AtSPL9 and AtSPL15 indicated their role in positive regulation of juvenile-to-adult growth phase transition and negative regulation of leaf initiation rate (plastochron), resulting in altered inflorescence architecture and enhanced branching (Schwarz et al. 2008; Wang et al. 2008). The closely related members AtSPL2, AtSPL10, and AtSPL11 control proper development of lateral organs in association with shoot maturation in the reproductive phase (Shikata et al. 2009). They also have functions in early embryonic patterning by regulating the morphogenesis-to-maturation transition (Nodine and Bartel 2010). AtSPL8 promotes sporogenous cell and parietal cell formation in early anthers, whereas the semisterile phenotype of AtSPL8 loss-of-function mutants revealed partial functional redundancy along with several other AtSPL genes (Zhang et al. 2007; Xing et al. 2010). AtSPL14 participates in the programmed cell death (PCD)-inducing fungal toxin fumonisin B1 (FB1) and regulates the development of normal plant architecture (Stone et al. 2005). Squamosa-promoter binding protein-box genes also affect anthocyanin biosynthesis and copper homeostasis in Arabidopsis (Yamasaki et al. 2009; Gou et al. 2011).
As with the function of several AtSPL genes in controlling flowering time and floral organs, SBP-box genes already have been implicated in regulation of flower and fruit development in crops; in other words, they affect the yield. In tomato, an SBP-box gene (LeSPL-CNR) is critical for normal ripening as demonstrated by positional cloning and virus-induced gene silencing (Manning et al. 2006). The most critical step in maize domestication was kernel liberation from the hardened, protective casing that surrounded the kernels in the maize progenitor (teosinte), and converted teosinte into a useful grain crop. This evolutionary step was controlled by teosinte glume architecture (tga1), an SBP-box gene (Wang et al. 2005). Another maize SBP-box gene (tasselsheath4, tsh4) plays an important role in inflorescence (the tassel and the ear) branch meristem initiation and maintenance (Chuck et al. 2010). In rice, OsSPL14 regulates the ideal plant architecture, including low tiller numbers with few unproductive tillers, more grains per panicle, and stronger culms. This pleiotropic gene significantly promotes panicle branching and higher grain productivity (Jiao et al. 2010; Miura et al. 2010). OsSPL16, at the GW8 locus, is a positive regulator of cell proliferation, and controls grain size, shape, and quality (Wang et al. 2012). We hypothesized that one or several SBP-box genes also play a role in wheat ear development. However, there has been no report on SBP-box genes in common wheat until now. In order to test the hypothesis, we isolated 10 novel SBP-box genes from wheat genome. Moreover: (i) a neighbor-joining (NJ) phylogenetic tree was constructed based on the conserved SBP domain of Arabidopsis, rice, and wheat; (ii) the diversity of motif composition was described; (iii) the expression pattern in different organs and the response to wheat ear development was examined; and (iv) nuclear localization of TaSPL17 was observed. The results suggest a potential role for wheat SBP-box genes in ear development.
RESULTS
Identification and cloning of SBP-box genes in wheat
To identify the wheat SBP-box genes, the draft sequences of the Aegilops tauschii (DD) genome were searched using a profile Hidden Molkov model (pHMM) of the SBP domain (Jia et al. 2013). The primary pHMM search revealed 20 sequences, and an E-value of less than 1 × 10−5 was used as the threshold: 19 were lower than 1 × 10−5; the E-value of the removed one was 0.9. There were no redundant sequences among them and each predicted gene was confirmed by a Pfam search for conserved SBP domains. Finally, 19 sequences were considered as putative wheat SBP-box genes and 10 were cloned in this study. As a final check, the full-length protein sequences of TaSPL were submitted to the National Center for Biotechnology Information to search the non-redundant protein sequences database. The result of BLASTP showed that none was reported in common wheat. However, six of them have been reported in A. tauschii, which were the predictions from whole genome shotgun sequence, and we designated the similar names (TaSPL1, TaSPL3, TaSPL6, TaSPL8, TaSPL15, TaSPL17) accordingly in wheat (Jia et al. 2013). The remaining four genes were named TaSPL20-TaSPL23 by consecutive nomenclature. The 10 TaSPL genes have been submitted to GenBank with accession numbers KF447877-KF447886 (Table S1). The identities of full-length protein sequences among the genes ranged 18.0%–86.9%, whereas the identities of the SBP domains ranged 58.2%–89.9%, indicating that the SBP domains encoded by the TaSPL genes displayed a high level of sequence conservation and the regions outside the domains were quite variable (Table1).
Table 1.
Identities (%) in protein sequence among wheat squamosa-promoter binding protein (SBP)-box genes
| SBP-box gene | 1 | 3 | 6 | 8 | 15 | 17 | 20 | 21 | 22 | 23 |
|---|---|---|---|---|---|---|---|---|---|---|
| TaSPL1 | 18.1 | 42.9 | 21.0 | 34.4 | 18.1 | 23.5 | 22.1 | 21.5 | 38.9 | |
| TaSPL3 | 58.2 | 18.0 | 20.6 | 20.3 | 31.1 | 23.0 | 19.4 | 21.3 | 36.0 | |
| TaSPL6 | 67.1 | 67.1 | 20.1 | 31.9 | 20.5 | 22.5 | 22.8 | 20.9 | 33.7 | |
| TaSPL8 | 62.0 | 69.6 | 68.4 | 25.3 | 25.5 | 28.0 | 26.8 | 27.3 | 46.0 | |
| TaSPL15 | 64.6 | 65.8 | 77.2 | 64.6 | 21.2 | 27.0 | 25.6 | 23.2 | 44.7 | |
| TaSPL17 | 62.0 | 70.9 | 70.9 | 70.9 | 65.8 | 26.0 | 23.4 | 24.9 | 48.8 | |
| TaSPL20 | 63.3 | 63.3 | 68.4 | 70.9 | 67.1 | 68.4 | 52.3 | 86.9 | 43.9 | |
| TaSPL21 | 59.5 | 64.6 | 69.6 | 73.4 | 62.0 | 63.3 | 82.3 | 52.6 | 44.0 | |
| TaSPL22 | 62.0 | 64.6 | 68.4 | 78.5 | 63.3 | 67.1 | 88.6 | 89.9 | 44.3 | |
| TaSPL23 | 62.0 | 67.1 | 67.1 | 70.9 | 68.4 | 65.8 | 67.1 | 69.6 | 70.9 |
Identities (%) in upper triangle region are based on full-length TaSPL protein sequences, whereas the data in the lower triangular matrix are based on sequences within the SBP domains.
Phylogenetic analysis of the SBP-box gene family in Arabidopsis, rice, and wheat
Based on sequence analysis, there are 17 SBP-box genes in Arabidopsis and 19 in the rice genome (Cardon et al. 1999; Xie et al. 2006; Yang et al. 2008). The gene functions of most members in Arabidopsis have been well characterized experimentally. In rice, OsSPL14 and OsSPL16 are also well known for promoting grain productivity (Jiao et al. 2010; Wang et al. 2012). In order to evaluate the phylogenetic relationships among the Arabidopsis, rice, and wheat, a total of 44 SBP-box gene family members were used for phylogenetic analysis based on their conserved SBP domains, including 16 from Arabidopsis, 18 from rice, and 10 from wheat. The 44 SBP-box genes were clustered in five groups, named G1–G5, and each group contained at least one member from Arabidopsis, rice, and wheat (Figure 1). There are several pairs of orthologous genes: AtSPL7 and OsSPL9, AtSPL8 and OsSPL8, and TaSPL8 in G1; OsSPL14/17 and TaSPL17 in G2; OsSPL6 and TaSPL6, OsSPL15 and TaSPL15 in G3; OsSPL1 and TaSPL1, OsSPL2 and TaSPL23 in G4; and OsSPL3/12 and TaSPL3 in G5. The results suggested that the main characteristics of the SBP-box gene family were established before the occurrence of speciation. Therefore, some of them should have similar biological functions. In other situations, paralogous genes were also recognized, such as AtSPL2/10/11 and OsSPL16/18, implying genes that evolved by gene duplication after separation of the three species.
Figure 1.
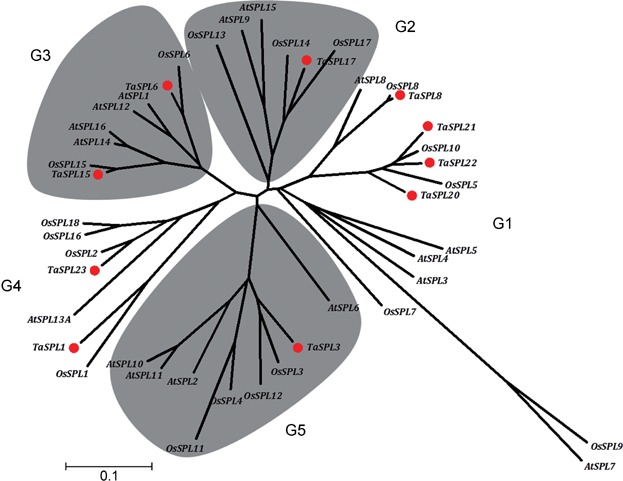
Phylogenetic analysis of squamosa-promoter binding protein (SBP)-box gene family based on the conserved SBP domain in Arabidopsis, rice, and wheat The tree was conducted by alignment of 16 Arabidopsis, 18 rice, and 10 wheat SBP-box genes by neighbor-joining methods with 1,000 bootstrap replicates. Species and gene names are indicated at the end of each branch; and the TaSPL genes are marked with red dots.
The phylogenetic tree also helped to predict the putative functions of the TaSPL genes based on the functions of SBP-box proteins in other species clustered in the same group. TaSPL8/20/21 clustered together with AtSPL8 and AtSPL3/4/5 in G1, implying that the functions of TaSPL8/20/21 may be related to flowering time and the development of floral organs (Wu and Poethig 2006; Kim et al. 2012). TaSPL17 grouped in G2 with rice OsSPL14 and Arabidopsis AtSPL9/15. OsSPL14 regulates ideal plant architecture and promotes panicle branching and higher grain productivity, and AtSPL9/15 alter inflorescence architecture and enhanced branching (Schwarz et al. 2008; Jiao et al. 2010). This is a strong indication that TaSPL17 may be involved in inflorescence branching.
Structural features of wheat SBP-box proteins within and outside the SBP domain
Squamosa-promoter binding protein-box gene proteins containing the SBP domain are suggested to be transcription factors involved in various aspects of plant development. The protein sequences of the SBP domain are highly conserved, whereas sequences outside the SBP domain are quite diverse in size and sequence (Cardon et al. 1999). In order to gain an insight into the sequence characters within and outside the SBP domain in wheat, the sequence logo for the SBP-box domain was conducted and protein motifs were searched based on the full-length of TaSPL proteins. In addition, the conserved motif structures of SBP-box genes in Arabidopsis and rice were also analyzed (Figures 2, 3).
Figure 2.
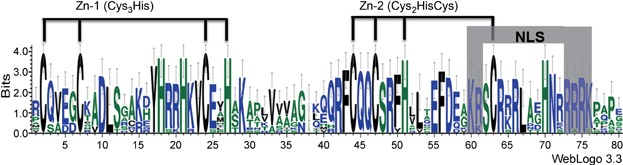
Sequence LOGO for the wheat squamosa-promoter binding protein (SBP)-box domain based on the TaSPL genes The overall height of the stack indicates the sequence conservation at that position, and the height of each letter represents the relative frequency of the corresponding amino acid at that position. The two zinc finger motifs (Zn-1 and Zn-2) and nuclear localization signal (NLS) are indicated.
Figure 3.
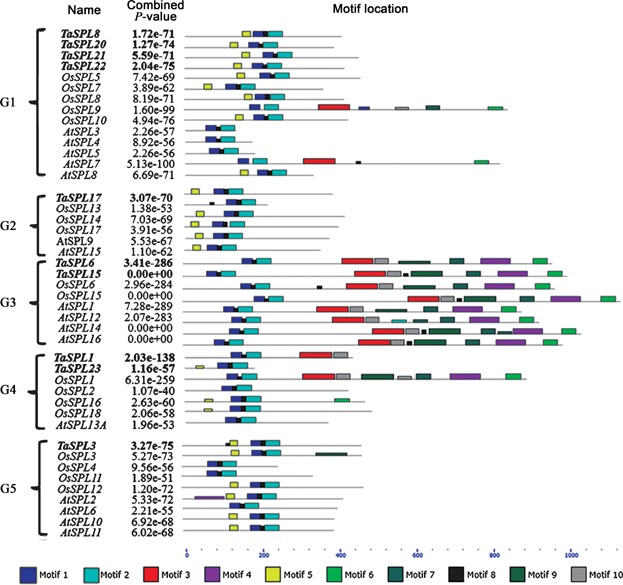
MEME motif search based on the full-length protein sequences of squamosa-promoter binding protein (SBP)-box genes in Arabidopsis, rice, and wheat Conserved motifs are indicated by numbered colored boxes.
Unlike the classic Zn-binding domain (Cys2His2 Zn finger), the wheat SBP domain consisted of two novel Zn-binding pockets, as reported for Zn finger patterns of SBP domain in other species, formed by eight conserved cystein and histidine residues (Cys3His and Cys2HisCys) that coordinates two Zn ions (Figure 2) (Mackereth et al. 2000; Yamasaki et al. 2004). As for transcription factors, a putative bipartite NLS was predicted at the C-terminal end of the wheat SBP domain (Figure 2), where covalent modification by nuclear proteins prevents or promotes their nuclear entry (Jans et al. 2000). The diverse combination of 10 motifs identified in wheat SBP-box genes demonstrated the diversity of sequence structures among the TaSPL genes, suggesting that they may have different roles in plant development (Figure 3). Motif 1 represents the first Zn-binding motif and motif 2 is the second Zn-binding motif. Motif 4 was an ankyrin repeat responsible for mediating protein-protein interactions and containing two alpha helices separated by loops (Mosavi et al. 2004). Moreover, the motif combinations in the same groups divided by phylogenetic analysis were mainly consistent with each other, revealing that the sequence features of SBP domains may also reflect the possibility of the sequence diversity outside the SBP domains in Arabidopsis, rice, and wheat.
Expression profiles of wheat SBP-box genes in different tissues
To investigate the spatial and temporal transcription patterns of the TaSPL genes, semiquantitative reverse transcription polymerase chain reaction (RT-PCR) was used to detect their expression in various wheat tissues, including root (SR, BR), shoot apical meristem (SC), leaf blade (SL, BL), node (BN), internode (BI), leaf sheath (BS), and ear (BE), at the seedling or late booting stages (Figure 4). Tissue-specific expression revealed gene functions related to some developmental processes. According to the phylogenetic tree, the five groups of TaSPL genes showed different expression patterns (Figure 4). TaSPL8/20/21 in G1 and TaSPL17 in G2 showed tissue-specific expression. TaSPL17/20/21 mainly expressed in the shoot apical meristem at seedling stage and the ear at late booting, indicating that these genes may have a functional role in regulating the shoot apical meristem to produce new leaves and spikelets. For example, TaSPL20 was abundantly expressed in the shoot apical meristem and ear, but was hardly detected in other tissues. In comparison with G1 and G2, genes in G3 (TaSPL6/15) generally showed the opposite situation, that is, they expressed lowly in the shoot apical meristem and ear, and were highly expressed in other tissues. The transcript levels of the genes in G4 (TaSPL1/23) were irregular among different tissues.
Figure 4.

Tissue-specific expression patterns of TaSPL genes in nine tissues The wheat tubulin gene was used as the control for equal cDNA concentrations. SR, SC, and SL, root, shoot apical meristem, and leaf blade at the seedling stage, respectively; BR, BN, BI, BS, BL, and BE, root, node, internode, leaf sheath, leaf blade, and ear at the late booting stage, respectively.
Wheat SBP-box genes involved in ear development assessed by the expression patterns
Because SBP-box genes are thought to be involved in flower and fruit development, various wheat tissues at different stages were collected to explore the expression profiles of the TaSPL genes during ear and grain development. The tissues used for semiquantitative RT-PCR included shoot apical meristems, ears, and grain at 15 stages from the four leaf stage to 21 d post-flowering (Figure 5). In general, the similar expression patterns of TaSPL8/20/21 in G1 and TaSPL17 in G2 reflected their functions involved in ear development; that is, they were gradually upregulated, reached their highest expression levels, and were downregulated at the end of the ear development. In accordance with the result of tissue-specific expression, the genes in these two groups (except TaSPL22) also exhibited tissue-specific expression, and they were barely detected in grains (Figures 4, 5). Interestingly, the expression of TaSPL6/15 in G3 increased with grain development, and TaSPL1/23 in G4 showed the completely opposite expression pattern (Figure 5). The gene in G5 (TaSPL3) was constantly expressed in all of the tested tissues (Figures 4, 5).
Figure 5.
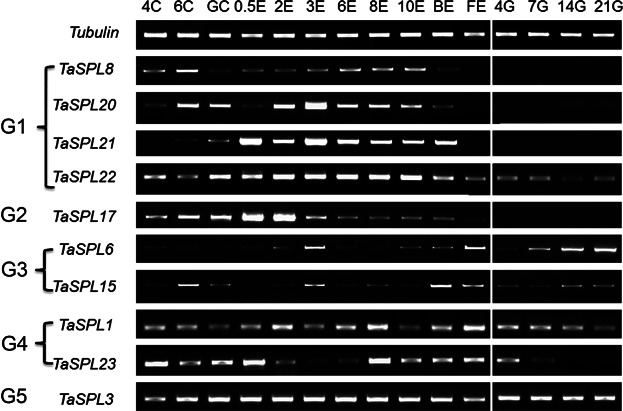
Expression analysis of the TaSPL genes in wheat during development of the shoot apical meristem, ear, and grain The wheat tubulin gene was used as a control for equal cDNA concentrations. 4C, 6C, and GC, shoot apical meristem at the four leaf, six leaf, and green-turning stages; 0.5E, 2E, 3E, 6E, 8E, and 10E, ears with different lengths, ranging 0.5–10 cm; BE and FE, ears at late booting stage and flowering stage, respectively; 4G, 7G, 14G, and 21G, grains at 4, 7, 14, and 21 d after flowering.
Real-time quantitative PCR of TaSPL17, TaSPL20, and TaSPL21 further confirming their functions involved in ear development
In order to check the expression profiles obtained by semiquantitative RT–PCR, TaSPL17, TaSPL20, and TaSPL21 were chosen as examples for illustrating the reliability. Because TaSPL17/20/21 mainly expressed in the shoot apical meristem and the ear, those wheat tissues at different stages were used for real-time quantitative (q)PCR. The expression levels revealed by the real-time PCR and semiquantitative RT–PCR were consistent with each other, further confirming their response to ear development (Figure 6).
Figure 6.
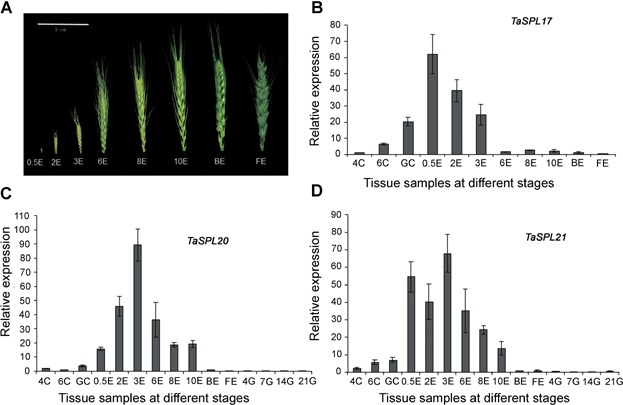
Real-time quantitative polymerase chain reaction analysis of TaSPL17 (B), TaSPL20 (C), and TaSPL21 (D) in wheat tissues at different stages 4C, 6C, and GC, shoot apical meristem at the four leaf, six leaf, and green-turning stages; 0.5E, 2E, 3E, 6E, 8E, and 10E, ears with different lengths, ranging 0.5–10 cm; BE and FE, ears at late booting stage and flowering stage (A); 4G, 7G, 14G, and 21G, grains at 4, 7, 14, and 21 d after flowering. Bar: 2 × SE.
When the young ear is approximately 0.5 cm (0.5E), the terminal spikelet initiated rapid growth of the completed ear and the stem commenced. Subsequently, the lateral spikelet and floret meristems start to initiate the different parts of the flower, such as stamens and carpel. When those occurred, the young ear length is approximately 3 cm (3E). TaSPL17/20/21 strongly expressed in 0.5E, 2E, and 3E. Their maximum expression levels differed from each other. The expression level of TaSPL17 peaked in 0.5E, with high level of expression in 2E. TaSPL20 strongly expressed in 2E, but the highest expression occurred in 3E. The double-peaked expression pattern of TaSPL21 was apparent in 0.5E and 3E (Figure 6). The high relative expression of TaSPL17/20/21 occurred during those periods suggested their functional role in the development of spikelet and floret. Around booting stage, meiosis occurs in the young ear (BE), almost simultaneously at the green anther stage. At the yellow anther stage, the florets are mature (FE) and anthesis is about to take place. TaSPL17/20/21 showed obvious low levels of expression in BE and FE as compared to 0.5E, 2E, and 3E (Figure 6).
Subcellular localization of the TaSPL17 protein
The deduced amino acid sequences of wheat SBP-box genes contained a putative bipartite NLS, a major characteristic of transcription factors. TaSPL17 protein was selected for validating the NLS experimentally. The recombinant construct of the TaSPL-green fluorescent protein (GFP) fusion gene was transiently expressed in living onion epidermal cells. As expected, TaSPL17-GFP protein (GFP) was localized in the cell nucleus (Figure 7).
Figure 7.
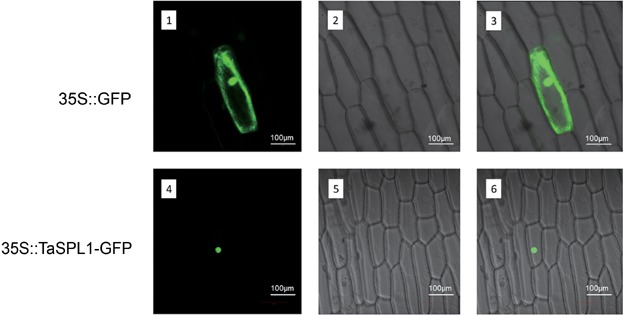
Subcellular localization of TaSPL17–green fluorescent protein (GFP) fusion proteins driven by the CaMV35S promoter The vector control (35S::GFP) and the fusion proteins (35S::TaSPL17–GFP) were introduced into onion epidermal cells and observed with a laser scanning confocal microscope. Images are in dark field (1, 4), bright field (2, 5), and combined (3, 6).
DISCUSSION
Diversity of wheat SBP-box gene family from sequence features to expression patterns
Squamosa-promoter binding protein-box genes have been reported in many plant species, such as Chlamydomonas (Kropat et al. 2005), Arabidopsis (Cardon et al. 1997, 1999), maize (Wang et al. 2005; Chuck et al. 2010), rice (Shao et al. 1999; Xie et al. 2006), tomato (Manning et al. 2006), Populus x canadensis (Wang et al. 2011), silver birch (Lännenpää et al. 2004), and Salvia miltiorrhiza (Zhang et al. 2013). However, few members were reported in wheat. From the draft sequences of the d genome of bread wheat, 19 sequences were identified as putative wheat SBP-box genes and 10 of them were cloned in this study. Based on protein sequences of the conserved SBP domains, phylogenetic analysis classified the TaSPL genes into five groups (Figure 1). The results of motif compositions and expression analyses supported this division. Furthermore, expression patterns of the five groups are distinctive; for example, TaSPL8/20/21 in G1 exhibited tissue-specific expressions and the gene in G5 (TaSPL3) expressed constitutively; TaSPL6/15 in G3 were upregulation and TaSPL1/23 in G4 were downregulation during grain development (Figures 4, 5). Thus, the variable protein sequences in the SBP domain, diversity motif compositions, and different expression patterns suggest that TaSPL is a diverse gene family. On the other hand, the consistency of the phylogenetic analysis, motif compositions, and expression patterns of 10 TaSPL genes revealed specific gene structures and functions. The TaSPL genes studied here obviously do not represent all members of the wheat SPL gene family, but they likely represent the diversity within the wheat SBP-box gene family.
Some wheat SBP-box genes involved in ear development
The diversity of SBP-box genes contributes to their regulation of diverse aspects of plant development, such as leaf shape and epidermal traits (Shikata et al. 2009; Wu et al. 2009), juvenile-to-adult phase transition and flowering (Schwarz et al. 2008; Wang et al. 2009), fertility (Xing et al. 2010), embryonic development (Nodine and Bartel 2010), anthocyanin biosynthesis (Gou et al. 2011), fruit ripening and grain yield (Manning et al. 2006; Jiao et al. 2010), and bract and ear glume development (Wang et al. 2005; Chuck et al. 2010). Numerous studies report that SBP-box genes play a critical role in regulation of flower and fruit development (Wang et al. 2005; Manning et al. 2006; Wu and Poethig 2006; Chuck et al. 2010; Jiao et al. 2010; Wang et al. 2012). In our study, TaSPL20/21 in G1 and TaSPL17 in G2 mainly expressed in the shoot apical meristem and the ear were barely detected in other tissues (Figure 4). Their expression levels responded to ear development (Figure 5). The temporal and specific expression patterns demonstrated that they may have a functional role in regulating the shoot apical meristem to produce new spikelets and affecting ear development. In addition, TaSPL8/20/21 clustered together with AtSPL8 and AtSPL3/4/5 in G1, and TaSPL17 grouped into G2 with rice OsSPL14 and Arabidopsis AtSPL9/15 (Figure 1). These genes in Arabidopsis and rice are involved in flowering time and inflorescence development (Wu and Poethig 2006; Schwarz et al. 2008; Jiao et al. 2010; Kim et al. 2012). OsSPL14 regulates rice plant architecture, including fewer tillers, stronger culms, and denser panicles (Jiao et al. 2010). TaSPL17, the presumed ortholog of OsSPL14, expressed predominantly in the root, shoot apical meristem, internode, and ear, quite consistent with the functions of OsSPL14 (Figures 1, 6). Therefore, the results of phylogenetic analysis and expression analysis of the SBP-box gene family further confirmed the putative functions of the wheat genes in G1 and G2.
Putative orthologous and paralogous SBP-box genes reveal both conserved and specialized functions
Orthologous genes originate from common ancestral genes and have the same functions (Fitch 1970; Mirny and Gelfand 2002), whereas paralogous genes typically diversify in function or undergo subfunctionalization (Adams and Wendel 2005). In higher plant evolution, the monocots (rice and wheat) and dicots (Arabidopsis) diverged from a common ancestor 200 million years ago (Wolfe et al. 1989). Squamosa-promoter binding protein-box genes, functioning specifically in plants, originated before divergence of green algae and ancestral land plants (Guo et al. 2008). Before the splits of Arabidopsis, rice, and wheat, the SBP-box gene family probably originated from the common ancestor of green plants, followed by duplication and divergence. We observed several pairs of orthologous and paralogous genes among AtSPL, OsSPL, and TaSPL genes in our study (Figure 1). The functions of orthologous genes were similar to each other. Arabidopsis AtSPL3 and orthologous genes in Antirrhinum (SBP1 and SBP2) and silver birch (BpSPL1) are involved in control of early flowering (Klein et al. 1996; Lännenpää et al. 2004; Wu and Poethig 2006). Arabidopsis AtSPL9 is involved in plastochron length, and the homologous OsSPL14 gene prolongs the plastochron and reduces tiller number in rice (Wang et al. 2008; Jiao et al. 2010). On the other hand, the functions of paralogous SBP-box genes display partial functional redundancy, although they may have their own specialized functions during plant development. In this study, similar expression patterns among genes in G1 and G2 (Figures 4, 5) suggested the occurrence of functional redundancy in wheat as well as in Arabidopsis (Huijser and Schmid 2011), but their transcription levels peaked at different stages implying specialized functions during ear development.
In this study, we cloned 10 wheat SBP-box genes, and presented a comprehensive analysis of their structures and functions. The diverse gene structures and different expression patterns suggested that wheat SBP-box genes have a wide range of functions. Phylogenetic and expression analysis suggest a potential role for wheat SBP-box genes in regulation of the shoot apical meristem to produce new spikelets or to affect ear development. Our results provide a significant beginning for detailed functional analysis of SBP-box genes in wheat.
MATERIALS AND METHODS
Plant materials and sample preparation
Common wheat (Triticum aestivum L.) cultivar Yanzhan 4110 was planted on the experimental farm (39°48′N, 116°28′E) of the Institute of Crop Science, the Chinese Academy of Agricultural Sciences, Beijing. Plants were sown at the beginning of October 2011 and harvested in mid-June 2012. For tissue-specific expression analysis, different tissue samples were collected from: (i) root (SR), shoot apical meristem (crown, SC), and leaf blade (SL) of seedlings; and (ii) root (BR), node (BN), internode (BI), leaf sheath (BS), leaf blade (BL), and ear (BE) at late booting. To detect dynamic expression patterns, samples at various developmental stages were collected, including three kinds of tissue samples. First, shoot apical meristem at the four leaf stage (4C), six leaf stage (6C), and green-turning stage after winter (GC). Second, ears with lengths ranging 0.5–10 cm (0.5E, 2E, 3E, 6E, 8E, and 10E), and ears at the late booting (BE) and flowering (FE) stages. Between 0.5E and 3E is the main phase for the spikelet and floret initiation, and the lateral spikelet and floret meristems start to initiate the different parts of the flower. Before booting stage, the immature ear (from 3E to BE) is completing the development of the male and female parts of the flower. Later, meiosis occurs then anthesis takes place (from BE to FE). Third, grains at 4 (4G), 7 (7G), 14 (14G), and 21 d (21G) post-flowering. Each of the above biological samples was represented by mixed tissue samples at the same stage from at least three different, randomly selected individuals.
Identification and cloning of SBP-box genes in wheat
The scaffolds of the Aegilops tauschii (DD, D genome donor species of common wheat) draft sequence were screened with a pHMM of the SBP domain (profile PF03110.9) to identify novel wheat SBP-box genes (Eddy 1998; Bateman et al. 2004; Jia et al. 2013). The HMMER 3.0 program was used to search the local database. An E-value of less than 1 × 10−5 was used as the cut-off in HMMER search. To exclude redundancy, we aligned all sequences with the ClustalW algorithm using the DNAStar software package and sequences that shared more than 95% matches were considered redundant (Zhang et al. 2012). Open reading frames were predicted by FGENESH (http://linux1.softberry.com/berry.phtml). All non-redundant protein sequences of putative wheat SBP-box genes were checked for the SBP domain by Pfam (http://pfam.sanger.ac.uk/).
For each wheat SBP-box gene, a pair of primers was designed based on the putative sequence with cDNA templates prepared from different tissue samples (Table S2). Polymerase chain reaction was performed in a 15 µL system using 0.3 U of TransStart FastPfu DNA polymerase (TransGen, Beijing, China) in a Veriti 96-Well Thermal Cycler (Applied Biosystems, Foster City, CA, USA). Amplified products were separated in a 1.2% agarose gel electrophoresis. The target bands were recovered, cloned into pEASY-Blunt vectors, and transformed to DH5α competent cell (TransGen). Twelve positive clones were sequenced for each candidate SBP-box gene with an ABI 3730 × L 96 capillary DNA analyzer (Applied Biosystems). Only the wheat SBP-box genes that were verified by sequencing were used in this study.
Phylogenetic analysis
Arabidopsis SBP-box genes were obtained from TAIR (http://www.arabidopsis.org/index.jsp) (Cardon et al. 1999). Rice SBP-box genes were downloaded from RGAP (Ouyang et al. 2007). The amino acid sequences of the conserved SBP domain were considered for phylogenetic analysis. SMART (http://smart.embl-heidelberg.de/) was used to identify the SBP domain sequences of SBP-box genes in Arabidopsis, rice, and wheat (Schultz et al. 1998; Letunic et al. 2012). Squamosa-promoter binding protein-domain sequences and accession numbers of SBP-box genes in Arabidopsis, rice, and wheat are presented in Table S3. The multiple sequence alignment was generated by the ClustalW algorithm within the software MEGA 4.1 (Thompson et al. 1994; Tamura et al. 2007). An NJ phylogenetic tree for Arabidopsis, rice, and wheat SBP-box gene family members was constructed, and the significance of the inferred relationships was determined by bootstrap analysis (1 000 replicates).
Gene structure analysis
The sequence logo of the wheat SBP domain sequences was generated by submitting the multiple alignment sequences to the online WebLogo 3 platform (http://weblogo.threeplusone.com/) (Schneider and Stephens 1990; Crooks et al. 2004). Conserved motif analysis of the whole sequences of wheat SBP-box genes was performed by MEME 4.9.0 with default settings, except that the minimum width was 10, the maximum width was 80, and the maximum number of motifs to find was 10 (Bailey et al. 2009).
Semiquantitative RT-PCR and real-time PCR
Total RNA was extracted using TRIZOL reagent, and DNA was removed by digestion with DNase I. Two micrograms of total RNA was used to synthesize first-strand cDNA using the SuperScriptII Reverse transcriptase (Invitrogen, San Diego, CA, USA). For semiquantitative RT–PCR, the wheat tubulin transcript was used to adjust the relative transcript level. Primer sequences amplifying wheat tubulin with 500 bp amplicon were: 5′-TGAGGACTGGTGCTTACCGC-3′ and 5′-GCACCATCAAACCTCAGGGA-3′. The gene-specific primers and the size of amplified fragments used in semiquantitative RT-PCR for wheat SBP-box genes are presented in Table S4. The primers were designed based on the cDNA sequence of wheat SBP-box genes. Polymerase chain reaction was performed using TransStart FastPfu DNA polymerase (TransGen). Amplified products were recovered from agarose gel and cloned into pEASY-Blunt vectors (TransGen). For each gene, 12 positive clones were randomly selected for sequencing. The primers that only amplified single, specific products were used in this study.
Various numbers of PCR cycles were tested to ensure that the reactions still had been in the exponential range of amplification. The PCR for the amplification of wheat tubulin was run for 23 cycles, which were tested from 20 to 30 cycles. For wheat SBP-box genes, PCR cycles were various depending on the expression levels of different genes, which were tested from 26 to 38 cycles. Ten microliters of the RT–PCR products were analyzed by 1.2% agarose gel electrophoresis and stained with ethidium bromide. Each RT–PCR was replicated three times.
For qRT–PCR, primer sequences amplifying wheat Actin were: 5′-CTCCCTCACAACAACAACCGC-3′ and 5′-TACCAGGAACTTCCATACCAAC-3′, which were used as a reference gene. The gene-specific qRT–PCR primers (5′-GTGTCTCGCAGGGGTCGC-3′ and 5′-CGGCGGCATGTAGTTACTCG-3′; 5′-CGGAGAATGACGACCAC-3′ and 5′-GTCGTTGTTCTGGTCGG-3′; 5′-CGCAGGGCAAGGACTC-3′ and 5′-CGACTTGTTGGTCCTGTGA-3′) were designed based on the coding region of TaSPL17, TaSPL20, and TaSPL21, respectively. Quantitative RT–PCR was performed in triplicate with an ABI Prism 7300 system using the SYBR Green PCR master mix kit (Applied Biosystems). The relative level of gene expression was analyzed by the 2−△△CT (Livak and Schmittgen 2001).
Subcellular localization of TaSPL17 protein
The coding region of TaSPL17 was fused upstream of the GFP gene under control of the CaMV 35S promoter and NOS terminator in the pJIT163–GFP expression vector. The primers used to amplify the full-length coding sequence added with restriction sites were: 5′-AGCGAAGCTTATGGAGATTGGAAGCGG-3′ (HindIII site underlined) and 5′-ACTCGGATCCCAGAGACCAGTTGGACGAG-3′ (BamHI site underlined). The constructed 35S::TaSPL17–GFP fusion protein was confirmed by restriction analysis and sequencing. Recombinant constructs were transformed into onion epidermal cells via a gene gun (Helios; Bio-Rad, Richmond, CA, USA). After incubation in Murashige–Skoog medium at 28 °C for 36–48 h, the transformed cells were observed with a laser scanning confocal microscope (Leika TCS-NT, Heidelberg, Germany).
Acknowledgments
We thank Professor Robert A. McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia) for revising the manuscript. This study was supported by the National High-tech R&D Program (2011AA100501) and the National Basic Research Program of China (2010CB951501).
Supporting Information
Additional supporting information may be found in the online version of this article:
Characteristics of wheat squamosa-promoter binding protein (SBP)-box genes
Primers for amplification squamosa-promoter binding protein (SBP)-box genes in wheat
Squamosa-promoter binding protein (SBP) domain sequences and accession numbers of SBP-box genes in Arabidopsis, rice, and wheat
Primers used in semiquantitative reverse transcription polymerase chain reaction (RT-PCR) for wheat squamosa-promoter binding protein (SBP)-box genes
REFERENCES
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005;352:585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/s0378-1119(99)00308-x. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang Z, Liu D, Zhang K, Li A, Mao L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J Integr Plant Biol. 2010;52:946–951. doi: 10.1111/j.1744-7909.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, Simmons BA, Pauly M, Hake S. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci USA. 2011;108:17550–17555. doi: 10.1073/pnas.1113971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Ye ZH. Regulation of plant biomass production. Curr Opin Plant Biol. 2010;13:298–303. doi: 10.1016/j.pbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Biol. 1970;19:99–113. [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu XC, Ge S, Yang J, Luo JC. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138:4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: A key control point in nuclear transport. BioEssays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer KFX, Li D, Pan S, Zheng F, Hu Q, Xia X, Li J, Liang Q, Chen J, Wicker T, Gou C, Kuang H, He G, Luo Y, Keller B, Xia Q, Lu P, Wang J, Zou H, Zhang R, Xu J, Gao J, Middleton C, Quan Z, Liu G, Wang J, Consortium IWGS, Yang H, Liu X, He Z, Mao L, Wang J. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–95. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depege N, Huijser P, Merchant S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lännenpää M, Jänönen I, Hölttä-Vuori M, Gardemeister M, Porali I, Sopanen T. A new SBP-box gene BpSPL1 in silver birch (Betula pendula L.) Physiol Plant. 2004;120:491–500. doi: 10.1111/j.0031-9317.2004.00254.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mackereth CD, Arrowsmith CH, Edwards AM, McIntosh LP. Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum. Proc Natl Acad Sci USA. 2000;97:6316–6321. doi: 10.1073/pnas.97.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Takeda Y, Hatano S, Matsuoka M, Hirano H. Rice genes encoding the SBP domain protein, which is a new type of transcription factor controlling plant development. Rice Genet Newsl. 1999;16:114. [Google Scholar]
- Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. Arabidopsis SBP-box genes SPL10SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009;50:2133–2145. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- Stone JM, Liang X, Nekl ER, Stiers JJ. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41:744–754. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS. miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Höhmann S, Berndtgen R, Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. SQUAMOSA promoter binding protein–like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell. 2009;21:347–361. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang X, Gu S, Hu Z, Xu H, Xu C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene. 2008;407:1. doi: 10.1016/j.gene.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schwarz S, Saedler H, Huijser P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol. 2007;63:429–439. doi: 10.1007/s11103-006-9099-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, Liu X, Kong X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot. 2012;63:203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu B, Zhao D, Li C, Shao F, Lu S. Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J Integr Plant Biol. 2013;56:38–50. doi: 10.1111/jipb.12111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of wheat squamosa-promoter binding protein (SBP)-box genes
Primers for amplification squamosa-promoter binding protein (SBP)-box genes in wheat
Squamosa-promoter binding protein (SBP) domain sequences and accession numbers of SBP-box genes in Arabidopsis, rice, and wheat
Primers used in semiquantitative reverse transcription polymerase chain reaction (RT-PCR) for wheat squamosa-promoter binding protein (SBP)-box genes


