Abstract
Intercellular communication is critical for the survival of unicellular organisms as well as for the development and function of multicellular tissues. Cell-to-cell signaling is also required to develop the interconnected mycelial network characteristic of filamentous fungi and is a prerequisite for symbiotic and pathogenic host colonization achieved by molds. Somatic cell–cell communication and subsequent cell fusion is governed by the MAK-2 mitogen activated protein kinase (MAPK) cascade in the filamentous ascomycete model Neurospora crassa, yet the composition and mode of regulation of the MAK-2 pathway are currently unclear. In order to identify additional components involved in MAK-2 signaling we performed affinity purification experiments coupled to mass spectrometry with strains expressing functional GFP-fusion proteins of the MAPK cascade. This approach identified STE-50 as a regulatory subunit of the Ste11p homolog NRC-1 and HAM-5 as cell-communication-specific scaffold protein of the MAPK cascade. Moreover, we defined a network of proteins consisting of two Ste20-related kinases, the small GTPase RAS-2 and the adenylate cyclase capping protein CAP-1 that function upstream of the MAK-2 pathway and whose signals converge on the NRC-1/STE-50 MAP3K complex and the HAM-5 scaffold. Finally, our data suggest an involvement of the striatin interacting phosphatase and kinase (STRIPAK) complex, the casein kinase 2 heterodimer, the phospholipid flippase modulators YPK-1 and NRC-2 and motor protein-dependent vesicle trafficking in the regulation of MAK-2 pathway activity and function. Taken together, these data will have significant implications for our mechanistic understanding of MAPK signaling and for homotypic cell–cell communication in fungi and higher eukaryotes.
Author Summary
Appropriate cellular responses to external stimuli depend on the highly orchestrated activity of interconnected signaling cascades. One crucial level of control arises from the formation of discrete complexes through scaffold proteins that bind multiple components of a given pathway. Central for our understanding of these signaling platforms is the archetypical MAP kinase scaffold Ste5p, a protein that is restricted to budding yeast and close relatives. We identified HAM-5, a protein highly conserved in filamentous ascomycete fungi, as cell–cell communication-specific scaffold protein of the Neurospora crassa MAK-2 cascade (homologous to the budding yeast pheromone pathway). We also describe a network of upstream acting proteins, consisting of two Ste20-related kinases, the small G-protein RAS-2 and the adenylate cyclase capping protein CAP-1, whose signals converge on HAM-5. Our work has implications for the mechanistic understanding of MAP kinase scaffold proteins and their function during intercellular communication in eukaryotic microbes as well as higher eukaryotes.
Introduction
Intercellular communication is critical for the survival of simple unicellular organisms such as bacteria and yeasts and is central for the development and function of multicellular plant and animal systems [1]–[4]. Cell-cell signaling and somatic cell fusion is also required to develop the interconnected mycelial network characteristic of filamentous fungi [5]. This feature is important for the fitness of the fungal colony by the shared use of information, nutrients and organelles between individual cells [6], [7]. Consequently, hyphal anastomosis is critical for host colonization and symbiotic interactions as well as for virulence of pathogenic species [8]–[12]. Hyphal fusion is comparable to homotypic cell fusion between genetically identical cells of higher eukaryotes, which results in the formation of multinucleate syncytia [13], [14]. Important examples for human biology are myoblast fusion during muscle differentiation, trophoblast fusion during placental development and osteoclast fusion during bone formation. Thus, fungal self-signaling may provide a powerful model for understanding molecular mechanisms of homotypic cell communication during animal and human tissue development.
In the ascomycete model mold Neurospora crassa, an unknown chemical ligand mediates chemotropic communication between genetically identical cells. Germinating spores mutually attract each other and subsequently fuse to generate an interconnected network of multinucleate cells that form the mycelial colony [5], [15]. This process of self-signaling is based on the oscillatory recruitment of the NRC-1–MEK-2–MAK-2 mitogen activated protein kinase (MAPK) cascade (homologous to the Ste11p-Ste7p-Fus3p mating pathway in budding yeast) and of SOFT, a protein of unknown molecular function, to the opposing tips of communicating germlings [16], [17]. The rapid alternation of these two different physiological states of “homing” cells likely reflects signal response and delivery, respectively. Although these findings resulted in a first qualitative model to describe the excitable behavior of the MAK-2 module [18], our understanding of oscillatory MAK-2 signaling is hampered by the fact that most components of the signaling machinery – including the postulated secreted signal and its cognate receptor(s), regulators of the MAPK cascade as well as most MAK-2 targets – are unknown.
N. crassa and other filamentous fungi possess G-protein coupled receptors, heterotrimeric G-proteins, STE20-related kinases, components of the cAMP machinery and Ras/Rho-type GTPase modules known to function upstream or in parallel of MAPK signaling in fungi and higher eukaryotes [19]–[22]. However, mutant analyses indicate that individual deletions of these components are dispensable for vegetative cell communication (summarized in [23]). We hypothesized that redundant functions between the mentioned proteins require additional approaches to classical mutant hunts in order to dissect MAK-2 signaling. In this study, we used a proteomics approach that allowed the identification of STE-50 as regulatory subunit of the Ste11p homolog NRC-1 and HAM-5 as cell-communication-specific scaffold protein of the MAPK cascade. Moreover we defined a network of proteins, consisting of two Ste20-related kinases, the small G-protein RAS-2 and the adenylate cyclase capping protein CAP-1, whose signals converge on the MAK-2 pathway.
Results
Composition of the MAK-2 cascade
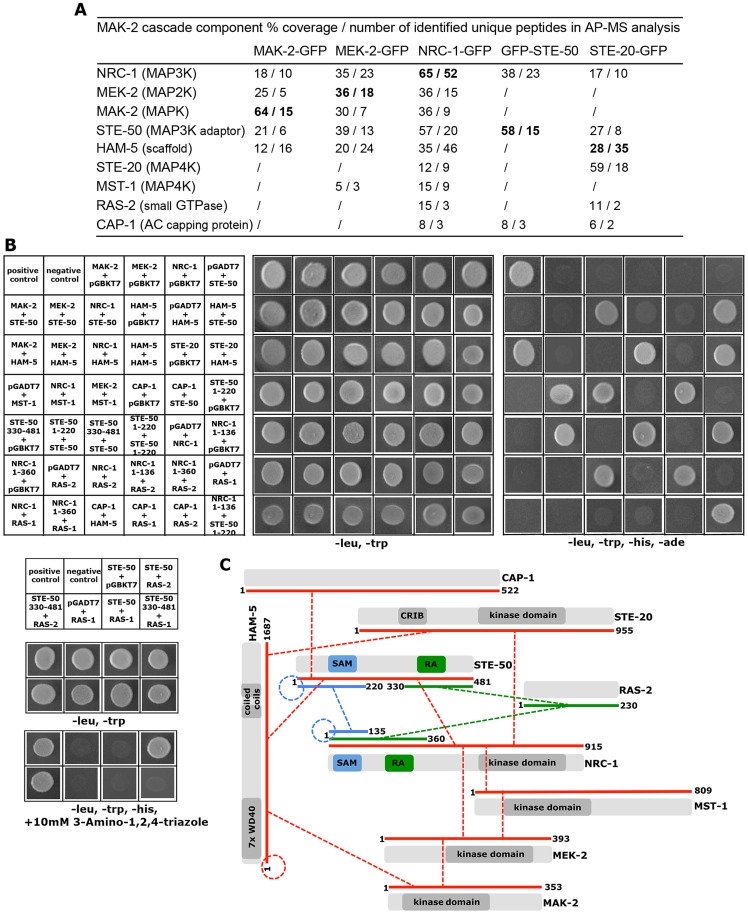
A high-quality interactome of the MAK-2 MAPK module was generated by affinity purification experiments coupled to mass spectrometry (AP-MS) with strains expressing functional GFP-fusion proteins of the three kinases of the MAK-2 cascade (Table S1). Each of the three bait proteins recovered the tripartite kinase cascade with high stringency in the two biological replicate purifications performed, indicating the suitability of the approach. Moreover, we identified two proteins, STE-50 and HAM-5, which displayed tight interactions with all three kinases (Figure 1 A).
Figure 1. Interaction network of MAK-2 pathway components.
(A) Proteins associated with the MAK-2 cascade were identified in affinity purification experiments coupled to mass spectrometry (AP-MS), and identified proteins were filtered against control purifications using GFP as bait (Table S1). Only proteins identified in two biological replicates and absent from the control data set are shown. Protein coverage by peptides and the number of identified total and unique peptides identified are given for the better of the two purifications. Bold numbers indicate the GFP-fusion protein used as bait. (B) Physical interactions between MAK-2 pathway components were mapped in yeast two-hybrid (Y2H) tests. The indicated constructs were co-expressed in strain AH109 and yeast growth was analyzed on the indicated selective media. The reciprocal Y2H assays are shown in Figure S2. (C) Summary schema of Y2H-based interactions of the indicated proteins and their domains. Color-coded lines below the protein schemas and dashed connectors indicate the used constructs and detected interactions, respectively.
The remaining hits that associated with all three kinases were only identified with poor coverage and/or in one of the two purifications and thus could constitute contaminants. However, among them were PPG-1, PP2A-A, HAM-3 and MOB-3, components of the striatin-interacting phosphatase 2A and kinase (STRIPAK) complex that is required for fungal cell-cell signaling [24]–[29]. This suggests that at least some of the additional hits may represent dynamically, and thus weakly interacting components. Thus, we assayed available mutants defective for additional candidate proteins that associated with the three kinases for tropic interaction defects and determined that the casein kinase 2 heterodimer CKA/CKB-1, the serine/threonine kinase YPK-1, type V myosin/NCU01440, and the hypothetical protein NCU06265 (designated HAM-13) were required for proper cell communication. Consistent with these defects, we also detected reduced MAK-2 phosphorylation levels in cell extracts of these five mutant strains (Table 1; Figure S1). The combined proteomics data and mutant characteristics indicate a specific involvement of these proteins in cell-cell communication despite the fact that they were identified only with poor coverage in the AP-MS analysis. However, further experiments are required to determine their mechanistic mode of action and to exclude indirect effects of the mutations on MAK-2 pathway functionality. In addition, mutants defective for the hypothetical proteins NCU00627, NCU02606, NCU02972, and NCU08957 that also interacted with the entire kinase cascade displayed inconspicuous communication patterns and were thus not tested for MAK-2 phosphorylation, because they likely represented contaminants.
Table 1. Mutant characteristics of MAK-2 pathway components.
| Strain (FGSC # or [reference]) | Annotation | Cell communication 1 2 | MAK-2 phosphorylation 2 |
| wild type (#987) | 95%±2 | 100% | |
| ΔNCU00455 (#17041×wt) | MAP3K regulator STE-50 | 0% | 0% |
| ΔNCU00627 (#16766) | hypothetical protein | 96%±2 | nd 3 |
| ΔNCU00772 [47] | STE kinase MST-1 | 95%±3 | 99%±10 |
| ΔNCU01440 (#11422) | type V myosin | 9%±3 * | 14%±8 |
| ΔNCU01789 (#15045) | IDC1 related protein HAM-5 | 9%±1 | 18%±9 |
| ΔNCU02606 (#17297) | hypothetical protein | 95%±2 | nd |
| ΔNCU02972 (#19746) | hypothetical protein | 95%±3 | nd |
| ΔNCU03124 (#17973 4) | casein kinase 2 alpha subunit CKA | 12%±4 | 28%±6 |
| ΔNCU05485 (#15567) | casein kinase 2 beta 1 subunit CKB-1 | 23%±2 * | 22%±8 |
| ΔNCU06265 (#11245) | hypothetical protein; HAM-13 | 33%±2 | 42%±5 |
| ΔNCU07280 (#13416 4) | protein kinase YPK-1 | 14%±2 * | nd |
| ypk-1(16-19) [79] | protein kinase YPK-1 | 54%±8 | 20%±8 |
| ΔNCU03616 (#12467) | Ras GTPase RAS-2/SMCO-7 | 11%±2 | 29%±10 |
| smco-7 [49] | Ras GTPase RAS-2/SMCO-7 | 9%±3 | 37%±7 |
| ΔNCU03894 (#11325) | STE kinase STE-20 | 77%±2 | 94%±5 |
| Δste-20;Δmst-1 (this study) | STE kinase double mutant | 69±5 | 93%±6 |
| ΔNCU06500 (#16014 4) | Ras GDP-GTP exchange factor | 14%±3 | 15%±9 |
| cdc-25(7-10) ([79]) | Ras GDP-GTP exchange factor | 5%±2 * | nd |
| Δste-20;Δras-2 (this study) | ste-20 ras-2 double mutant | 5%±3 | 21%±5 |
| ΔNCU08008 (#12371) | adenylate cyclase capping protein CAP-1 | 28%±5 | 63%±8 |
| ΔNCU08377 (#11514×wt) | adenylate cyclase CR-1 | 34%±5 * | 37%±6 |
| ΔNCU08957 (#19203) | hypothetical protein | 97%±2 | nd |
assayed at 6 h post inoculation except for strains marked with *, which were analyzed 8 h post inoculation due to reduced germination rate and abnormal growth.
in %±SEM.
not determined.
heterokaryotic deletion strain.
Interestingly, we also determined that MAK-2, yet not the two upstream kinases interacted with multiple components of the nuclear import/export machinery (i.e. importin alpha, importin beta-1, importin beta-3, exportin-1, nuclear pore protein NCU01702; Table S1) and the two transcriptional regulators PP-1 and RCO-1, which were previously identified as key effectors of the MAK-2 pathway [30]–[32].
STE-50 is required for NRC-1 activation
Budding yeast Ste50p functions as adaptor protein of the MAP3K Ste11p that connects heterotrimeric G-proteins and small GTPases with various MAPK cascades [33], [34]. This is achieved through the modular structure of Ste50p. The protein consists of an N-terminal protein interaction domain called the sterile alpha motif (SAM) [35], [36] and a C-terminal Ras association (RA) domain that can bind to small Ras and Rho-type GTPases and is required for membrane delivery of the Ste11p/Ste50p complex [37], [38]. Homologs in filamentous fungi are not as well characterized, but have been proposed to function as scaffold proteins in MAPK cascades [39]–[42]. Yeast two hybrid (Y2H) assays confirmed the physical interaction of STE-50 with itself and with NRC-1, yet not MEK-2 and MAK-2, through the SAM domains present in the two proteins (Figure 1 B, C; Figure S2). The presence of a stable STE-50/NRC-1 complex was further supported by the reciprocal AP-MS analysis of GFP-STE-50 interacting proteins, which revealed a stable interaction of STE-50 with NRC-1, but not other components of the MAPK cascade (Figure 1 A).
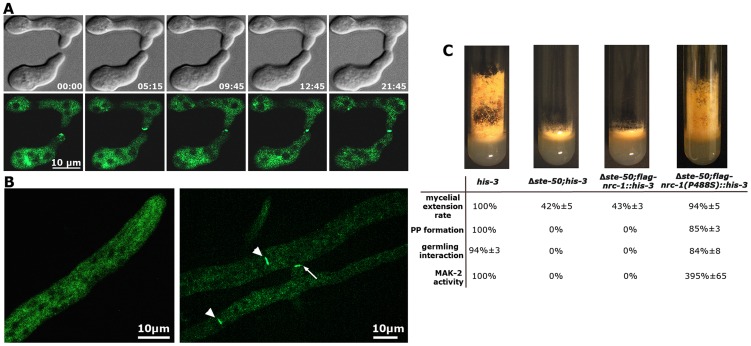
Strains expressing gfp-ste-50 under its native promoter complemented the defects of the deletion strain, but barely showed any fluorescence, which was consistent with the weak expression profile of its binding partner NRC-1 [17]. Thus, we analyzed strains expressing the fusion construct under the control of the Pccg-1 promoter. GFP-STE-50 displayed a subcellular dynamics similar to that previously described for the three kinases of the MAK-2 cascade [16], [17] and accumulated at the future fusion site after the two germlings had established physical contact (Figure 2 A). Consistent with the expression profile and localization pattern described for NRC-1 [17], we observed only weak cytoplasmic localization of GFP-STE-50 in mature hyphae and exclusion of the fusion protein from nuclei. However, GFP-STE-50 accumulated at septa and contact sites of fusing hyphae within an established colony (Figure 2 B).
Figure 2. STE-50 functions as regulatory subunit of NRC-1.
(A) GFP-STE-50 localizes in a dynamic manner to opposing tips of two communicating germlings and the site of cell-cell contact. The oscillation period is approximately three to five minutes. See Video S1 for time course. (B) GFP-STE-50 localizes in a diffuse cytosolic manner in non-communicating hyphae and is excluded from nuclei (left image). Moreover, GFP-STE-50 accumulates at septa (arrowheads) and the contact point (arrow) of communicating hyphal tips (right image). (C) Δste-50 fully phenocopies Δmak-2 defects (see Figure S3 for comparative characterization of mutant defects), and the mutant characteristics are complemented by expression of a constitutive active NRC-1(P488S) allele in Δste-50. The slant images represent macroscopic appearance and conidiation pattern, while the Table summarizes rates of mycelial extension, protoperithecia (PP) formation, germling communication and MAK-2 activities of the indicated strains.
We determined that a Δste-50 deletion mutant fully phenocopied all defects described for defective MAK-2 signaling [31], [43], [44] and lacked detectable MAK-2 kinase activity (Table 1; Figure 2 C; Figure S3). Based on these characteristics of the mutant and the tight interaction of STE-50 with NRC-1, we hypothesized that STE-50 may function as regulatory subunit of the MAP3K. To test this, we expressed a recently generated [17], constitutive active NRC-1 version NRC-1(P488S) in Δste-50. This construct fully complemented all defects of the deletion strain and confirmed our hypothesis (Figure 2 B).
HAM-5 functions as cell communication-specific scaffold of the MAK-2 module
Ham-5 was originally identified in a genetic screen for N. crassa mutants that fail to undergo cell fusion [30]. Its closest homolog, Podospora anserina IDC1, is required for NADPH-oxidase-dependent nuclear accumulation of the cell wall integrity MAPK Mpk1 [45], but the mechanistic basis underlying this observation remains unresolved. MAK-2 activity was reduced to ca. 1/10 of wild type in the Δham-5 mutant (Table 1; Figure S3), indicating significantly compromised, but not abolished MAK-2 pathway function in this strain. We observed increased mycelial extension rates of Δham-5 when compared to Δste-50 or Δmak-2, residual tropic interactions (≤10% of wild type) and the retained capacity to produce low amounts of infertile protoperithecia (ca. 20% of wild type; Figure S3). Co-immunoprecipitation experiments were used to confirm the interaction of HAM-5 with all three kinases of the MAK-2 pathway Figure S2). In order to further dissect the physical interaction pattern of these proteins, we performed Y2H tests. In these assays, we determined that HAM-5, which contained seven N-terminally located WD40 repeats and two short coiled coil regions in the C-terminal region, was able to homo-dimerize and also interacted with STE-50 and with MAK-2, yet not with MEK-2 and NRC-1 (Figure 1 B, C; Figure S2).
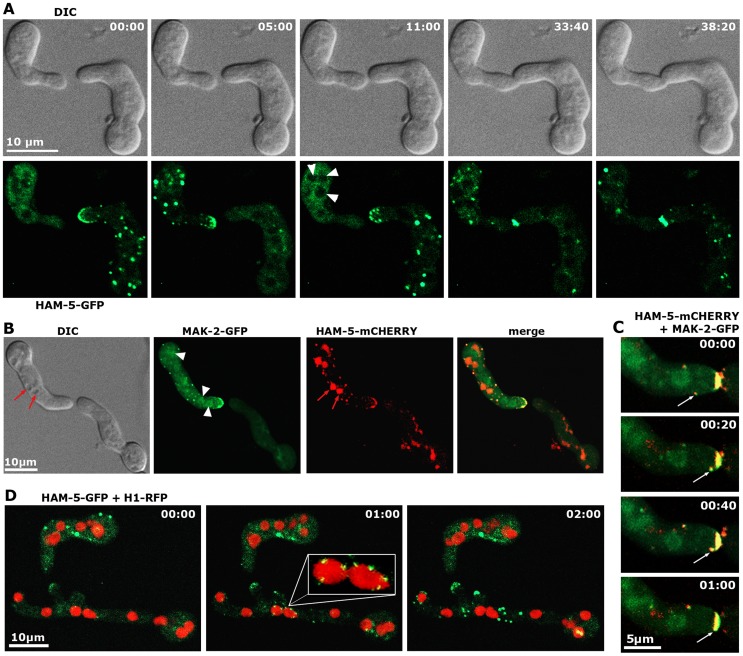
Functional Pccg-1-driven HAM-5-GFP displayed the predicted dynamic subcellular localization during germling communication and strictly co-localized with MAK-2 as dynamically forming intracellular complexes that associated with the communicating tips of the two cells with an oscillation period of three to five minutes (Figure 3 A, B). At least some of the tip-associated signal was generated by recruitment of cytosolic HAM-5- and MAK-2-containing puncta to the apex (Figure 3 C). Diffuse HAM-5-GFP label was excluded from nuclei, and bright HAM-5 puncta were not obviously associated with nuclei. However, we detected weakly labeled HAM-5 puncta that decorated nuclear envelopes (preferentially marking those nuclei that localized closely to the cell tip) simultaneously with the first appearance of tip-associated HAM-5 signal. (Figure 3 D).
Figure 3. HAM-5 and MAK-2 dynamically co-localize in communicating germlings.
(A) Motile HAM-5-GFP puncta appear in a dynamic manner at opposing tips and in the cell body of two communicating germlings with an oscillation period of approximately three to five minutes and subsequently accumulate at the contact point of the two tips. See Video S2 for time course. Note that the diffuse cytoplasmic label of HAM-5-GFP decreased during the formation of HAM-5 puncta. Arrowheads indicate nuclei, from which HAM-5 is excluded. (B) HAM-5 and MAK-2 co-localize to puncta at the cell tip and within the germling (marked by arrows). MAK-2, but not HAM-5 also accumulates in nuclei (marked by arrowheads). Note that mCHERRY-tagged HAM-5 is strongly targeted to vacuoles for degradation (marked by red arrows). See Video S3 for time course. (C) Movement of a HAM-5/MAK-2 double-labeled spot towards the cell tip. See Video S3 for time course. (D) HAM-5 puncta can associate with nuclear envelopes (enlarged insert). See Video S4 for time course. (C) Movement of a HAM-5/MAK-2 double-labeled spot towards the cell tip. See Video S3 for time course.
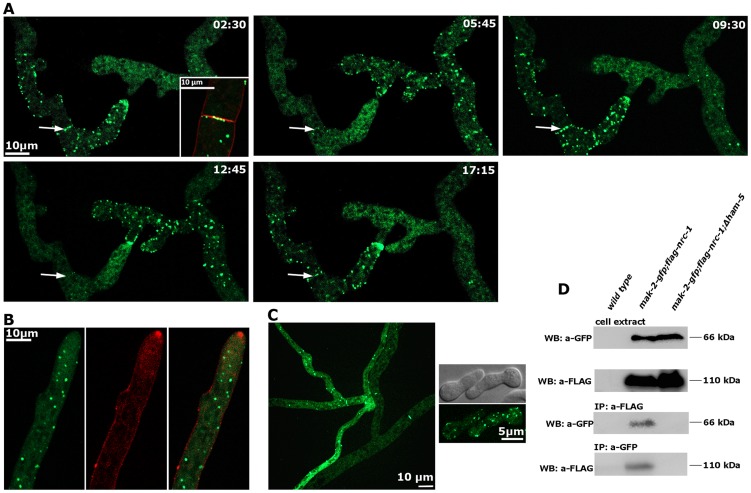
Dynamic co-recruitment of HAM-5 and MAK-2 to communicating hyphal tips, septa and intracellular puncta was also observed in the mature colony (Figure 4 A, Figure S4). Moreover, HAM-5 and SOFT oscillated with opposing recruitment phases in communicating hyphae (Figure S4), indicating that the molecular machinery required for germling communication is also operating during hyphal anastomosis within the mature colony. In contrast to MAK-2 [17], HAM-5 was not detected at the apex of non-communicating hyphae (Figure 4 B). We therefore asked if MAK-2 activity is required for HAM-5 aggregation during cell signaling. When we localized HAM-5-GFP in Δmak-2 germlings, we observed the presence of HAM-5-GFP puncta, which remained stable over long time periods (Figure 4 C). Moreover, we noticed that intracolonial Δmak-2 hyphae that had (by chance) established physical contact grew in parallel over longer distances with HAM-5-GFP puncta formed primarily in one of the two hyphae, while septal pores were strongly labeled in the second cell (Figure 4 C). Quantification of this phenomenon revealed that 72±7% (n = 141) of the analyzed intracolonial hyphal pairs displayed this HAM-5 distribution. Thus, MAK-2 is essential to regulate the subcellular dynamics of HAM-5, but complex formation does not require the MAPK.
Figure 4. HAM-5 is a cell communication-specific scaffold of the MAK-2 module.
(A) Dynamic HAM-5-GFP oscillation is observed in approaching tips within the established mycelium. Dynamic localization of HAM-5 to septa is indicated by arrows, and septum-associated HAM-5-GFP puncta are enlarged in the first image (insert co-labeled with FM4-64 to show plasma membrane and septum). See Video S5 for time course. (B) HAM-5 is not enriched at hyphal tips of non-communicating hyphae, although HAM-5-GFP spots are visible throughout the cytoplasm of the growing hypha. Plasma membrane and Spitzenkörper are labeled with FM4-64. (C) HAM-5-GFP puncta are also formed in Δmak-2 hyphae (left panel) and germlings (right panel), although the protein does not concentrate at germling tips. Note the distinct localization patterns of HAM-5-GFP in the two contacting hyphae in a Δmak-2 colony: multiple HAM-5 puncta are visible the approaching (left) hypha, while primarily the septal pores are labeled in the contacted (right) hypha and only few puncta are visible. (D) Reciprocal co-immunoprecipitation experiments of the tagged kinases flag-NRC-1 and MAK-2-GFP indicate interaction of the MAPK module in wild type but not in the Δham-5 mutant.
We reasoned that the interaction pattern of HAM-5 with multiple MAK-2 pathway components and its localization dynamics were consistent with a scaffold function of HAM-5 for the MAK-2 cascade. We tested this hypothesis by performing co-immunoprecipitation experiments (Figure 4 D). NRC-1 and MAK-2 co-precipitated when tagged versions of both proteins were co-expressed in wild type, while we were unable to detect interactions between the two kinases in a Δham-5 background, indicating that HAM-5 is critical for maintaining the integrity of the kinase cascade.
A network of Ste20-related kinases, RAS-2 and the adenylate cyclase capping protein CAP-1 signals upstream of the MAK-2 pathway
Our AP-MS analysis identified several proteins that specifically interacted with NRC-1, yet not MEK-2 or MAK-2 and thus could constitute upstream components of the MAK-2 pathway. Among them were the MAP4K STE-20, the small GTPase RAS-2/SMCO-7 and the capping protein CAP-1/NCU08008 of the adenylate cyclase (AC) complex (Table S1). A fully functional STE-20-GFP fusion construct (Figure S5) recovered STE-50, NRC-1, HAM-5 and RAS-2 in reciprocal AP-MS experiments (Figure 1 A), and Y2H assays confirmed physical interactions of STE-20 with HAM-5, NRC-1 and STE-50 (Figure 1 B, C; Figure S2). Moreover, NRC-1 and weakly also STE-50 interacted with RAS-2 through the Ras-association domains present in both proteins in Y2H tests. We did not detect any Y2H interaction of STE-50 and NRC-1 with the second Ras-type GTPase, RAS-1/NCU08823 (also called BAND; [46]), present in N. crassa, underscoring the specificity of the MS and Y2H analyses. We also detected MST-1, a recently identified accessory Ste20-related kinase of the septation initiation network, which localized to spindle pole bodies and septa [47], [48] as NRC-1- and MEK-2-interacting protein by AP-MS and Y2H analysis (Table S1; Figure 1 B). However, communication frequencies and MAK-2 activities of Δmst-1 and of a generated Δste-20;Δmst-1 double mutant indicated that MST-1 is not critically required for MAK-2 signaling (Table 1).
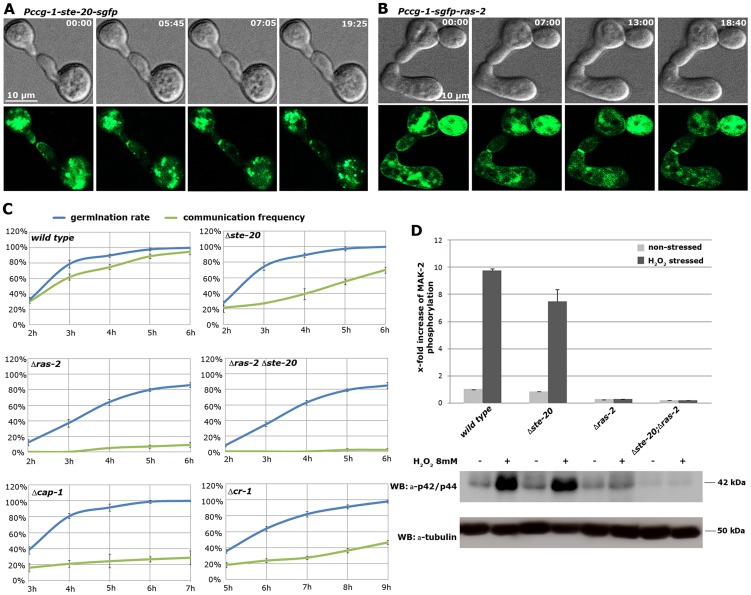
Consistent with an involvement in cell-cell communication, we found that STE-20-GFP formed membrane-associated apical crescents in mature hyphae, was enriched in a stable manner at both communicating germling tips and localized at the contact point of interacting cells in addition to its association with septa in germlings as well as mature hyphae (Figure 5 A; Figure S5). Moreover, a functional GFP-RAS-2 construct distributed along the entire plasma membrane in germlings and hyphae and localized to the contact point of interacting germlings (Figure 5 B; Figure S5). As predicted for components that signal upstream of the MAK-2 cascade, we observed reduced MAK-2 activities and cell communication frequencies in Δste-20, in Δras-2, the previously described ras-2 allele smco-7 [49], and in two mutants affecting the predicted RAS-activating GDP-GTP-exchange factor CDC-25 (Table 1; Figure 5 C). Only residual levels of germling communication and MAK-2 activity were detected in a Δste-20;Δras-2 double mutant, highlighting the joint importance of RAS-2 and STE-20 for self-signaling. The MAK-2 pathway is also induced by other external stimuli, such as reactive oxygen species [17], [28], and we asked if the identified proteins were also required for stress-induced activation of MAK-2. We determined that H2O2-induced activation of the MAK-2 pathway was abolished in Δras-2 and in the Δste-20;Δras-2 double mutant (Figure 5 D), indicating that both proteins are general components of the MAK-2 pathway.
Figure 5. The MAK-2 pathway elements STE-20 and RAS-2 are important for cell-cell communication.
(A) STE-20-GFP localizes in a stable manner to the apices of two communicating germlings and marks the site of contact. See Video S6 for time course. (B) GFP-RAS-2 associates with the entire plasma membrane of germinating spores and localizes at the contact point of two communicating cells. See Video S7 for time course. (C) Time course experiments to determine rates of germination and of chemotropic interactions of the indicated mutants. Note that due to delayed germination Δcr-1 and Δcap-1 strains are assayed at later time points than the other mutants. (D) Quantification of basal and stress-induced MAK-2 phosphorylation levels (detected with p42/44 antibodies) in cell extracts of exponentially growing liquid cultures of the indicated strains (n = 3). A representative Western blot is depicted below; tubulin was used as loading control.
We also tested for binding of these components to the AC capping protein CAP-1 and detected a positive Y2H interaction with STE-50, but not with any of the other proteins of the MAK-2 pathway (Figure 1 B). This may indicate that our AP-MS-based identification of CAP-1 in precipitates of STE-50, NRC-1 and STE-20 (Table S1; Figure 1 A) was mediated through STE-50. Nevertheless, we observed altered cell communication frequency and reduced MAK-2 activity in Δcap-1, consistent with a functional involvement of CAP-1 in MAK-2 signaling (Table 1). Significantly, we also detected reduced tropic germling interactions in the AC mutant Δcr-1 (Figure 5 C), indicating that cAMP signaling is involved in, but is not essential for cell-cell communication. These data are consistent with a previous report, which showed that cr-1 germlings are able to generate conidial anastomosis tubes, although no quantitative analysis was performed in this study [50].
Discussion
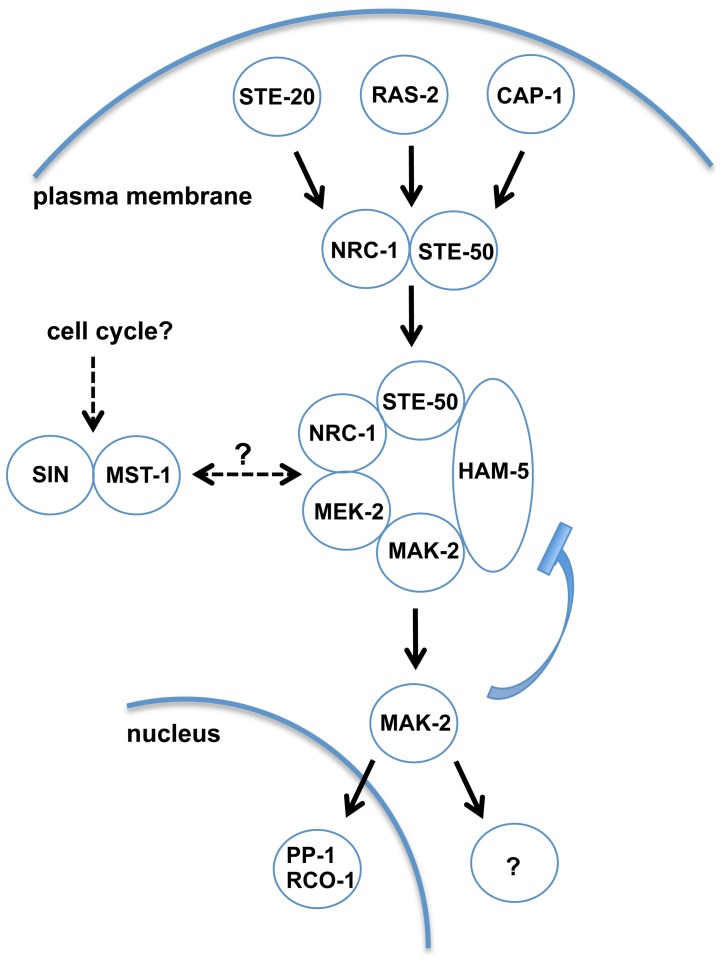
Despite considerable progress in recent years, our mechanistic understanding of oscillatory MAK-2 behavior during homotypic cell communication is hampered by the fact that many components of the signal transduction machinery are still unknown and that the molecular functions of proteins known to be required for signaling are only poorly understood. One important finding of this study is the identification of STE-50 and HAM-5 as central components of the MAK-2 pathway (Figure 6). We propose STE-50 as tightly associated, regulatory subunit of NRC-1 and HAM-5 as scaffold protein of the MAK-2 cascade. Based on our data, STE-50 may have both NRC-1-activating as well as targeting functions, and we currently cannot rule out any of these hypotheses. Nevertheless, RAS-2 interacts with both NRC-1 and STE-50 in Y2H assays, and thus STE-50 may not be essential for membrane targeting, yet full complementation of Δste-50 by expression of constitutive active NRC-1(P488S) indicates that STE-50 is critical for activation of the MAP3K. We do not have any evidence for a scaffold function of STE-50 in N. crassa, contrasting data obtained in other filamentous fungi, which have suggested interactions of STE-50 homologs with other kinases in addition to the MAP3K [40], [42], [51].
Figure 6. Model depicting network organization of the MAK-2 pathway and putative regulatory mechanisms.
Ligand-induced activation of an unknown receptor may be transmitted through plasma membrane-associated STE-20, RAS-2 and CAP-1, which signal toward the NRC-1/STE-50 complex and recruit the MAPK cascade through activation and clustering of the scaffold HAM-5 at intracellular puncta. MAK-2 activation triggers nuclear gene expression through interaction with the transcription factor PP-1 and the RCO-1/RCM-1 complex and the cytosolic activation of the secretory pathway and cell polarity machineries to coordinate pulsed signal release and chemotrophic growth towards the partner cell, respectively. MAK-2 activity is also required for termination of the receiver phase, potentially through negative feedback phosphorylation of the MAPK and disassembly of the MAK-2/HAM-5 module. MAK-2 pathway function may also be regulated through the STRIPAK complex, the CK2 heterodimer, membrane lipid composition, the septum-associated septation initiation network SIN and motor protein-dependent vesicle trafficking.
Central for our understanding of scaffold proteins is the archetypical MAP kinase scaffold Ste5p of the yeast mating pathway [52], [53]. However, Ste5p is restricted to budding yeast and close relatives [20], [54]. In contrast, homologs of HAM-5 are detected in all sequenced members of the Pezizomycotina subphylum (the group of filament-forming ascomycete fungi), but are absent from the genomes of unicellular ascomycete fungi (Figure S6). Thus, we propose HAM-5 as scaffold of the N. crassa MAK-2 cascade and of homologous MAPK modules in other filamentous fungi. This hypothesis is further strengthened by an accompanying study, which also identified HAM-5 as scaffold protein of the MAK-2 pathway [55]. We hypothesize that HAM-5 and MAK-2 are co-recruited to intracellular puncta in the presumed signal receiver phase [15], [16] and that some these puncta are subsequently targeted to the apical region of communicating cells. This process may potentially reflect the predicted auto-amplification of the incoming signal through the MAK-2 cascade or priming of the receiving cell for signal release during the next phase of communication [18]. The formation of HAM-5 puncta in the Δmak-2 mutant indicates that MAK-2 is not required for complex formation of HAM-5. HAM-5 puncta remain stable over long time periods in Δmak-2, and hyphae in contact frequently display distinct HAM-5 localization patterns, suggesting that the two cells are locked into two distinct signaling modes. Thus, complex dispersion and the switch from signal receiving to signal sending require MAK-2 activity. One possible mechanism for complex disassembly and termination of the signal receiving phase of the cell may involve accumulating phosphorylation of the HAM-5 scaffold through MAK-2 and/or additional unknown kinases as proposed in [55]. This hypothesis is also in line with previous work reporting that unregulated (both reduced as well as increased) MAK-2 activity allows reasonable tip growth of vegetative hyphae, while cell-cell communication requires the regulated on/off switch of the MAK-2 cascade [17], [30]. In contrast to MAK-2 [17], HAM-5 did not accumulate at the apex of non-communicating hyphae, and residual MAK-2 pathway functionality is retained in Δham-5 allowing reasonable rates of mycelial extension. These data imply a cell communication-specific function of HAM-5 for the MAK-2 cascade. In addition, previous work had identified the kinase adapter HYM-1/MO25, which associates with multiple Ste20-related kinases in N. crassa (this study; [17], [47]) and higher eukaryotes [56] as general platform that is required for MAK-2-dependent intercellular communication and for basic growth-associated functions of the MAK-2 pathway.
We identified STE-20 and RAS-2 that together are critical for signal input of the MAK-2 pathway. Moreover, the AC capping protein CAP-1 is also involved in cell-cell communication. The yeast homolog of CAP-1, Srv2p, was identified as part of a RAS-responsive AC complex in S. cerevisiae [57], [58]. CAP-1 homologs also play a critical role in regulating actin dynamics and cell polarity in various fungi as well as higher eukaryotes [59]–[62]. Thus, the association of STE-50 with CAP-1 may link upstream components of the MAK-2 pathway with Ras/cAMP signaling and with cell morphogenesis through regulation of actin dynamics. We did not detect the second STE-20-related kinase CLA-4 as MAK-2 pathway-associated protein in our proteomics analysis, consistent with previous Y2H data that indicate no physical interaction between NRC-1 and CLA-4 [17]. Thus, CLA-4, which was recently implicated in self-signaling in N. crassa [63], may function as part of another module, such as the predicted BEM-1/CDC-42/RAC/CDC-24/CLA-4 complex that regulates cell polarity and potentially also chemotrophic growth [64]-[66]. Intriguingly, we also identified the Ste20-related kinase MST-1 as NRC-1- and MEK-2-interacting protein (this study; [47]). Although the significance of these interactions is currently unclear, the entire MAK-2 cascade including STE-50 and HAM-5 associates with septa ([16], [17]; this study). The dynamic localization of HAM-5 at septa during intra-colony communication and its strong septal pore association may therefore indicate that MAK-2-dependent signals can originate from septa and/or that incoming signals are integrated by the MAPK pathway at these sites. This speculation is supported by accumulating evidence of central functions of septal pores as signaling hubs within the fungal colony [67]–[69].
Finally, the association of the casein kinase 2 holoenzyme and the PP2A heterotrimer with the MAK-2 cascade opens the intriguing possibility for activity regulation of the MAPK cascade. Analogous results were obtained for the mammalian casein kinase 2, which also associates with phosphatase 2A and down-regulates the PP2A substrate MEK1 [70], [71]. A similar, regulatory role may also be attributed to the recently defined STRIPAK complex [27], [28], [72]. Significantly, we did not detect the STRIPAK subunits HAM-2 and HAM-4 in our MS analysis, and thus only a sub-complex consisting of the PP2A heterotrimer and the kinase adaptor protein MOB-3 may associate with and regulate the MAK-2 cascade. MAK-2 pathway regulation may also occur through modulation of lipid composition and thus membrane dynamics, which is central for the organization and dynamic localization of multiple signal transduction pathways, including the yeast pheromone pathway [73], and has also been implicated in self-signaling in N. crassa [74]. Yeast homologs of the N. crassa kinases YPK-1 and NRC-2 function as regulators of so-called flippase complexes [75], [76], which are primarily localized to the plasma membrane at sites of polarized growth, and phospholipid flipping has been shown to regulate Cdc42p signaling during polarized growth in yeast [77]. Because N. crassa ypk-1 and nrc-2 strongly phenocopy vegetative and developmental traits of MAK-2 pathway mutants [78], [79], the association of YPK-1 with NRC-1 and the cell communication defects detected in deletion and temperature-sensitive ypk-1 mutants will be of particular interest for dissecting oscillatory MAK-2 signaling and the chemotropic behavior of communicating cells.
Materials and Methods
Strains, media and tagged protein constructs
Strains and oligonucleotides used in this study are listed in Tables S2 and S3, respectively. General genetic procedures and media used in the handling of N. crassa are available through the Fungal Genetic Stock Center (www.fgsc.net). Fusion proteins were ectopically expressed under the control of the ccg-1 or gpd-1 promoters at the his-3 locus. The ORFs of ste-50, ham-5, ste-20 and ras-2 were amplified by PCR as annotated by the N. crassa database and introduced into pMF272, pNGFP and pJV15-2 [80]–[82]. For co-expression of tagged proteins, the pccg-1-sgfp sequence of the vector pNGFP [81] was replaced with the inversely oriented ccg-1 and gpd-1 promoters amplified from the vector pBiFC [83] to generate the vector pCCG1-pGPD1, which allowed insertion of fusion-constructs via SgsI/SpeI (mak-2-sgfp) and SwaI/EcoRI (3xflag-nrc-1) amplified from the template plasmids pMF272-mak2 and pFLAG-nrc1 [17]. Resulting plasmids were transformed into his-3 and/or his-3;Δ strains. Complementation of tropic interaction, growth and developmental defects of the deletion strain was used to confirm functionality of the constructs.
Protein methods
Co-immunoprecipitation experiments were performed as described [28], [84]. Conditions and plasmids used for the Y2H assays are specified in [83], [85]. Basal and stress-induced MAK-2 activity of exponentially growing N. crassa liquid cultures was determined using polyclonal rabbit α-Phospho-p44/42 MAPK (Cell Signaling Technology, USA) and goat α-rabbit IgG-HRP (Santa Cruz, USA) as primary and secondary antibodies, respectively as described [86], [87]. Briefly, exponentially growing, liquid cultures were harvested gently by filtration using a Büchner funnel and ground in liquid nitrogen. The frozen mycelial powder was incubated in 95% ethanol at -20°C for ≥12 h, the supernatant removed after centrifugation and the pellet vacuum-dried in a SpeedVac concentrator (Thermo Fisher Scientific, USA). extraction buffer (50 mM Tris/HCL pH 7,5, 100 mM KCl, 10 mM MgCl2, 0.15% NP-40, 5 mM NaF, 1 mM PEFA, 1 mM Na3VO4, 25 mM β-glycerophosphate, 2 mM benzamidine, 2 ng/µl pepstatin A, 10 ng/µl aprotinin, 10 ng/µl leupeptin) was added, the samples mixed and incubated at 80°C for 5 min and the supernatant collected after centrifugation. After a second round of extraction, the supernatants pooled, subjected to another centrifugation step, and the protein concentration determined using a Nanodrop spectrophotometer (ND-1000, Peqlab, Germany). Sample volumes corresponding to 75 µg total protein per lane were subjected to SDS polyacrylamide gel electrophoresis. ≥3 biological replicates were quantified for each experiment. For quantification of MAK-2 phosphorylation levels, exposed films were scanned at a resolution of 600 dpi and densitometry was performed on the resulting tif files employing the AIDA Image Analyzer (version 4.22; raytest Isotopenmessgeräte, Germany) in transmission mode. Intensity values [arbitrary units] measured within a region of interest of fixed size containing the MAK-2 protein bands were corrected by subtraction of local background, normalized to the protein amount loaded and used for further evaluation.
GFP-trap experiments, mass spectrometry and database analysis were performed as described [28], [88]. Pulverized mycelium was mixed 1∶1 with extraction buffer and centrifuged (1 h, 10,000 rpm, Sorvall SS34 rotor) in order to obtain crude cell extracts. Cell extracts were incubated with 2 µl GFP-trap beads (Chromotek, Germany) per 15 ml cell extract on a rotator for 2 h at 4°C. The beads were washed three times with IP buffer, associated proteins were recovered by boiling them in Laemmli buffer and separated by SDS polyacrylamide gel electrophoresis analysis. Peptides of in-gel trypsinated proteins were extracted from Coomassie-stained gel slices. Peptides of 5 µl sample solution were trapped and washed with 0.05% trifluoroacetic acid on an Acclaim PepMap 100 column (75 µm×2 cm, C18, 3 µm, 100 Å, P/N164535 Thermo Scientific, USA) at a flow rate of 4 µl/min for 12 min. Analytical peptide separation by reverse phase chromatography was performed on an Acclaim PepMap RSLC column (75 µm×15 cm, C18, 3 µm, 100 Å, P/N164534 Thermo Scientific, USA) running a gradient from 96% solvent A (0.1% formic acid) and 4% solvent B (acetonitrile, 0.1% formic acid) to 50% solvent B within 25 min at a flow rate of 250 nl/min. Peptides eluting from the chromatographic column were on-line ionized by nano-electrospray using the Nanospray Flex Ion Source (Thermo Scientific, USA) and transferred into the mass spectrometer. Full scans within m/z 300–1850 were recorded by the Orbitrap-FT analyzer at a resolution of 60,000 at m/z 400. Each sample was analyzed using two different fragmentation techniques applying a data-dependent top 5 experiment: collision-induced decay with multistage activation and readout in the LTQ Velos Pro linear ion trap, and higher energy collision dissociation and subsequent readout in the Orbitrap-FT analyzer. LC/MS method programming and data acquisition was performed with XCalibur 2.2 (Thermo Scientific, USA). Orbitrap raw files were analyzed with Proteome Discoverer 1.3 (Thermo Scientific, USA) using the Mascot and Sequest search engines against the N. crasssa protein database with the following criteria: peptide mass tolerance 10 ppm, MS/MS ion mass tolerance 0.8 Da, and up to two missed cleavages allowed.
Microscopy
Spinning disc confocal microscopy was performed as described [47] using an inverted Axio Observer Z1 microscope (Zeiss, Germany) equipped with a CSU-22 confocal scanner unit and a CCD camera (Axiocam MRm Rev.3). ZEN Blue 2012 software (Zeiss, Germany) was used for image/video acquisition and image analysis. Plasma membrane was stained with FM4-64 (1 mg/ml-1), and time-lapse imaging was performed at capture intervals of 20 s for periods up to 40 min using a C-Apochromat 63x/1.2 W objective. Image series were converted into movies (*.mov).
Tropic germling interactions of ≥100 germlings in each of 2-3 biological replicates per experiment were quantified as described [28], [65]. In brief, strains were grown on Vogel's minimal media slants for 7 days at 26°C. Conidia were harvested with 1 to 2 ml H2O, and the conidial suspension was filtered through cheesecloth. A total number of 5×106 fresh spores were spread out on a minimal media plate, incubated at 30°C and analyzed after the indicated time points using a Zeiss Axiophot 2 microscope with a Zeiss Plan-Apochromat 63x/1.40 oil immersion objective. Only germlings that had produced germ-tubes of at least 2 µm length and were localized within a distance smaller than 10 µm to another germling were included in the trophic interaction analysis.
Supporting Information
Mutant characteristics of MAK-2 pathway components. (A) Pictures of communicating germlings of the indicated strains were taken after 4-6 h germination at 30°C. (B) MAK-2 phosphorylation levels were determined with p42/44 antibodies in cell extracts of exponentially growing liquid cultures of the indicated strains. Tubulin was used as loading control.
(PDF)
Interaction network of the MAK-2 pathway. (A) Physical interactions between MAK-2 pathway components were mapped in yeast two-hybrid (Y2H) tests. The indicated constructs were co-expressed in strain AH109 and yeast growth was analyzed on the indicated selective media. (B) Reciprocal co-immunoprecipitation experiments from cell extracts co-expressing the functionally tagged proteins NRC-1, MEK-2 and MAK-2 and HAM-5 indicate interaction of all three kinases with HAM-5.
(PDF)
Comparative characterization of Δste-50 and Δham-5. Analysis of macroscopic appearance (A), mycelial extension rate (B), sexual development (C), germling communication frequency (D), and basal/stress-stimulated MAK-2 activity level (E) indicates that Δste-50 fully phenocopies Δmak-2 defects, while Δham-5 retains residual MAK-2 pathway functionality.
(PDF)
Co-localization of HAM-5 with the SOFT/MAK-2 cell communication machinery in communicating hyphae within the established mycelium. (A) MAK-2-GFP and HAM-5-mCherry co-localize in a dynamic manner to opposing tips of two communicating hyphae. Note the dynamic nuclear accumulation of MAK-2, yet not HAM-5 in the presumed signal receiver phase. (B) Dynamic recruitment to communicating hyphal tips is also observed for SOFT. (C) HAM-5-mCherry and SO-1-GFP oscillate with strictly opposing recruitment phases in communicating hyphae and do not co-localize.
(PDF)
Characterization of ste-20 (left) and ras-2 (right panels). (A) Macroscopic appearance and mycelial extension rates of the indicated deletion mutants and complemented strains. (B) Localization of GFP fusion constructs in mature hyphae. See Videos S8 and S9 for time courses. Plasma membrane and Spitzenkörper are labeled with FM4-64. H1-RFP was used to label nuclei (left panel only).
(PDF)
Phylogram of fungal HAM-5 homologs. The tree was generated by using ClustalX 2.1 with bootstrap support (111 random number generator seed and 1000 bootstrap trials) and predicted protein sequences from selected ascomycete proteins. Note that only the WD40 domains of each protein were included in this analysis in order to use the WD40 domain of N. crassa HAM-3 as outgroup member (similar tree topologies were obtained with full length sequences; multiple alignment parameters: gap opening 10.0, gap extension 0,2, delay divergent sequences 30%, protein weight matrix gonnet series).
(PDF)
AP-MS analysis of the MAK-2 pathway.
(XLSX)
N. crassa strains used in this study.
(DOCX)
Oligonucleotides used in this study.
(DOCX)
Time-course of GFP-STE-50 localization during germling communication and chemotrophic growth. GFP-STE-50 is recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5-GFP localization during germling communication and chemotrophic growth. HAM-5-GFP is recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5-mCHERRY and MAK-2-GFP co-localization during germling communication and chemotrophic growth. Both proteins are co-recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5 puncta formation at the nuclear envelope during germling communication. HAM-5 and histone H1 are tagged with GFP and RFP, respectively.
(MOV)
Time-course of HAM-5-GFP localization in the established colony. HAM-5-GFP is recruited in an oscillatory manner to the tips and septa of communicating hyphae.
(MOV)
Time-course of STE-20-GFP localization during germling communication and chemotrophic growth. STE-20-GFP is enriched at both approaching tips and accumulates at the contact point of the two cells.
(MOV)
Time-course of GFP-RAS-2 localization during germling communication and chemotrophic growth. GFP-RAS-2 associates with the plasma membrane and strongly accumulates at the contact point of the two cells.
(MOV)
Time-course of STE-20-GFP localization at the tip of mature hyphae. STE-20-GFP form a dynamic apical crescent and subapical collar at the hyphal tip.
(MOV)
Time-course of GFP-RAS-2 localization at the tip of mature hyphae. GFP-RAS-2 associates with the entire plasma membrane in a non-specific manner in established hyphae.
(MOV)
Acknowledgments
We thank Klaus Palme and the Institute of Biology II for providing lab space for the group.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research project was financially supported through the DFG Heisenberg fellowship SE1054/6-1 and the DFG grant SE1054/9-1. The Fungal Genetics Stock Center (www.fgsc.net) and the Neurospora program grant PO1 GM068087 “Functional analysis of a model filamentous fungus” are acknowledged for strains. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Van Norman JM, Breakfield NW, Benfey PN (2011) Intercellular communication during plant development. Plant Cell 23: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Celiker H, Gore J (2013) Cellular cooperation: insights from microbes. Trends Cell Biol 23: 9–15. [DOI] [PubMed] [Google Scholar]
- 3. Kornberg TB, Roy S (2014) Communicating by touch - neurons are not alone. Trends Cell Biol 24: 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloemendal S, Kuck U (2013) Cell-to-cell communication in plants, animals, and fungi: a comparative review. Naturwissenschaften 100: 3–19. [DOI] [PubMed] [Google Scholar]
- 5. Leeder AC, Palma-Guerrero J, Glass NL (2011) The social network: deciphering fungal language. Nat Rev Microbiol 9: 440–451. [DOI] [PubMed] [Google Scholar]
- 6. Simonin A, Palma-Guerrero J, Fricker M, Glass NL (2012) Physiological significance of network organization in fungi. Eukaryot Cell 11: 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richard F, Glass NL, Pringle A (2012) Cooperation among germinating spores facilitates the growth of the fungus, Neurospora crassa . Biol Lett 8: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Craven KD, Velez H, Cho Y, Lawrence CB, Mitchell TK (2008) Anastomosis is required for virulence of the fungal necrotroph Alternaria brassicicola . Eukaryot Cell 7: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charlton ND, Shoji JY, Ghimire SR, Nakashima J, Craven KD (2012) Deletion of the fungal gene soft disrupts mutualistic symbiosis between the grass endophyte Epichloe festucae and the host plant. Eukaryot Cell 11: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roca MG, Weichert M, Siegmund U, Tudzynski P, Fleissner A (2012) Germling fusion via conidial anastomosis tubes in the grey mould Botrytis cinerea requires NADPH oxidase activity. Fungal Biol 116: 379–387. [DOI] [PubMed] [Google Scholar]
- 11. Eaton CJ, Cox MP, Scott B (2011) What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci 180: 190–195. [DOI] [PubMed] [Google Scholar]
- 12. Ishikawa FH, Souza EA, Shoji JY, Connolly L, Freitag M, et al. (2012) Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS One 7: e31175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oren-Suissa M, Podbilewicz B (2010) Evolution of programmed cell fusion: common mechanisms and distinct functions. Dev Dyn 239: 1515–1528. [DOI] [PubMed] [Google Scholar]
- 14. Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, et al. (2013) Genetic basis of cell-cell fusion mechanisms. Trends Genet 29: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Read ND, Lichius A, Shoji JY, Goryachev AB (2009) Self-signalling and self-fusion in filamentous fungi. Curr Opin Microbiol 12: 608–615. [DOI] [PubMed] [Google Scholar]
- 16. Fleissner A, Leeder AC, Roca MG, Read ND, Glass NL (2009) Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc Natl Acad Sci U S A 106: 19387–19392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dettmann A, Illgen J, Maerz S, Schurg T, Fleissner A, et al. (2012) The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa . PLoS Genet 8: e1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goryachev AB, Lichius A, Wright GD, Read ND (2012) Excitable behavior can explain the “ping-pong” mode of communication between cells using the same chemoattractant. Bioessays 34: 259–266. [DOI] [PubMed] [Google Scholar]
- 19. Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, et al. (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, et al. (2009) Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol 46: 287–298. [DOI] [PubMed] [Google Scholar]
- 21. Qi M, Elion EA (2005) MAP kinase pathways. J Cell Sci 118: 3569–3572. [DOI] [PubMed] [Google Scholar]
- 22. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa . Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- 23. Read ND, Goryachev AB, Lichius A (2012) The mechanistic basis of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biology Reviews 26: 1–11. [Google Scholar]
- 24. Xiang Q, Rasmussen C, Glass NL (2002) The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa . Genetics 160: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maerz S, Dettmann A, Ziv C, Liu Y, Valerius O, et al. (2009) Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa . Mol Microbiol 74: 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simonin AR, Rasmussen CG, Yang M, Glass NL (2010) Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa . Fungal Genet Biol 47: 855–868. [DOI] [PubMed] [Google Scholar]
- 27. Bloemendal S, Bernhards Y, Bartho K, Dettmann A, Voigt O, et al. (2012) A homologue of the human STRIPAK complex controls sexual development in fungi. Mol Microbiol 84: 310–323. [DOI] [PubMed] [Google Scholar]
- 28. Dettmann A, Heilig Y, Ludwig S, Schmitt K, Illgen J, et al. (2013) HAM-2 and HAM-3 are central for the assembly of the Neurospora STRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol Microbiol 90: 796–812. [DOI] [PubMed] [Google Scholar]
- 29. Fu C, Iyer P, Herkal A, Abdullah J, Stout A, et al. (2011) Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa . Eukaryot Cell 10: 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aldabbous MS, Roca MG, Stout A, Huang IC, Read ND, et al. (2010) The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa . Microbiology 156: 2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li D, Bobrowicz P, Wilkinson HH, Ebbole DJ (2005) A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa . Genetics 170: 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leeder AC, Jonkers W, Li J, Glass NL (2013) Early colony establishment in Neurospora crassa requires a MAP kinase regulatory network. Genetics 195: 883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen RE, Thorner J (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae . Biochim Biophys Acta 1773: 1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saito H (2010) Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol 13: 677–683. [DOI] [PubMed] [Google Scholar]
- 35. Qiao F, Bowie JU (2005) The many faces of SAM. Sci STKE 2005: re7. [DOI] [PubMed] [Google Scholar]
- 36. Grimshaw SJ, Mott HR, Stott KM, Nielsen PR, Evetts KA, et al. (2004) Structure of the sterile alpha motif (SAM) domain of the Saccharomyces cerevisiae mitogen-activated protein kinase pathway-modulating protein STE50 and analysis of its interaction with the STE11 SAM. J Biol Chem 279: 2192–2201. [DOI] [PubMed] [Google Scholar]
- 37. Truckses DM, Bloomekatz JE, Thorner J (2006) The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae . Mol Cell Biol 26: 912–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M (2006) Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20: 734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schamber A, Leroch M, Diwo J, Mendgen K, Hahn M (2010) The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea . Mol Plant Pathol 11: 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park G, Xue C, Zhao X, Kim Y, Orbach M, et al. (2006) Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea . Plant Cell 18: 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu J, Mares C, Lizcano A, Liu Y, Wickes BL (2011) Insertional mutagenesis combined with an inducible filamentation phenotype reveals a conserved STE50 homologue in Cryptococcus neoformans that is required for monokaryotic fruiting and sexual reproduction. Mol Microbiol 79: 990–1007. [DOI] [PubMed] [Google Scholar]
- 42. Bayram O, Bayram OS, Ahmed YL, Maruyama J, Valerius O, et al. (2012) The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet 8: e1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pandey A, Roca MG, Read ND, Glass NL (2004) Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa . Eukaryot Cell 3: 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maerz S, Ziv C, Vogt N, Helmstaedt K, Cohen N, et al. (2008) The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa . Genetics 179: 1313–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jamet-Vierny C, Debuchy R, Prigent M, Silar P (2007) IDC1, a pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina . Fungal Genet Biol 44: 1219–1230. [DOI] [PubMed] [Google Scholar]
- 46. Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, et al. (2007) The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev 21: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heilig Y, Dettmann A, Mourino-Perez RR, Schmitt K, Valerius O, et al. (2014) Proper actin ring formation and septum constriction requires coordinated regulation of SIN and MOR pathways through the germinal centre kinase MST-1. PLoS Genet 10: e1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heilig Y, Schmitt K, Stephan Seiler (2013) S (2013) Phospho-regulation of the Neurospora crassa septation initiation network. PLOS One 8: e79464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kana-uchi A, Yamashiro CT, Tanabe S, Murayama T (1997) A ras homologue of Neurospora crassa regulates morphology. Mol Gen Genet 254: 427–432. [DOI] [PubMed] [Google Scholar]
- 50. Roca MG, Arlt J, Jeffree CE, Read ND (2005) Cell biology of conidial anastomosis tubes in Neurospora crassa . Eukaryot Cell 4: 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kramer B, Thines E, Foster AJ (2009) MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea . Fungal Genet Biol 46: 667–681. [DOI] [PubMed] [Google Scholar]
- 52. Dard N, Peter M (2006) Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. Bioessays 28: 146–156. [DOI] [PubMed] [Google Scholar]
- 53. Witzel F, Maddison L, Bluthgen N (2012) How scaffolds shape MAPK signaling: what we know and opportunities for systems approaches. Front Physiol 3: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cote P, Sulea T, Dignard D, Wu C, Whiteway M (2011) Evolutionary reshaping of fungal mating pathway scaffold proteins. MBio 2: e00230–00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jonkers W, Leeder AC, Ansong C, Yang F, Camp II DG, et al. (2014) HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa . PLoS Genet 10: e1004783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Filippi BM, de los Heros P, Mehellou Y, Navratilova I, Gourlay R, et al. (2011) MO25 is a master regulator of SPAK/OSR1 and MST3/MST4/YSK1 protein kinases. EMBO J 30: 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gourlay CW, Ayscough KR (2006) Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae . Mol Cell Biol 26: 6487–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Field J, Vojtek A, Ballester R, Bolger G, Colicelli J, et al. (1990) Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 61: 319–327. [DOI] [PubMed] [Google Scholar]
- 59. Zhou X, Zhang H, Li G, Shaw B, Xu JR (2012) The Cyclase-associated protein Cap1 is important for proper regulation of infection-related morphogenesis in Magnaporthe oryzae . PLoS Pathog 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, et al. (2001) Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans . Mol Biol Cell 12: 3631–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bertling E, Quintero-Monzon O, Mattila PK, Goode BL, Lappalainen P (2007) Mechanism and biological role of profilin-Srv2/CAP interaction. J Cell Sci 120: 1225–1234. [DOI] [PubMed] [Google Scholar]
- 62. Moriyama K, Yahara I (2002) Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci 115: 1591–1601. [DOI] [PubMed] [Google Scholar]
- 63. Lichius A, Goryachev AB, Fricker MD, Obara B, Castro-Longoria E, et al. (2014) CDC-42 and RAC-1 regulate opposite chemotropisms in Neurospora crassa . J Cell Sci 127: 1953–1965. [DOI] [PubMed] [Google Scholar]
- 64. Frieser SH, Hlubek A, Sandrock B, Bolker M (2011) Cla4 kinase triggers destruction of the Rac1-GEF Cdc24 during polarized growth in Ustilago maydis . Mol Biol Cell 22: 3253–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schuerg T, Brandt U, Adis C, Fleissner A (2012) The Saccharomyces cerevisiae BEM1 homologue in Neurospora crassa promotes co-ordinated cell behaviour resulting in cell fusion. Mol Microbiol 86: 349–366. [DOI] [PubMed] [Google Scholar]
- 66. Araujo-Palomares CL, Richthammer C, Seiler S, Castro-Longoria E (2011) Functional characterization and cellular dynamics of the CDC-42 - RAC - CDC-24 module in Neurospora crassa . PLoS One 6: e27148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Riquelme M, Yarden O, Bartnicki-Garcia S, Bowman B, Castro-Longoria E, et al. (2011) Architecture and development of the Neurospora crassa hypha — a model cell for polarized growth. Fungal Biol 115: 446–474. [DOI] [PubMed] [Google Scholar]
- 68. Seiler S, Justa-Schuch D (2010) Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol Microbiol 78: 1058–1076. [DOI] [PubMed] [Google Scholar]
- 69. Jedd G, Pieuchot L (2012) Multiple modes for gatekeeping at fungal cell-to-cell channels. Mol Microbiol 86: 1291–1294. [DOI] [PubMed] [Google Scholar]
- 70. Heriche JK, Lebrin F, Rabilloud T, Leroy D, Chambaz EM, et al. (1997) Regulation of protein phosphatase 2A by direct interaction with casein kinase 2alpha. Science 276: 952–955. [DOI] [PubMed] [Google Scholar]
- 71. Lebrin F, Bianchini L, Rabilloud T, Chambaz EM, Goldberg Y (1999) CK2alpha-protein phosphatase 2A molecular complex: possible interaction with the MAP kinase pathway. Mol Cell Biochem 191: 207–212. [PubMed] [Google Scholar]
- 72. Goudreault M, D'Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, et al. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics 8: 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J (2010) Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci U S A 107: 11805–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mahs A, Ischebeck T, Heilig Y, Stenzel I, Hempel F, et al. (2012) The essential phosphoinositide kinase MSS-4 is required for polar hyphal morphogenesis, localizing to sites of growth and cell fusion in Neurospora crassa . PLoS One 7: e51454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J (2010) A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci U S A 107: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell 19: 1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saito K, Fujimura-Kamada K, Hanamatsu H, Kato U, Umeda M, et al. (2007) Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev Cell 13: 743–751. [DOI] [PubMed] [Google Scholar]
- 78. Kothe GO, Free SJ (1998) The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa . Genetics 149: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Seiler S, Plamann M (2003) The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa . Mol Biol Cell 14: 4352–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND (2004) GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa . Fungal Genet Biol 41: 897–910. [DOI] [PubMed] [Google Scholar]
- 81. Honda S, Selker EU (2009) Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Verdin J, Bartnicki-Garcia S, Riquelme M (2009) Functional stratification of the Spitzenkorper of Neurospora crassa . Mol Microbiol 74: 1044–1053. [DOI] [PubMed] [Google Scholar]
- 83. Maerz S, Dettmann A, Seiler S (2012) Hydrophobic Motif Phosphorylation Coordinates Activity and Polar Localization of the Neurospora crassa Nuclear Dbf2-Related Kinase COT1. Mol Cell Biol 32: 2083–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Seiler S, Vogt N, Ziv C, Gorovits R, Yarden O (2006) The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol Biol Cell 17: 4080–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Richthammer C, Enseleit M, Sanchez-Leon E, Marz S, Heilig Y, et al. (2012) RHO1 and RHO2 share partially overlapping functions in the regulation of cell wall integrity and hyphal polarity in Neurospora crassa . Mol Microbiol 85: 716–733. [DOI] [PubMed] [Google Scholar]
- 86. Maddi A, Dettman A, Fu C, Seiler S, Free SJ (2012) WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa . PLoS One 7: e42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fu C, Ao J, Dettmann A, Seiler S, Free SJ (2014) Characterization of the Neurospora crassa cell fusion proteins HAM-6, HAM-7, HAM-8, HAM-9, HAM-10, AMPH-1 and WHI-2. PLoS ONE 9: e107773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Riquelme M, Bredeweg EL, Callejas-Negrete O, Roberson RW, Ludwig S, et al. (2014) The Neurospora crassa exocyst complex tethers Spitzenkorper vesicles to the apical plasma membrane during polarized growth. Mol Biol Cell 25: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutant characteristics of MAK-2 pathway components. (A) Pictures of communicating germlings of the indicated strains were taken after 4-6 h germination at 30°C. (B) MAK-2 phosphorylation levels were determined with p42/44 antibodies in cell extracts of exponentially growing liquid cultures of the indicated strains. Tubulin was used as loading control.
(PDF)
Interaction network of the MAK-2 pathway. (A) Physical interactions between MAK-2 pathway components were mapped in yeast two-hybrid (Y2H) tests. The indicated constructs were co-expressed in strain AH109 and yeast growth was analyzed on the indicated selective media. (B) Reciprocal co-immunoprecipitation experiments from cell extracts co-expressing the functionally tagged proteins NRC-1, MEK-2 and MAK-2 and HAM-5 indicate interaction of all three kinases with HAM-5.
(PDF)
Comparative characterization of Δste-50 and Δham-5. Analysis of macroscopic appearance (A), mycelial extension rate (B), sexual development (C), germling communication frequency (D), and basal/stress-stimulated MAK-2 activity level (E) indicates that Δste-50 fully phenocopies Δmak-2 defects, while Δham-5 retains residual MAK-2 pathway functionality.
(PDF)
Co-localization of HAM-5 with the SOFT/MAK-2 cell communication machinery in communicating hyphae within the established mycelium. (A) MAK-2-GFP and HAM-5-mCherry co-localize in a dynamic manner to opposing tips of two communicating hyphae. Note the dynamic nuclear accumulation of MAK-2, yet not HAM-5 in the presumed signal receiver phase. (B) Dynamic recruitment to communicating hyphal tips is also observed for SOFT. (C) HAM-5-mCherry and SO-1-GFP oscillate with strictly opposing recruitment phases in communicating hyphae and do not co-localize.
(PDF)
Characterization of ste-20 (left) and ras-2 (right panels). (A) Macroscopic appearance and mycelial extension rates of the indicated deletion mutants and complemented strains. (B) Localization of GFP fusion constructs in mature hyphae. See Videos S8 and S9 for time courses. Plasma membrane and Spitzenkörper are labeled with FM4-64. H1-RFP was used to label nuclei (left panel only).
(PDF)
Phylogram of fungal HAM-5 homologs. The tree was generated by using ClustalX 2.1 with bootstrap support (111 random number generator seed and 1000 bootstrap trials) and predicted protein sequences from selected ascomycete proteins. Note that only the WD40 domains of each protein were included in this analysis in order to use the WD40 domain of N. crassa HAM-3 as outgroup member (similar tree topologies were obtained with full length sequences; multiple alignment parameters: gap opening 10.0, gap extension 0,2, delay divergent sequences 30%, protein weight matrix gonnet series).
(PDF)
AP-MS analysis of the MAK-2 pathway.
(XLSX)
N. crassa strains used in this study.
(DOCX)
Oligonucleotides used in this study.
(DOCX)
Time-course of GFP-STE-50 localization during germling communication and chemotrophic growth. GFP-STE-50 is recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5-GFP localization during germling communication and chemotrophic growth. HAM-5-GFP is recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5-mCHERRY and MAK-2-GFP co-localization during germling communication and chemotrophic growth. Both proteins are co-recruited in an oscillatory manner to the tips of communicating germlings.
(MOV)
Time-course of HAM-5 puncta formation at the nuclear envelope during germling communication. HAM-5 and histone H1 are tagged with GFP and RFP, respectively.
(MOV)
Time-course of HAM-5-GFP localization in the established colony. HAM-5-GFP is recruited in an oscillatory manner to the tips and septa of communicating hyphae.
(MOV)
Time-course of STE-20-GFP localization during germling communication and chemotrophic growth. STE-20-GFP is enriched at both approaching tips and accumulates at the contact point of the two cells.
(MOV)
Time-course of GFP-RAS-2 localization during germling communication and chemotrophic growth. GFP-RAS-2 associates with the plasma membrane and strongly accumulates at the contact point of the two cells.
(MOV)
Time-course of STE-20-GFP localization at the tip of mature hyphae. STE-20-GFP form a dynamic apical crescent and subapical collar at the hyphal tip.
(MOV)
Time-course of GFP-RAS-2 localization at the tip of mature hyphae. GFP-RAS-2 associates with the entire plasma membrane in a non-specific manner in established hyphae.
(MOV)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.