Abstract
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of an array of pathogenic autoantibodies, including high-affinity anti-dsDNA IgG antibodies, which plays an important role in disease development and progression. Lupus preferentially affects women during their reproductive years. The pathogenesis of lupus is contributed by both genetic factors and epigenetic modifications that arise from exposure to the environment. Epigenetic marks, including DNA methylation, histone post-translational modifications and microRNAs (miRNAs), interact with genetic programs to regulate immune responses. Epigenetic modifications influence gene expression and modulate B cell functions, such as class switch DNA recombination (CSR), somatic hypermutation (SHM) and plasma cell differentiation, thereby informing the antibody response. Epigenetic dysregulation can result in aberrant antibody responses to exogenous antigens or self-antigens, such as chromatin, histones and dsDNA in lupus. miRNAs play key roles in the post-transcriptional regulation of most gene-regulatory pathways and regulate both the innate and the adaptive immune responses. In mice, dysregulation of miRNAs leads to aberrant immune responses and development of systemic autoimmunity. Altered miRNA expression has been reported in human autoimmune diseases, including lupus. The dysregulation of miRNAs in lupus could be the result of multiple environmental factors, such as sex hormones and viral or bacterial infection. Modulation of miRNA is a potential therapeutic strategy for lupus.
Keywords: activation-induced cytidine deaminase (AID), antibody, autoantibody, autoimmunity, B cell, class switch DNA recombination (CSR), epigenetics, microRNA, somatic hypermutation (SHM), systemic lupus erythematosus (SLE)
Introduction
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of an array of pathogenic autoantibodies against nuclear self-antigens, including high-affinity anti-dsDNA, anti-histone, and anti-ribonucleoprotein autoantibodies, which mediate widespread tissue and organ injury (1). These autoantibodies are mutated and class-switched, mainly to IgG, and are secreted by a large number of plasma cells, indicating that immunoglobulin (Ig) somatic hypermutation (SHM), class switch DNA recombination (CSR) and plasma cell differentiation are important in their generation (2, 3). Epigenetic modulation by DNA methylation, histone post-translational modifications and microRNAs (miRNAs) allows regulation of B cell mechanisms, such as SHM, CSR and plasma cell differentiation (4–6). Dysregulation of epigenetic elements or mediators, including miRNAs, can result in aberrant immune responses, including dysregulated antibody production, and compound genetic susceptibility to mediate autoimmunity.
The development and clinical manifestations of lupus are determined by genetic and environmental factors (1, 7). The role of genetic dysregulations has been extensively investigated in lupus, revealing widespread polymorphisms throughout of genome elements correlated with the disease. Although studies investigating common genetic variants in lupus have revealed a number of susceptibility loci, the cumulative effect size of these loci accounts for a small fraction of disease heritability (8). Epigenetic dysregulation likely compounds genetic susceptibility (9) in the generation of autoantibodies and disease (10–14), evidenced by low lupus penetrance (25–45%) in monozygotic twins (15), alterations of histone modifications and DNA methylation in lupus patients, and autoimmunity in mice with miRNA dysregulation (16–20).
miRNAs are small (~22 nucleotides), evolutionarily conserved noncoding RNAs derived from much larger primary transcripts encoded by their “host genes”. miRNAs bind to complementary sequences within the 3′ untranslated region (3′ UTR) of their target mRNAs and negatively regulate protein expression at the post-transcriptional level through inhibition of translation and/or reduction of mRNA stability (21). The mammalian genome encodes thousands of miRNAs that collectively affect the expression of more than half of protein-coding genes. In addition, miRNAs have been implicated as fine-tuning regulators controlling diverse biological processes at post-transcriptional level (22). They can potentially regulate every aspect of cellular activity, from differentiation and proliferation to apoptosis, as well as modulate a large range of physiological and pathological processes. Expression of these short RNA molecules is regulated by transcription of their host genes (e.g., Bic for miR-155 (23) and Rtl1 for miR-127 (24)), followed by sequential processing of long primary transcripts (pri-miRNA) by the RNase III enzyme Drosha, and processing of miRNAs precursors (pre-miRNA) by Dicer (25). In mammals, some of these miRNAs are expressed in a tissue-specific and developmental stage-specific manner and play an important role in regulation and maintenance of the immune system response to various environmental stimuli. Dysregulation of miRNAs has been linked to diverse pathological processes, including autoimmunity that causes lupus.
Epigenetic marks/changes are induced in B cells by signals triggered by CD40, Toll-like receptors (TLRs) and cytokine receptors to regulate CSR/SHM and plasma cell differentiation (5, 6, 26–34). They inform the antibody response to exogenous (e.g., microbial) antigens in healthy subjects or self-antigens in autoimmune patients (5). miRNAs also cross-regulate with DNA methylation and histone modifications (35). These cross-regulations would contribute to unique epigenetic landscapes associated with distinct cell developmental and differentiation stages, thereby controlling stage-specific gene expression and function (36–38).
miRNA biogenesis and functions
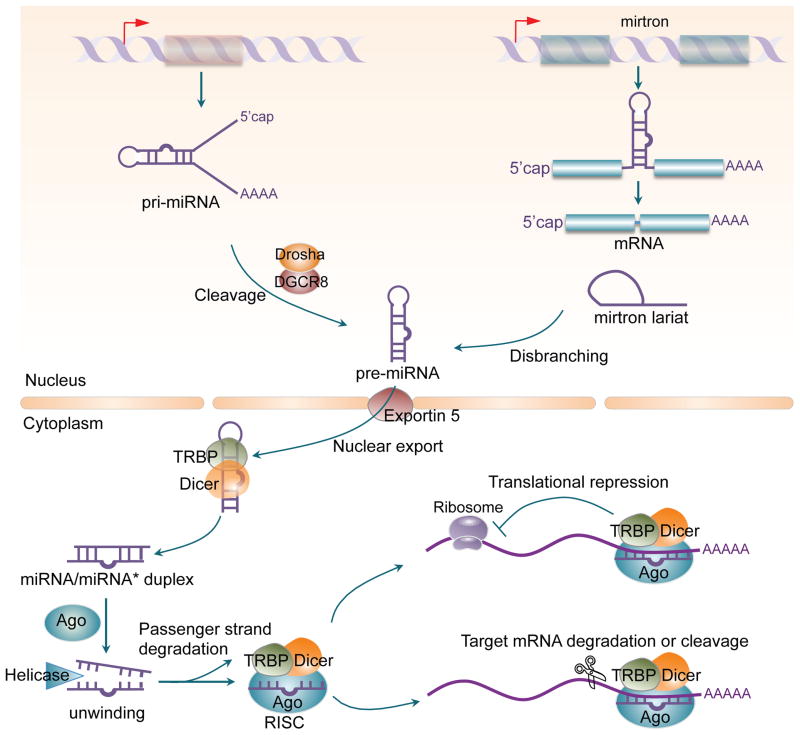
miRNAs possess cellular and functional specificity that results from a highly regulated biogenesis process (39) (Figure 1). They are produced either from their own unique genes, such as miR-146a, from intronic sequences within protein and non-protein-coding genes, i.e. miR-126, or from exons of non-protein-coding transcripts. This variability in encoding matrixes allows for multiple possibilities in their regulation, such as alternative processing or miRNA processing. The canonical miRNA biogenesis pathway involves transcription from these genes by RNA polymerase II. Some miRNAs are co-regulated with their host gene as a part of transcriptional regulation during the development of immune cells. miRNA-containing primary transcripts can produce a single miRNA, or a cluster of multiple and different miRNAs located on a single transcript, typically working in concert (e.g. the miR-17-92 family, which include miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1).
Figure 1. miRNA biogenesis and function.
Mature miRNAs are generated from endogenous hairpin-shaped transcripts and derived from the processing of long primary miRNA transcripts (pri-miRNA) by a Drosha/DGCR8 microprocessor in the nucleus to produce precursor miRNAs (pre-miRNAs). Alternatively, in a noncanonical miRNA biogenesis pathway, “mirtrons,” a new class of miRNA precursors bypasses Drosha processing and are spliced by the spliceosome to yield branched pre-mitrons, which then go through lariat-mediated debranching to generate pre-miRNAs. After being exported from the nucleus into the cytoplasm by Exportin-5, pre-miRNAs are processed by the RNAIII enzyme Dicer, yielding imperfectly matched miRNA/miRNA* duplexes which are loaded into the Argonaute (Ago) protein to generate the RNA-induced silencing complex (RISC). The guided strand of the miRNA/miRNA* duplex remains in the RISC as a mature miRNA, and the complementary strand (the passenger strand miRNA*) is degraded. Once loaded onto RISC, the mature miRNA will interact with the 3′ UTR of its target mRNA to silence the mRNA strand, thereby, dampening gene expression.
Most miRNAs derive from longer, intramolecular double-stranded primary miRNA gene transcripts (pri-miRNA) that are sequentially cleaved into shorter intermediates by specialized ribonuclease III (RNase III) enzymes that partner with specific double-stranded RNA-binding proteins. The pri-miRNAs comprise either a monocistronic or polycistronic precursor cluster (40–42). They are processed by the microprocessor complex in the nucleus, which consists of the Drosha RNase III endonuclease and DiGeorge syndrome critical region protein 8 (DGCR8) (43), to produce a 60–70 nt stem-loop intermediate, precursor-miRNA (pre-miRNA) hairpin. The pre-miRNA are subsequently exported from the nucleus to the cytoplasm by exportin 5 (44). Once transported to the cytoplasm, the pre-miRNA is cleaved by the RNase III endonuclease Dicer, to generate a double-stranded RNA duplex, which contains both the mature (guide) and an antisense passenger (*) strand (39). The double-stranded miRNA duplex is then unwound and incorporated into the RNA-induced silencing complex (RISC), by strand selection based on thermodynamic properties. Of the two strands that could potentially serve as a functional miRNA, only one strand of the duplex is selected as the guide strand to be packaged into the RISC complex, which comprise of Argonaute2 (Ago2) and other proteins. Once the miRNA strand has been selected and loaded onto the RISC complex, it will target the 3′UTR of the mRNA which shares a 6–8 nucleotide partial complementarity seed region at the 5′ end of the miRNA and downregulate the expression of the gene by degradation or destabilization (45). Although miRNA repression of its individual target is generally mild (usually less than 2-fold), it may have a powerful effect on the overall function of the cells in which it acts on. One reason for this is that miRNAs tend to be highly pleiotropic; each miRNA may target several mRNAs. Thus, a miRNA can target several members of a given signaling pathway or different targets in converging pathways to have a constitutive effect on the cell. miRNAs are also highly redundant, with multiple individual miRNAs converging on the same target mRNAs.
In addition to canonical miRNA biogenesis, a number of alternative non-canonical pathways have been identified (46). The first and most widely characterized is the mirtron pathway (47). In this particular pathway, miRNAs are encoded in short intronic genes with hairpin potential. The intron is spliced out into a lariat form that later adopts a pre-miRNA structure due to a lariat debranching enzyme and existing mRNA splicing machinery. In this manner, the mimicked pre-miRNA structure bypasses the canonical microprocessor complex (48). A Dicer-independent miRNA biogenesis pathway has also been reported (49, 50). This pathway utilizes the catalytic activity of Ago2 to directly cleave miRNA hairpins (51–53). miR-451, which is upregulated in the patients with lupus or rheumatoid arthritis (RA), is one example of a well characterized miRNA produced independent of Dicer (49, 51–54). The unusual short stem structure of pre miR-451 promotes the binding and processing by Ago2 (55).
Regulation of miRNAs
miRNAs are tightly regulated by a range of mechanisms involving protein-protein and protein-RNA interactions affecting primary miRNA transcription, miRNA biogenesis, function and degradation. These processes are essential for the specific functions of miRNAs, as alterations are commonly associated with different disease states (56). Abundance of miRNA expression could result from: (i) the transcription of the miRNA host gene, (ii) the processing of the transcribed pri-miRNA into the mature miRNA, and (iii) the stability of the miRNA.
Similar to protein-encoding genes, microRNA expression can be regulated at the transcriptional and the post-transcriptional level. While some miRNAs are constitutively expressed, others display cell type- or developmental stage-specific patterns. Regulation of miRNA host gene transcription, which can be modulated by DNA methylation and histone post-translational modifications, is important for the tissue or development stage specific expression of miRNAs. As with many other cells, immune cells employ miRNAs to regulate lineage commitment, proliferation, migration and differentiation. Like the genes that encode proteins, the expression of miRNA-producing transcripts also changes during the immune response.
Processing of miRNAs is regulated in multiple steps and results in either elevated or decreased miRNA levels. Altered miRNA levels may be caused by regulatory proteins that influence miRNA processing, acquired variations in the miRNA transcript, or changes in the nuclear export efficiency. A number of proteins that regulate miRNA processing have been suggested to function as key elements in defining the unique expression patterns of miRNAs in different tissues, cell types, and pathological conditions (57). These proteins can be subdivided into three groups: Drosha binding/associated proteins, Dicer binding proteins, and proteins that bind to the terminal loop of the pri- and/or pre-miRNAs. Regulatory proteins are a dominant factor in the regulation of Drosha-mediated pri-miRNA processing. It is likely that more Drosha-associated proteins regulate miRNA processing, and, as such, the balance between positive and negative regulators may determine the efficiency of miRNA processing. Various signaling pathways enhance or reduce the efficiency of this step. In addition to these regulatory mechanisms, single nucleotide polymorphisms (SNPs) that are associated with many diseases can also have a pronounced effect on the efficiency of the miRNA processing machinery.
Most miRNAs appear to be stable with half-lives of a few days but display differential stability in human cells (58). Specific modifications and exonucleases can profoundly influence miRNA existence. miRNA stability or abundance can be affected by both cis- and trans-acting factors. In contrast to their precursor pri-miRNAs, most miRNAs and pre-miRNAs appear to have 5′ and 3′ unprotected ends that may render them accessible to exoribonucleases. The post-transcriptional nucleotide(s) addition to the 3′ ends of pre-miRNAs or mature miRNAs significantly affect miRNA stability.
By controlling different stages of miRNA biogenesis and localization, degradation and activity, RNA-binding proteins (RBPs) function as key components in the determination of miRNA function. Alteration of RBP function in any of the crucial steps of the miRNA pathway can lead to impairment (59). Like miRNA, RBPs can regulate mRNA stability and translation. They bind to mRNAs and facilitate or counteract miRISC activity. Furthermore, RBPs that enhance repression via miRNAs could facilitate or stabilize miRISC binding. Enhanced silencing could also occur by strengthening interactions between miRISC components and downstream effectors. This may involve translational modifications of protein components. Recruitment of translational repressors or deadenylation factors by RBPs independent of miRISC also increases target repression. RBPs counteracting with miRISC function can either prevent miRISC binding or displace miRISC from mRNA. Alternatively, they may also promote post-translational modification of miRISC components. Other proteins could also interfere with the interaction between miRISC components and downstream effectors.
miRNA in immune responses
miRNAs that are important for the immune response have been the focus of extensive study (18). miRNAs are essential in both adaptive and innate immunity, including controlling the differentiation of various immune cell subsets as well as their immunological functions (60–62) (Table 1 and Figure 2). miRNAs are essential for B cell development and immune function, particularly in the germinal center (GC) response. In T cells, microRNAs function as key regulators of the lineage induction pathways, and play an important role in the activation, function and maintenance of the regulatory T-cell lineage.
Table 1.
miRNAs involved in immune responses
| miRNA | Target gene(s) | Cells | Processes | Ref. |
|---|---|---|---|---|
| miR-155 | Aicda | B cell | CSR and SHM | (29, 30, 76) |
| Pu.1 | B cell | (137) | ||
| Shp1 | B cell | GC response | (113, 138) | |
| STAT1 | T cell | (139) | ||
| IL-17 | Th17 | Th17 development | (140) | |
| miR-181b | Aicda | B cell | CSR and SHM | (31) |
| miR-93 | Human AICDA | B cell | CSR and SHM | (78) |
| miR-361 | Aicda | B cell | CSR and SHM | (77) |
| miR-30a | Prdm1 | B cell | Plasma cell differentiation | Zan, et al, unpublished |
| miR-125b | Irf4, TNFα, Prdm1 | B cells | B cell differentiation | (82) |
| miR-146a |
Traf6, Irak1 IRF5 STAT1 |
T cell | TLR signaling IFN signaling |
(141) |
| miR-150 | c-Myb | B cell | B cell differentiation | (69) |
| miR-21 | NFκB | TLR signaling | ||
| Foxp3 | Treg | Treg development | (142) | |
| miR-9 | NFKB | Monocyte, neutrophil | TLR signaling | (143) |
| miR-10a | Bcl-6 | T cell | Th cell plasticity and fate | (144) |
| miR-17-92 | Bim | B and T cell precursors | B and T cell development | (17) |
| PTEN | Memory CD8 T cell | (145) | ||
| miR-210 | Oct-2 | B cells | B cell activation | (79) |
| miR-221 | Multiple targets | Pre-B cells | B cell development | (146) |
| miR-34a | Foxp1 | B cell precursors | B cell development | (68) |
| miR-326 | Ets-1 | TH17 | Promote TH17 differentiation | (147) |
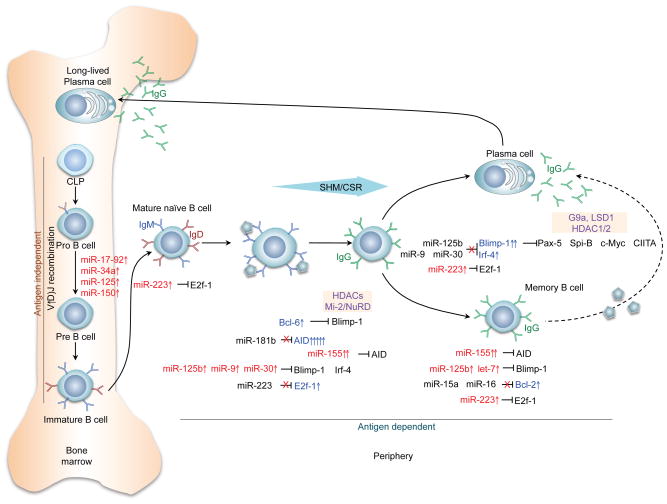
Figure 2. miRNA in immune and autoimmune responses.
Select miRNAs specifically regulate genes that are critical at different stages of peripheral B cell differentiation. miRNAs play an important role in gene regulation through direct targeting of the 3′ UTR transcripts of those genes, thereby regulating sequential B cell differentiation stages. Naïve mature B cells in peripheral lymphoid organs are activated by primary stimuli to undergo SHM and CSR (e.g., to IgG, depicted) and to differentiate into plasma cells or memory B cells. Several miRNAs (miR-17-92, miR-34a, miR-125, and miR-150) are involved in the pro-B to pre-B cell transition and regulate the generation of B cells in the bone marrow during the early stages of development. Other miRNAs, miR-181b and miR-155 target AID, while miR-125b, miR-9, and miR-30 target Blimp-1 and Irf4, which mediate plasma cell differentiation. Upregulation (red arrow) or downregulation (black arrow) of miRNAs results in alterations in the expression of these key genes. E2f-1, E2 promoter binding factor-1; AID, activation-induced cytidine deaminase; Blimp-1, B lymphocyte-induced maturation protein-1; HDAC, histone deacetylase; Irf4, interferon regulatory factor 4; NuRD, nucleosome remodeling and histone deacetylase; Pax-5, Paired box protein-5; Spi-B, spleen focus-forming virus integration site; CIITA, major histocompatibility complex class II transactivator.
During primary antibody diversification, B cell differentiation occurs in the bone marrow via somatic DNA rearrangement of immunoglobulin (Ig) genes by V(D)J (V, variable; D, diversity; J, joining) recombination (63), thereby ensuring that the developing B cell will express a B cell receptor (BCR) for antigens with unique specificities. In this process, developing B cells in the bone marrow assemble a DHJH and VHDHJH joint in progenitor B (pro-B) cells from the germline IgH locus. If the pro-B cell produces a VHDHJH rearrangement with a correct reading frame, an IgH chain of class μ (Igμ) will be expressed together with surrogate light chains λ5 and VpreB, as the surface receptor complex identified as pre-BCR. A phase of vigorous clonal expansion follows; the resulting precursor B (pre-B) cells undergo subsequent rearrangement of VL and JL gene segments in the IgL locus, give rise to immature B cells with a BCR. These express a BCR at a high Igμ/Igδ ratio, and are ready to leave the bone marrow to colonize peripheral secondary lymphoid organs.
Transition of Immature B cells into peripheral lymphoid organs, such as the spleen, lymph nodes and Peyer’s patches, for terminal differentiation. During this transition, the B cells undergo multiple intermediate stages, eventually giving rise to either follicular (FO) or marginal zone (MZ) B cells. The two B cell subsets differ in that the FO B cells are localized in B cell follicles and can freely recirculate, whereas MZ B cells reside within the MZ of the spleen (64). The fate decision between the two subsets have been linked to BCR signaling, but it has also been shown that B cells with autoreactive BCRs could be driven towards a MZ fate (65). When a FO B cell is engaged by antigen, it undergoes a germinal center (GC) reaction involving SHM and CSR. SHM inserts mainly point-mutations into the Ig variable region loci, providing the antibody with a structural substrate for the selection of higher affinity antibody mutants. CSR replaces the CH exon cluster with distinct constant heavy-chain (CH) regions in the Ig heavy chain locus, thereby generating class-switched antibodies with different biological effector functions without altering the antigen-binding variable region. The initiation of SHM and CSR are mediated by activation-induced cytidine deaminase (AID, encoded by Aicda).
Ablation of Dicer during early B cell development in mb1-Creki/+ Dicerfl/fl mice led to a dramatic block of B cell differentiation at the pro-B to pre-B transition, suggesting that miRNAs are critical for B cell development (66). In these mice, the miR-17-92 cluster is critically downregulated in both pro- and pre-B cells. Germline deletion of miR-17-92 also results in a block of pro-B to pre-B transition, suggesting that miR-17-92 plays a critical role in B cell development (67). Accordingly, Bim, a proapoptotic gene known to be involved in lymphocyte development and enriched with the miR-17-92 complementary seed sequence in the 3′ UTR, is highly upregulated in both Dicer-deficient and miR-17-92 deficient mice (67). B cell development could be partially rescued by ablation of Bim or transgenic expression of the prosurvival protein Bcl-2, indicating a critical role of miR-17-92 in regulating B cell development and Bim-induced apoptosis. The pro- to pre-B cell transition in B cell development is also regulated by other miRNAs, including miR-34a, miR-125 and miR-150. Constitutive expression of miR-34a led to a block in B cell development at the pro-B to pre-B cell transition, leading to a reduction in mature B cells (68). This block is mediated primarily by inhibited expression of the transcription factor Foxp1. miR-125b targets the Lin28A 3′UTR to upregulate the number of common myeloid progenitors while inhibit the development of pre-B cells. miR-150 modulates B-cell differentiation and function through the regulation of c-Myb (69), a transcription factor controlling multiple steps of lymphocyte development. Similarly, B cell development can also be regulated by miR-181a, which targets Bim1, Lin28A and Mcl-1.
Dicer, and therefore, miRNA deletion at later stages of B cell development (using CD19-Cre) showed that miRNAs also specifically promote the generation of FO B cells, while transitional and MZ B cell numbers were increased. Dicer deletion at this stage also resulted in a skewed BCR repertoire with hallmarks of autoreactivity correlated with high titers of autoantibodies and autoimmune features in females, suggesting that miRNAs prevent the generation of autoantibodies (70).
GCs are specialized structures that form within secondary lymphoid organs during T-dependent antibody responses. They are the functional site of antigen-specific B cell proliferation and selection events that lead to robust high-affinity antibody responses and B cell memory. miRNAs modulate antibody response, partially by mediating GC B cell formation. Aicda-Cre Dicerflox/flox mice, in which Dicer was selectively deleted in (Aicda expressing) active B cell, fail to produce high-affinity class-switched antibodies and generate memory B and long-lived plasma cells in response to T–dependent antigens (71). In the absence of Dicer, GC B cell formation and memory B cell as well as plasma cell differentiations, are significantly compromised as a result of defects in cell proliferation and survival. Dicer-deficient GC B cells express higher levels of cell cycle inhibitor genes and the proapoptotic protein Bim. Ablation of Bim could partially rescue the defect in GC B cell formation in Dicer-deficient mice. These findings reveal that miRNAs play a key regulatory role in controlling the expression levels of proteins, both temporally and spatially, to maintain proper functionality of the immune response.
miR-155 is one of the most important miRNAs in function of the immune system. It is expressed in a variety of immune cell types, including B cells, T cells, macrophages, dendritic cells, progenitor/stem cell populations and plays an important role in hematopoiesis (72). miR-155 expression is rapidly upregulated by a variety of immune stimuli, such as antigen, TLR ligands, and inflammatory cytokines, in most of these cells types. miR-155 is highly expressed in GC B cells, and miR-155-deficient and miR-155-overexpressing mice revealed important roles for this miRNA in several aspects of the GC response, including GC size and number, CSR, SHM, affinity maturation, antibody production, and cytokine production by Th cells. Defective GC formation and CSR has been observed in miR-155 knockout mice following infection or vaccination.
The intrinsic requirement of miR-155 in B cells for a fully functional extrafollicular and GC response is suggested to be due to the targeting of the transcription factor purine-rich box-1 (PU.1, encoded by Sfpi1) and the SH2 domain-containing inositol 5′-phosphatase 1 (SHIP-1). miR-155 directly targets PU.1 and SHIP-1 3′UTRs in activated B cells. PU.1 has been suggested to play a role in early B cell differentiation and in the BCR signaling pathway. Overexpression of PU.1 in B cells impairs CSR induction and plasma cell differentiation. SHIP-1 negatively regulates BCR-mediated B cell activation and proliferation.
Antibody and autoantibody responses are critically modulated by AID. AID initiates CSR and SHM by catalyzing targeted deamination of deoxycytidine (dC) residues in DNA resulting in dU:dG mismatches, which are processed into point-mutations in SHM or double-strand breaks (DSBs) in CSR (4, 73–75). As a potent DNA mutator, AID expression must be tightly regulated at transcriptional and post-transcriptional levels (76). AID is another important direct target of miR-155. miR-155 is the top miRNA predicted to target AID by miRNA target-prediction algorithms (30). It contributes to the repression of AID in naïve B cells and in B cells that have completed CSR and SHM. B cells from the knock-in mice, in which the conserved miR-155 binding site in the 3′ UTR of Aicda was mutated, or B cells from a transgenic mice with mutated miR-155-binding site, display an increased Aicda expression, demonstrating that miR-155 directly targets AID (29, 30). Accordingly, these B cells showed increased CSR and c-Myc/IgH translocation upon stimulation of LPS plus IL-4, a stimulus that induces AID expression and CSR to IgG1 (29). The miR-155-binding site mutation also results in increased AID expression in vivo and higher levels of CSR to IgG3, IgG1 and IgG2a in B cells upon stimulation with LPS, LPS and IL-4, or LPS and IFN-γ, respectively (30). In addition, the dysregulated expression of AID in these mice was associated with impaired affinity maturation and increased the mutation rate in an off-target gene (30). As miR-155 promotes the GC B cell response, it is interesting that, at the same time, it inhibits AID. In human B cells, BCL6, a transcriptional repressor which has emerged as a critical GC B cell regulator, inhibits the expression of several miRNAs, including miR-155 and miR-361, another AID-targeting miRNA (77). The coordinated spatiotemporal regulation of BCL6, miR-155 and miR-361 expression in GC B cells, suggests a coordinated activity of BCL6 in sustaining high levels of AID expression in GC B cells undergoing SHM and CSR of their Ig genes.
AID is also downregulated by other miRNAs, including miR-181b and miR-93 (human B cells only), that target different sites in the 3′ UTR of Aicda transcripts (31, 76, 78). miR-181b expression is downregulated upon B-cell activation. AID and miR-181b have complementary expression profiles, with AID levels low in resting B cells and increasing sharply in CSR-induced B cells, whereas miR-181b expression is highest in non-stimulated B cells and drops upon stimulation, suggesting that high expression of miR-181b prevents the accumulation of AID transcripts (31). Like miR-155, miR-181b also impairs CSR in activated B cells by targeting AID. Overexpression of miR-181b in activated B cells greatly reduced CSR (31). Unlike miR-181b, which is downregulated in B cells undergoing CSR, miR-210 is upregulated in activated B cells (79). miR-210 is under the control of the transcription factor, Oct-2. Although there is no single gene dramatically dysregulated in miR-210 knockout B cells, it negatively regulates CSR and prevents autoimmunity. In the mice, miR-210-deficiency led to spontaneous production of high levels of autoantibodies.
Many other miRNAs have also been suggested to regulate different aspects of mature B cell biology, such as GC response and plasma cell differentiation. miR-150 is highly expressed in naïve B cells and limits the magnitude of the GC response (33, 80). In addition to its role in B cell development, miR-150 is also involved in mature B cell function (69). miR-150-deficiency led to enhanced expression of c-Myb in GC B cells and increased secretion of antigen-specific antibodies in response to a T-dependent antigen. In contrast, overexpression of miR-125b, which is specifically expressed in a subset of GC B cells, promotes GC B cell differentiation by directly targeting and inhibiting B lymphocyte-induced maturation protein-1 (Blimp-1, encoded by Prdm1) and interferon regulatory factor-4 (Irf4), two transcription factors that drive post-GC plasma cell differentiation (81–83). Overexpression of miR-125b in B cells inhibited plasma cell differentiation and Ig secretion in vitro (82).
Specific deletions of Dicer in the T cell lineage also result in defective T cell development and irregular helper T cell differentiation and cytokine production (84–86). miRNAs are necessary for proper T cell function (86). Treg are important for preventing autoimmunity. The generation and function of Tregs is reliant on the Dicer-dependent miRNA biosynthesis pathway (86, 87). Mice that have a conditional deletion of Dicer or Drosha in Treg cells show an early onset of autoimmunity, similar to that caused by inactivation of Foxp3, the master regulator of Treg differentiation (84, 86, 88). In these mice, Dicer-deficient Treg cells completely lose suppressor capacity (86). Foxp3 regulates the expression of miR-155 in Tregs and deficiency of miR-155 results in decreased proportions, proliferation and viability of Treg cells (87). In T cells, miR-155 promotes differentiation toward the Th1 subset. In dendritic cells, miR-155 is necessary for proper activation of responder T cells in the context of antigen presentation. Likewise, miR-146a-deficient hematopoietic cells failed to rescue Foxp3-deficient T-cell-mediated autoimmunity (89).
miR-146a plays a role in the pathogenesis of lupus. It represses the function of IFN (type one interferon) and TLR signaling, both of which are important for SLE, by repressing target genes such as Traf6/Irak1, Stat1 and Tlr7 or Tlr9 (90, 91). In mice, miR-146a is expressed predominantly in immune tissues, and its expression can be induced in immune cells upon cell activation or maturation (19). Ablation of miR-146a expression in mice results in several immune-related phenotypes, correlating well with its localization of expression. Deficiency of miR-146a results in hyper-responsiveness of macrophages to bacterial LPSs and leads to an exaggerated inflammatory response in endotoxin-challenged mice. In contrast, overexpression of miR-146a in monocytes has the opposite effect. Aged miR-146a deficient mice develop a severe lupus-like autoimmune disorder characterized by splenomegaly, lymphadenopathy, multi-organ inflammation, and greatly increased autoantibody production. These mice produce about 60 fold higher amounts of anti-dsDNA autoantibodies than their wild type littermates. The autoimmune phenotype in miR-146a deficient mice is consistent with the finding of elevated amounts of activated T cells in the periphery, but may also be dependent on increased activation of B cells (19). Thus, miRNAs play pivotal roles in the regulation of both cell development and function in the innate and adaptive immune systems. miRNA dysregulation can contribute to dysregulation of the immune system and autoimmunity.
Dysregulation of miRNA in lupus
miRNAs have emerged as crucial mediators of a variety of human diseases, including autoimmune diseases (13). They play a key role in fine-tuning the function of different types of cells to modulate the immune response. Dysregulation of miRNAs has been associated with autoimmune traits (92–94). Aberrantly expressed miRNAs have been observed in different cell types and tissues and play an important role in the progression of lupus (Table 2).
Table 2.
miRNAs involved in lupus
| miRNAs | Target gene | Cells | Functions | Expression in lupus | Correlation with lupus activity | Ref. |
|---|---|---|---|---|---|---|
| miR-146a | Traf6, Irak1 | PBMC, CD4+ T cells, Th17 cells | Negatively regulates NFKB | ↓ | Inverse | (148) |
| miR-23b | IL-17 | Suppresses autoimmune inflammation | ↓ | Inverse | (20) | |
| Blimp1 | Regulates autoantibody production | Zan, et al, unpublished | ||||
| miR-125a | KLF13 | T cells | Inhibit inflammatory chemokine RANTES | ↓ | Inverse | (149) |
| miR-21 | DNMT1 | CD4+ T cells | Induces DNA hypomethylation and the expression autoimmune-associated genes | ↑ | Positive | (114) |
| miR-126 | DNMT1 | Induces DNA hypomethylation and the expression autoimmune-associated genes | ↑ | Positive | (115) | |
| miR-142 | SAP, CD84, IL-10 | T cells | Inhibit T cell activity | ↓ | Inverse | (93) |
| miR-148a | DNMT1 | CD4+ T cells | Regulates DNA methylation in lupus T cells | ↑ | Positive | (114) |
| miR-150 | c-Myb | B cells, CD4+ T cells, Th1 cells | Inhibit B/T cell activation and proliferation | ↓ | Inverse | (97) |
| miR-17-92 | Bim | T cells | Upregulation leads to abnormal B cell activation, enhanced GC responses, and abnormal inflammatory T cell production | ↑ | Positive | (97) |
| miR-155 | Aicda | B cells, Treg cells | Inhibit CSR/SHM | ↑ | Negative | (76) |
| CD62L | Regulate Treg phenotype | (87) | ||||
| SHIP1 | B cells | Regulate B cell activation and survival | Positive | (113) | ||
| miR-182-96-183 |
MITF Foxo1 |
B cells, T cells | B cell activation T cell activation |
↑ | Positive | (97) |
B cells contribute to autoimmune diseases, mainly through their primary function of antibody production. Mice with a conditional deletion of Dicer in mature B cells develop abnormal B cell subsets, have high autoantibody titers, and develop autoimmune disease with end-organ damage. This phenotype is particularly strong in female mice. Similarly, B cell-specific Dicer deletion, which abolishes mature miRNAs, results in a skewed BCR repertoire with hallmarks of autoreactivity, suggesting that select miRNAs in B cells prevent the generation of self-reactive antibodies (66). The effect of miRNA on B cell homeostasis is directed towards preventing the generation of autoreactive antibodies (30, 70). Dicer-deficient B lymphocytes produced increased numbers of autoreactive antibodies, suggesting that miRNAs play a role in terminal B cell differentiation and the establishment of B cell tolerance.
Several miRNAs are upregulated or downregulated in autoimmune diseases, such as lupus (92, 95–100) and rheumatoid arthritis (101–104). miR-150 expression is downregulated in B cells from lupus-prone MRL/Faslpr/lpr mice. This likely stems from decreased histone acetyltransferase activity, resulting in impaired acetylation and decreased transcription of the miR-150 host gene (97). A B cell-specific conditional knock-in dominant negative p300 acetyltransferase mutant results in the production of class-switched anti-dsDNA autoantibodies and the development of lupus-like traits in C57BL/6 129Sv F1 mice (105). By contrast, increased expression of certain miRNAs may contribute to the development of autoimmunity, as suggested by increased numbers of germinal center B cells in the spleen and peripheral lymph nodes in miR-17-92 transgenic mice (17) upon silencing of miR-21 (106). Therefore, dysregulated expression of miRNAs can contribute to the pathogenesis of autoimmune diseases.
The innate immune system is involved in the pathogenesis of systemic autoimmune diseases, such as lupus. Nucleic acid-containing immune complexes activate the innate response by engaging specific TLRs and promoting autoantibody generation (Figure 3). TLR signaling is tightly regulated to avoid excessive inflammation. Numerous miRNAs are induced by TLR activation in innate immune cells. These and other miRNAs target the 3′ UTRs of mRNAs encoding the TLRs or components of the TLR signaling system. miRNAs also prove to be an important link between the innate and adaptive immune systems and their dysregulation might have a role in the pathogenesis of autoimmune diseases. CSR, SHM and plasma cell differentiation can be induced by TLR signaling, which induces expression of not only proteins but also multiple miRNAs, including miR-155, miR-146a, and miR-21. These miRNAs undergo two paradigms of function: the pro-inflammatory miRNAs, e.g., miR-155, precisely regulate the levels of their targets to promote the immune response, while the negative feedback regulators, typified by miR-146a and miR-21, mute the immune response.
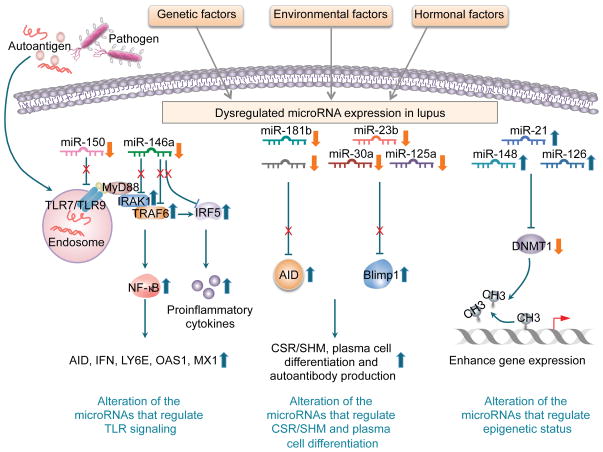
Figure 3. Modulation of miRNA in lupus.
Genetic, environmental and hormonal factors all contribute to miRNA dysregulation in lupus. For example, miR-146a contributes to lupus pathogenesis by repressing the function of type I IFN and TLR signaling, through downregulating target genes Traf6/Irak1, and Tlr7 or Tlr9 and inflammatory cytokine expression. miR-181 impairs CSR in activated B cells by targeting AID, whereas miR-23b, miR-30a, miR-125a regulate autoantibody production by repressing Blimp-1 expression and plasma cell differentiation. In lupus, miR-21, miR-148 and miR-126, which target Dnmt1, are all upregulated suggesting that hypomethylation in lupus is mediated by those miRNAs. Conversely, histone modifications and DNA methylation also affect miRNAs in lupus T cells.
miR-146a plays a dominant role in innate immunity, as it negatively regulates signal transduction pathways leading to NF-κB activation by targeting Traf6 and Irak1 (19), and is downregulated in association with lupus pathogenesis (99, 107). Decreased miR-146a expression leads to hyperactivation of type I interferon pathway that is a hallmark of SLE (99). Genome-wide studies have identified specific genetic variants in the promoter or intergenic region of miR-146a that account for miR-146a down-regulation and could be responsible for the genetic susceptibility of lupus (108). By contrast, miR-146a is consistently upregulated in RA (101), where it negatively regulates pro-inflammatory cytokines such as TNF-α (109) and IL-17 (110). Deficiency of miR-146a results in autoimmunity, including the production of anti-dsDNA autoantibodies (19). In addition, miR-23b is also downregulated in patients with lupus or RA, as well as mouse models of these autoimmune diseases (20). While miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α, it also regulates autoantibody production by repressing Blimp-1 expression, as the 3′UTR of Prdm1 transcripts contains two putative evolutionally conserved miR-23b target sites.
TLR7 and TLR9 have been suggested to play pathogenic and protective roles, respectively, in the development of lupus. In lupus-prone B6.Nba2 mice, enhanced disease is associated with functionally upregulated expression of TLR7, as documented by an increased TLR7-dependent activation of B cells and plasmacytoid dendritic cells. In lupus-prone yaa mice, pathogenic autoimmunity is associated with a translocation of Tlr7 from the X chromosome onto the yaa Y chromosome. This translocation results in a twofold increased Tlr7 expression in male mice bearing yaa, which is sufficient to dysregulate TLR7-mediated activation of innate immune responses (111). In humans, the G or C allele of rs3853839 at the 3′UTR of TLR7 mRNA is associated with elevated expression of TLR7 transcripts and protein leading to increased risk for lupus. It has been shown recently that TLR7 3′UTR sequence bearing the G/C allele of rs3853839 matches a predicted binding site of miR-3148, suggesting that this miRNA may regulate TLR7 expression. Indeed, miR-3148 levels were inversely correlated with TLR7 transcript levels in peripheral blood mononuclear cells (PBMCs) from SLE patients (112). Further, the 3′UTR of human and mouse TLR7/Tlr7 likely contains target sites of miR-150, which is downregulated in lupus B cells (Zan, et al, unpublished).
As mentioned earlier, miR-155 regulates antibody responses and subsequent B cell effector functions to exogenous antigens. A recent study showed that ablation of miR-155 in lupus prone B6 faslpr/lpr mice reduced autoantibody responses, accompanied by a decrease in serum IgG but not IgM anti-dsDNA antibodies and a reduction of kidney inflammation (113). miR-155 deletion in B6 faslpr/lpr B cells restored the reduced SH2 domain-containing inositol 5′-phosphatase 1 to normal levels. In addition, co-aggregation of the Fcγ receptor IIB with the B-cell receptor in miR-155−/−B6 faslpr/lpr B cells resulted in decreased ERK activation, proliferation and production of switched antibodies compared with miR-155 sufficient faslpr/lpr B cells. Thus, by controlling the levels of SH2 domain-containing inositol 5′-phosphatase 1, miR-155 in part maintains an activation threshold that allows B cells to respond to antigens.
Different epigenetic marks can undergo reciprocal regulation in both the development and disease activity of lupus. DNA methylation occurs at the 5′ position of cytosine in the context of CpG dinucleotides. CpG methylation may result in transcriptional repression. T cells from patients with active lupus display global DNA hypomethylation. The reduced DNA methylation in lupus is associated with decreased enzymatic activity of DNA methyltransferases (DNMTs). In lupus patients and lupus mice, miR-21, miR-126 and miR-148a, which target Dnmt1, are all upregulated (106, 114, 115), suggesting that hypomethylation in lupus is at least partially mediated by these miRNAs. Conversely, histone modifications and DNA methylation also affect miRNAs in lupus T cells. Lymphocyte-specific miRNAs are either tightly controlled by polycomb group-mediated H3K27me3 or maintained in a semi-activated epigenetic state prior to full expression (33). In lupus CD4+ T cells, miR-142-3p and miR-142-5p are decreased, and result in overexpression of the SLAM-associated protein (SAP), IL-10, and CD84 (93). miR-142 reduction is associated with increased H3K27 methylation levels in the putative miR-142 regulatory regions (93). The three CpG pairs closest to the miR-142 transcription start site have been also found to be hypermethylated. Further, treatment with HDAC inhibitors downregulates the level of DNMTs, alters DNMT1 nuclear dynamics and its interactions with chromatin, and reduces overall DNA methylation (116). Thus, reduced DNMTs in lupus could be from the increased levels of miRNAs that target Dnmt genes.
Environmental and hormonal regulation of miRNAs in lupus
Lupus is caused by a combination of genetic and environmental factors (Figure 3). Environmental triggers include ultraviolet (UV) exposure, bacterial and viral infections, antibiotics and other medications, diet, and stress. Gene expression can be altered in response to such environmental factors. These changes can be modulated by specific miRNAs. For example, air pollution metal-rich particulate matter (PM) greatly enhances the expression of miR-21 (117), which was found to be upregulated in lupus, and strongly correlated with disease activity (98). UV light triggers lupus flares and can promote DNA hypomethylation in T cells and lead to self-immune reaction (118). The reduced DNA methylation can alter the expression of mRNAs and miRNAs. Indeed, alteration of miRNAs has been found in cells exposed to UV (119).
In humans and most animal models of autoimmunity, females show a much higher incidence of disease. Women are ten times more likely to develop lupus than men, predominantly during their childbearing years, consistent with a role of the female hormone estrogen in the development of lupus autoantibodies. Upon estrogenic activation, the estrogen receptors alpha and beta (ERα and ERβ) regulate the transcription of target genes by directly binding to their DNA. miRNAs modulated by ER have the potential to fine tune these regulatory systems and provide an alternate mechanism that could impact estrogen-dependent developmental and pathological systems. Sex hormones, such as 17β-estradiol (E2), the most abundant and potent natural estrogen in vertebrates, suppress or enhance miRNA expression in many cell types by inducing ER-binding to miRNA host gene promoters. In addition, activated ERα attenuates the processing of specific primary miRNAs into pre-miRNAs through estrogen-dependent association with the Drosha complex, resulting in an increased stabilization of the miRNA-targeting transcript. This suggests that estrogen can post-transcriptionally control gene expression through regulation of the miRNAs, which target the 3′UTRs of these genes (120, 121).
Estrogen promotes the secretion of pro-inflammatory cytokines such as IFN-γ and IL-17, which are involved in lupus pathogenesis, either through the modification of key transcription factors in inflammation or through the regulation of miRNA expression. A negative TLR signaling regulator miR-146a, which, as discussed above, is downregulated in lupus, was decreased in freshly isolated splenic lymphocytes from mice treated with estrogen (122). The estrogen-mediated reduction of miR-146a results in increased LPS-induced IFN-γ and iNOS expression. Estrogen promotes AID expression that leads to increased antibody and autoantibody responses in females (76, 123). In addition to enhancing transcription of the Aicda gene by activating the promoter of HOXC4/HoxC4, which is important for Aicda induction (76, 123, 124), estrogen can modulate AID expression by regulating miRNAs, specifically miRNA-155 and miRNA-181b, which target the 3′UTR of Aicda mRNA. Estrogen-ERα has been shown to inhibit miRNA-155 and miRNA-181b expression in human breast cancer cells (125, 126). It has also been shown to modulate the expression of miR-181a, miR-125a and miR-21 (120, 125, 127).
Similarly, the vitamin A metabolite, all-trans-retinoic acid (ATRA), belongs to a class of retinoids that exert immunomodulatory and anti-inflammatory functions, and can suppress the development of lupus nephritis (128, 129). The effect of retinoids is mediated by nuclear retinoid receptors (RARs and RXRs), which are ligand-activated transcription factors that are able to bind to retinoic acid response elements (RAREs) located in the promoter regions of target genes. Interestingly, RAR and ERα binding sites are highly coincident, resulting in a widespread crosstalk of RA and estrogen signaling. Retinoic acid and estrogen exert antagonistic regulations on transcription of not only coding genes, but also the miRNA host genes (130). For example, miR-23a is reduced in patients with lupus and downregulated by estrogen, but upregulated by retinoic acid. Conversely, estrogen increases expression of the miR-17-92 cluster, which is positively involved in lupus, yet reduced by retinoic acid (130).
Although sex differences in immune and autoimmune responses are largely due to the direct influence of estrogen, a number of recent reports suggest the potential impact of dosage effects of genes or miRNAs encoded on the X chromosome (131, 132). miRNAs are disproportionately localized to the X chromosome, where 10% of all human and mouse miRNAs are located, while the Y chromosome has no identifiable miRNAs. The “double” set of miRNAs encoded by the two X chromosomes in female may together with higher estrogen levels contribute to the strong female predisposition to autoimmune diseases, particularly lupus.
miRNAs as therapeutic targets in lupus
The immune system integrates miRNAs into the complex network of gene regulation necessary for immunity. The role of miRNAs in immune responses and their dysregulation in immunopathogenesis of lupus and autoimmunity make these miRNAs potentially important therapeutic targets. miRNAs that have been demonstrated to be necessary for normal lymphocyte function could be antagonized to produce a more specific and moderate immune suppression. The ability to target multiple functionally related protein-coding genes and pathways also make miRNA-based therapeutics a powerful tool for the treatment of lupus.
Based on the biological functions of individual miRNAs, it is possible to either inhibit or overexpress miRNAs to modulate immune functions. Specific miRNAs can be administered to downregulate target genes, while blocking others can be utilized as a strategy to increase expression of target genes. Inhibition of specific miRNA can be performed using antagomirs, chemically engineered antisense oligonucleotides targeting the miRNA guide strand to block the interaction between the miRNA recognition elements within the 3′-UTR of the target mRNA genes. Antagomirs can be utilized to correct and stabilize miRNA levels to normal expression levels or increase expression of genes that suppress autoimmune diseases (133). Another approach utilized to antagonize miRNAs is seed region targeting locked nucleic acid (LNA) oligonucleotides, which inhibit miRNAs that share the same seed sequence (134). For example, inhibition of overexpressed miR-21 by LNA in lupus-prone B6.Sle1,2,3 mice led to amelioration of autoimmune splenomegaly (106). The first miRNA-targeting drug, Miravirsen, entered phase 2 clinical trials. Miravirsen is an LNA-antagomir that specifically inhibits hsa-miR-122, a liver-specific miRNA that the hepatitis C virus (HCV) requires for its replication. To overexpress miRNAs, the cells are transfected with synthesized miRNAs (miRNA mimics) or plasmids or viral vectors that express pre-miRNAs (23). Lastly, epigenetic modification of dysregulated genes or sequences that upregulate miRNA which modulate expression of these genes can also be potential therapeutic targets in autoimmune diseases (135), although such approach is still premature.
Concluding Remarks
miRNAs function as key modulators in gene expression and play an important role in the immune response. Proper regulation of miRNA expression is crucial for disease prevention and aberrant miRNA expression in the immune system would result in disease states, including lupus. Although miRNAs constitute only a small portion of total human genes, it is known that miRNAs are capable of regulating over 40% of human mRNAs encoding immune genes including key targets in B cell and autoantibody responses, such as Aicda (AID), Prdm1 (Blimp1), Smad7, and Bcl6 (34, 136). The identification and characterization of the role miRNAs play in pathogenesis of lupus need to be further elucidated. Determining how given therapeutics alters pathogenic miRNAs would provide a viable screening tool for specific, targeted therapy in lupus. The ability of miRNAs to combinatorially regulate key mRNAs containing multiple miRNA-binding sites in their 3′ UTR may lead to an even greater level of target mRNA repression than that mediated by a single miRNA, and could have important physiological and disease relevance. The fact that a single miRNA can repress multiple target mRNAs simultaneously may reveal new gene networks. Overall, a better understanding of miRNAs that are implicated in lupus pathogenesis may further our knowledge of the molecular mechanisms underlying autoimmune disease and lead to the development of potential therapies.
Acknowledgments
We apologize that owing to space limitations only a fraction of the relevant literature was cited in this review article. This work was supported by the Arthritis National Research Foundation grant (ANRF 55384) to Hong Zan, the U.S. National Institutes of Health grants AI 105813, AI 079705 and AI 060573, and the Alliance for Lupus Research Target Identification in Lupus Grant (ALR 295955) to Paolo Casali.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Zan H, Zhang J, Ardeshna S, Xu Z, Park S-R, Casali P. Lupus-prone MRL/Faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: Concurrent upregulation of somatic hypermutation and class switch DNA recombination. Autoimmunity. 2009;42:89–103. doi: 10.1080/08916930802629554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White CA, Seth Hawkins J, Pone EJ, Yu ES, Al-Qahtani A, Mai T, Zan H, Casali P. AID dysregulation in lupus-prone MRL/Faslpr/lpr mice increases class switch DNA recombination and promotes interchromosomal c-Myc/IgH loci translocations: modulation by HoxC4. Autoimmunity. 2011;44:585–598. doi: 10.3109/08916934.2011.577128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Zan H, Xu Z, Casali P. Epigenetics of the antibody response. Trends Immunol. 2013;34:460–470. doi: 10.1016/j.it.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, White CA, Lam T, Pone EJ, Tran DC, Hayama KL, Zan H, Xu Z, Casali P. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 2013;5:702–714. doi: 10.1016/j.celrep.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow MK. Collaboration, genetic associations, and lupus erythematosus. N Engl J Med. 2008;358:956–961. doi: 10.1056/NEJMe0800096. [DOI] [PubMed] [Google Scholar]
- 8.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6:461–479. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia BA, Busby SA, Shabanowitz J, Hunt DF, Mishra N. Resetting the epigenetic histone code in the MRL-lpr/lpr mouse model of lupus by histone deacetylase inhibition. J Proteome Res. 2005;4:2032–2042. doi: 10.1021/pr050188r. [DOI] [PubMed] [Google Scholar]
- 11.Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol. 2011;7:263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- 12.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Trans Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes T, Sawalha AH. The role of epigenetic variation in the pathogenesis of systemic lupus erythematosus. Arthritis Res Ther. 2011;13:245, 241–211. doi: 10.1186/ar3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 19.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, Shi Y, Harley JB, Shen N, Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 2012;18:1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 25.Ruegger S, Grosshans H. MicroRNA turnover: when, how, and why. Trends Biochem Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Jolly CJ, Neuberger MS. Somatic hypermutation of immunoglobulin kappa transgenes: association of mutability with demethylation. Immunol Cell Biol. 2001;79:18–22. doi: 10.1046/j.1440-1711.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- 28.Fraenkel S, Mostoslavsky R, Novobrantseva TI, Pelanda R, Chaudhuri J, Esposito G, Jung S, Alt FW, Rajewsky K, Cedar H, Bergman Y. Allelic ‘choice’ governs somatic hypermutation in vivo at the immunoglobulin kappa-chain locus. Nat Immunol. 2007;8:715–722. doi: 10.1038/ni1476. [DOI] [PubMed] [Google Scholar]
- 29.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Yébenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea J, Rajewsky K, Casellas R. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Yebenes VG, Bartolome-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol Rev. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 36.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 40.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 41.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 42.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 43.Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 44.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 45.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 46.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 47.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernando TR, Rodriguez-Malave NI, Rao DS. MicroRNAs in B cell development and malignancy. J Hematol Oncol. 2012;5:7. doi: 10.1186/1756-8722-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. Rna. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 60.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ansel KM. RNA regulation of the immune system. Immunol Rev. 2013;253:5–11. doi: 10.1111/imr.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danger R, Braza F, Giral M, Soulillou JP, Brouard S. MicroRNAs, major players in B cells homeostasis and function. Front Immunol. 2014;5:98. doi: 10.3389/fimmu.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 64.Garraud O, Borhis G, Badr G, Degrelle S, Pozzetto B, Cognasse F, Richard Y. Revisiting the B-cell compartment in mouse and humans: more than one B-cell subset exists in the marginal zone and beyond. BMC Immunol. 2012;13:63. doi: 10.1186/1471-2172-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 66.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 67.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 70.Belver L, de Yébenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 72.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casali P, Zan H. Class switching and Myc translocation: how does DNA break? Nat Immunol. 2004;5:1101–1103. doi: 10.1038/ni1104-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zan H, White CA, Thomas LM, Mai T, Li G, Xu Z, Zhang J, Casali P. Rev1 recruits ung to switch regions and enhances du glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012;2:1220–1232. doi: 10.1016/j.celrep.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, Dalla-Favera R. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med. 2012;209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borchert GM, Holton NW, Larson ED. Repression of human activation induced cytidine deaminase by miR-93 and miR-155. BMC Cancer. 2011;10:347. doi: 10.1186/1471-2407-11-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mok Y, Schwierzeck V, Thomas DC, Vigorito E, Rayner TF, Jarvis LB, Prosser HM, Bradley A, Withers DR, Martensson IL, Corcoran LM, Blenkiron C, Miska EA, Lyons PA, Smith KG. MiR-210 is induced by Oct-2, regulates B cells, and inhibits autoantibody production. J Immunol. 2013;191:3037–3048. doi: 10.4049/jimmunol.1301289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baumjohann D, Ansel KM. MicroRNA regulation of the germinal center response. Curr Opin Immunol. 2014;28C:6–11. doi: 10.1016/j.coi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O’Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Divekar AA, Dubey S, Gangalum PR, Singh RR. Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J Immunol. 2011;186:924–930. doi: 10.4049/jimmunol.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan EK, Ceribelli A, Satoh M. MicroRNA-146a in autoimmunity and innate immune responses. Ann Rheum Dis. 2013;72(Suppl 2):ii90–ii95. doi: 10.1136/annrheumdis-2012-202203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.So AY, Zhao JL, Baltimore D. The Yin and Yang of microRNAs: leukemia and immunity. Immunol Rev. 2013;253:129–145. doi: 10.1111/imr.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang D, Shen N. MicroRNA involvement in lupus: the beginning of a new tale. Curr Opin Rheumatol. 2012;24:489–498. doi: 10.1097/BOR.0b013e3283563363. [DOI] [PubMed] [Google Scholar]
- 93.Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, Wang Y, Yin H, Zhang P, Zhang Q, Lu Q. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2953–2963. doi: 10.1002/art.34505. [DOI] [PubMed] [Google Scholar]
- 94.Amarilyo G, La Cava A. miRNA in systemic lupus erythematosus. Clin Immunol. 2012;144:26–31. doi: 10.1016/j.clim.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 95.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 96.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, Bruner GR, Harley JB, Ojwang JO. Identification of unique microRNA signature associated with lupus nephritis. PloS one. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, Ahmed SA. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010;5:e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, Iliopoulos D, Boumpas DT. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis. 2011;70:1496–1506. doi: 10.1136/ard.2010.139857. [DOI] [PubMed] [Google Scholar]
- 99.Löfgren SE, Frostegård J, Truedsson L, Pons-Estel BA, D’Alfonso S, Witte T, Lauwerys BR, Endreffy E, Kovács L, Vasconcelos C, Martins da Silva B, Kozyrev SV, Alarcón-Riquelme ME. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13:268–274. doi: 10.1038/gene.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen N, Liang D, Tang Y, de Vries N, Tak PP. MicroRNAs--novel regulators of systemic lupus erythematosus pathogenesis. Nat Rev Rheumatol. 2012;8:701–709. doi: 10.1038/nrrheum.2012.142. [DOI] [PubMed] [Google Scholar]
- 101.Ceribelli A, Nahid MA, Satoh M, Chan EK. MicroRNAs in rheumatoid arthritis. FEBS lett. 2011;585:3667–3674. doi: 10.1016/j.febslet.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wittmann J, Jack HM. microRNAs in rheumatoid arthritis: midget RNAs with a giant impact. Ann Rheum Dis. 2011;70(Suppl 1):i92–196. doi: 10.1136/ard.2010.140152. [DOI] [PubMed] [Google Scholar]
- 103.Filkova M, Jungel A, Gay RE, Gay S. MicroRNAs in rheumatoid arthritis: potential role in diagnosis and therapy. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2012;26:131–141. doi: 10.2165/11631480-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 104.Duroux-Richard I, Jorgensen C, Apparailly F. What do microRNAs mean for rheumatoid arthritis? Arthritis Rheum. 2012;64:11–20. doi: 10.1002/art.30651. [DOI] [PubMed] [Google Scholar]
- 105.Forster N, Gallinat S, Jablonska J, Weiss S, Elsasser HP, Lutz W. p300 protein acetyltransferase activity suppresses systemic lupus erythematosus-like autoimmune disease in mice. J Immunol. 2007;178:6941–6948. doi: 10.4049/jimmunol.178.11.6941. [DOI] [PubMed] [Google Scholar]
- 106.Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, Obad S, Petri A, Kauppinen S, Kiriakidou M. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol Med. 2011;3:605–615. doi: 10.1002/emmm.201100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ceribelli A, Sato H, Chan EK. MicroRNAs and autoimmunity. Curr Opin Immunol. 2012;24:686–691. doi: 10.1016/j.coi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, Qian X, Tang Y, Lau YL, de Vries N, Tak PP, Tsao BP, Shen N. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, Suzuki O, Adachi N, Ochi M. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord. 2010;11:209. doi: 10.1186/1471-2474-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, Schultz RA, Wakeland EK. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng Y, Zhao J, Sakurai D, Kaufman KM, Edberg JC, Kimberly RP, Kamen DL, Gilkeson GS, Jacob CO, Scofield RH, Langefeld CD, Kelly JA, Ramsey-Goldman R, Petri MA, Reveille JD, Vila LM, Alarcon GS, Vyse TJ, Pons-Estel BA, Freedman BI, Gaffney PM, Sivils KM, James JA, Gregersen PK, Anaya JM, Niewold TB, Merrill JT, Criswell LA, Stevens AM, Boackle SA, Cantor RM, Chen W, Grossman JM, Hahn BH, Harley JB, Alarcomicronn-Riquelme ME, Brown EE, Tsao BP. MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 2013;9:e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thai TH, Patterson HC, Pham DH, Kis-Toth K, Kaminski DA, Tsokos GC. Deletion of microRNA-155 reduces autoantibody responses and alleviates lupus-like disease in the Fas(lpr) mouse. Proc Natl Acad Sci USA. 2013;110:20194–20199. doi: 10.1073/pnas.1317632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 115.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, Yin H, Lu Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 116.Arzenani MK, Zade AE, Ming Y, Vijverberg SJ, Zhang Z, Khan Z, Sadique S, Kallenbach L, Hu L, Vukojevic V, Ekstrom TJ. Genomic DNA hypomethylation by histone deacetylase inhibition implicates DNMT1 nuclear dynamics. Mol Cell Biol. 2011;31:4119–4128. doi: 10.1128/MCB.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect. 2010;118:763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Z, Li X, Qin H, Zhu X, Xu J, Shi W. Ultraviolet B enhances DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus via inhibiting DNMT1 catalytic activity. J Dermatol Sci. 2013;71:167–173. doi: 10.1016/j.jdermsci.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 119.Kraemer A, Chen IP, Henning S, Faust A, Volkmer B, Atkinson MJ, Moertl S, Greinert R. UVA and UVB irradiation differentially regulate microRNA expression in human primary keratinocytes. PLoS One. 2013;8:e83392. doi: 10.1371/journal.pone.0083392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O’Malley BW, Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 121.Dai R, McReynolds S, Leroith T, Heid B, Liang Z, Ahmed SA. Sex differences in the expression of lupus-associated miRNAs in splenocytes from lupus-prone NZB/WF1 mice. Biol Sex Differ. 2013;4:19. doi: 10.1186/2042-6410-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dai R, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA. Suppression of LPS-induced Interferon-gamma and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: a novel mechanism of immune modulation. Blood. 2008;112:4591–4597. doi: 10.1182/blood-2008-04-152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mai T, Zan H, Zhang J, Hawkins JS, Xu Z, Casali P. Estrogen receptors bind to and activate the promoter of the HoxC4 gene to potentiate HoxC4-mediated AID induction, immunoglobulin class-switch DNA recombination and somatic hypermutation. J Biol Chem. 2010;285:37797–37810. doi: 10.1074/jbc.M110.169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K, Sahlberg N, Kristensen VN, Borresen-Dale AL, Saviranta P, Perala M, Kallioniemi O. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 126.Zhang C, Zhao J, Deng H. 17β-estradiol up-regulates miR-155 expression and reduces TP53INP1 expression in MCF-7 breast cancer cells. Mol Cell Biochem. 2013;379:201–211. doi: 10.1007/s11010-013-1642-6. [DOI] [PubMed] [Google Scholar]
- 127.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 128.de Lema Perez G, Lucio-Cazana FJ, Molina A, Luckow B, Schmid H, de Wit C, Moreno-Manzano V, Banas B, Mampaso F, Schlondorff D. Retinoic acid treatment protects MRL/lpr lupus mice from the development of glomerular disease. Kidney Int. 2004;66:1018–1028. doi: 10.1111/j.1523-1755.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- 129.Nozaki Y, Yamagata T, Yoo BS, Sugiyama M, Ikoma S, Kinoshita K, Funauchi M, Kanamaru A. The beneficial effects of treatment with all-trans-retinoic acid plus corticosteroid on autoimmune nephritis in NZB/WF mice. Clin Exp Immunol. 2005;139:74–83. doi: 10.1111/j.1365-2249.2005.02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saumet A, Vetter G, Bouttier M, Antoine E, Roubert C, Orsetti B, Theillet C, Lecellier CH. Estrogen and retinoic acid antagonistically regulate several microRNA genes to control aerobic glycolysis in breast cancer cells. Mol Biosyst. 2012;8:3242–3253. doi: 10.1039/c2mb25298h. [DOI] [PubMed] [Google Scholar]
- 131.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 132.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 133.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 134.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Santis M, Selmi C. The therapeutic potential of epigenetics in autoimmune diseases. Clin Rev Allergy Immunol. 2012;42:92–101. doi: 10.1007/s12016-011-8293-8. [DOI] [PubMed] [Google Scholar]
- 136.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 137.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy AJ, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 139.Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry EJ, Turner M, Katsikis PD. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A, Akl H, Mourtada M, El Rifai M, Burny A, Romero P, Martiat P, Badran B. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- 143.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O’Shea JJ. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Khan AA, Penny LA, Yuzefpolskiy Y, Sarkar S, Kalia V. MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood. 2013;121:4473–4483. doi: 10.1182/blood-2012-06-435412. [DOI] [PubMed] [Google Scholar]
- 146.Knoll M, Simmons S, Bouquet C, Grun JR, Melchers F. miR-221 redirects precursor B cells to the BM and regulates their residence. Eur J Immunol. 2013;43:2497–2506. doi: 10.1002/eji.201343367. [DOI] [PubMed] [Google Scholar]
- 147.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 148.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 149.Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, Luo X, Huang X, Li J, Chen S, Shen N. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]