Abstract
We utilized RNAseq analysis of the Aspergillus fumigatus response to early hypoxic condition exposure. The results show that more than 89% of the A. fumigatus genome is expressed under normoxic and hypoxic conditions. Replicate samples were highly reproducible; however, comparisons between normoxia and hypoxia revealed that greater than 23% and 35% of genes were differentially expressed after 30 and 120 minutes of hypoxia exposure, respectively. Consistent with our previous report detailing transcriptomic and proteomic responses at later time points, the results here show major repression of ribosomal function and induction of ergosterol biosynthesis, as well as activation of alternate respiratory mechanisms at the later time point. RNAseq data was used to define thirty-two hypoxia-specific genes, which are not expressed under normoxic conditions. Transcripts of a C6 transcription factor and a histidine kinase response regulator were found only in hypoxia. In addition, several genes involved in the phosphoenyl pyruvate (PEP) and D-glyceraldehyde-3-phosphate (G3P) metabolism were only expressed in hypoxia. Interestingly, a 216 bp ncRNA Afu-182 in the 3’ region of insA (AFUB_064770), was significantly repressed under hypoxia with a 40-fold reduction in expression. A detailed analysis of Afu-182 showed similarity with several genes in the genome, many of which were also repressed in hypoxia. The results from this study show that hypoxia induces very early and widely drastic genome-wide responses in A. fumigatus that include expression of protein-coding and ncRNA genes. The role of these ncRNA genes in regulating the fungal hypoxia response is an exciting future research direction.
INTRODUCTION
Aspergillus fumigatus is a fungal pathogen capable of causing severe disease, such as invasive aspergillosis (IA). Molecular mechanisms of IA pathogenesis and other forms of aspergillosis are still relatively poorly understood. The ability of the fungus to persist in extreme environments may select for traits that give the fungus the ability to colonize the human host [1]. In particular, we are focused on understanding the fungal response to hypoxia. In the host lung, areas of infection have been shown to be hypoxic, defined as conditions below ~1.5% available oxygen [2]. As the infection site microenvironments in the lung of IA are characterized in part by hypoxia [2,3], the ability to adapt to hypoxia is a key virulence attribute of A. fumigatus and other fungi [4,5]. We have recently reported the rapid response to hypoxia by A. fumigatus using transcriptomics and proteomics [2,6–8]. Our studies showed a decrease in TCA cycle, ribosome biogenesis and purine metabolism and an increase in oxidative stress response, fermentation, iron metabolism and cell wall biosynthesis. Most importantly, our studies highlighted the interaction between sterol metabolism and hypoxia in A. fumigatus [9]. Here, we performed RNAseq to better understand very early hypoxia transcriptional responses, and thus provide finer detail on the molecular mechanisms of hypoxia adaptation. Novel to this study, we observed specific down-regulation of an INSIG (insulin-induced gene) -associated ncRNA under hypoxia. INSIG genes are well studied in mammals in relation to SREBP regulation [10–12] and ncRNA regulation of sterol and lipid metabolism by miR-33 and miR-96 of INSIG-1 was recently described in humans [13,14]. However, ncRNAs are not well studied in A. fumigatus [15] and their potential roles in gene regulation are not known. A better understanding of the molecular mechanisms of hypoxia adaptation in this human pathogenic filamentous fungus will facilitate a greater understanding of aspergillosis and is expected to reveal potential areas to exploit for improving IA patient outcomes [16].
METHODS
Strains and growth conditions
A. fumigatus A1163 conidia (1×106per ml) in 100 ml LGMM (liquid glucose minimal medium, pH 6.5) were grown for 16 hours shaking at 37°C under ambient light at which point the baffle flask was either harvested as the normoxia condition, or transferred to hypoxia (5% CO2 and 1% O2) and incubated with shaking for 30 or 120 minutes. Hypoxia LGMM was preconditioned in the hypoxia chamber. Mycelia were harvested at the indicated time via Buchner funnel and snap frozen in liquid nitrogen. Frozen tissue was lyophilized and RNA was extracted as described previously [9]. Total RNA was measured by Nanodrop-1000 (Thermo Fisher). Triplicates of each condition and time point were analyzed.
RNA sample preparation and Illumina sequencing (RNASeq)
To identify transcriptionally active genes, extracted RNA samples were DNased using the RNeasy kit (Qiagen), and sequencing libraries were generated using the ScriptSeq kit v2 (Epicenter) following manufacturer’s directions with a median insert size of 580 bp. All libraries were sequenced (7 sample per lane) using the Illumina HiSeq2000 instrument (www.illumina.com) (Table S1) using paired-end 2×100 bp reads. The reads were trimmed based on quality (>Q30). Transcripts were assembled and expression levels were estimated using the TopHat, Bowtie, and Cufflinks packages [17–20] and CLC Genomics Workbench (CLC Bio). Reads were mapped at 90% length, 90% identity and reads that mapped to more than one location in the genome were excluded. A minimum of 4 reads in each condition were required to analyze gene expression, over 88% of genes had over 4 reads in every condition. The ERCC controls (Invitrogen) were used to normalize gene expression values between samples following manufacturer’s instructions. ERCC transcripts showed high correlation between samples (R2= 0.92) demonstrating high reproducibility in sample preparation and sequencing. Differentially expressed genes between 30 min or 120 min and normoxia were determined in triplicate data using SAM with a false discovery rate of 0 as implemented in MeV [21]. GO term enrichment analysis was done using EASE [21] in MeV or GOEAST server [22].
Improvement of gene annotations
Existing gene models were improved by comparing RNAseq-predicted transcripts using PASA [23]. Only modifications that were supported by 4 or more reads were accepted. 5’ and 3’ UTR were detected both by comparing de novo transcripts using PASA as above and also by analyzing coverage upstream and downstream of the predicted start and stop codons of each annotated ORF. The longest continuous chromosomal regions that were covered with at least 3 reads were considered part of the UTRs. When candidate UTRs extended into neighboring annotated ORFs or UTRs, they were considered overlapping and were not included.
Accession numbers and data availability
The RNAseq data was deposited to SRA under accession number SRP007489.
RESULTS
Hypoxia shows differential expression of a large fraction of the genome
We studied transcript levels of Aspergillus fumigatus genes after a switch from normoxia to hypoxic conditions after 30 and 120 minutes. We obtained between 16 and 30 million reads per sample, with ~90% of reads mapped to the A1163 genome, and 66% of the reads mapped directly to genes, including the ribosomal locus. We performed de novo transcript identification using at least three different tools and merging their predictions. We found that the correct start and/or stop boundaries of 597 genes (6%) could be detected. Similarly, internal exon/intron boundary structures were improved for 157 genes (1.6%), some being either novel introns, or refuting the presence of a predicted intron. Interestingly, in our small sampling we found evidence for alternatively spliced isoforms of 11 genes. We used the transcripts to identify both 5’ and 3’ UTRs for non-overlapping genes. The average UTR length was close to 600 bp, but ranged from 3 to >1000 bp. In addition to structural variations in protein-coding genes, we found evidence of transcription for all predicted ncRNAs greater than 200 bp [15].
The RNAseq results showed that over 88% of protein-coding genes were expressed, approximately 10% of the genes with low expression (< 4 RPKM) (Table S2). Transcript levels spanned 4 orders of magnitude (10 – 20,000 RPKM), showing a wide dynamic range of transcript abundances. Samples of the same time point were significantly reproducible (R2> 0.9) whereas comparison of any hypoxic sample with the normoxic condition was dissimilar (R2=0.72). Expression analysis data showed drastic, genome-wide changes at both the 30 min and 120 min hypoxia time points, with roughly 23% and 35% of the genes significantly differentially expressed, respectively. As a general rule, if genes were repressed at 30 min their expression continued to be repressed at 120 min (Figure 1), though the magnitude of the change was significantly different (see below). Functional enrichment of gene ontology (GO) terms showed repression of transcription, protein synthesis and amino acid biosynthesis at both time points (Table 1). Pathway analysis showed that glycolysis, TCA cycle, and steroid biosynthesis were increased in response to hypoxia (Table S3).
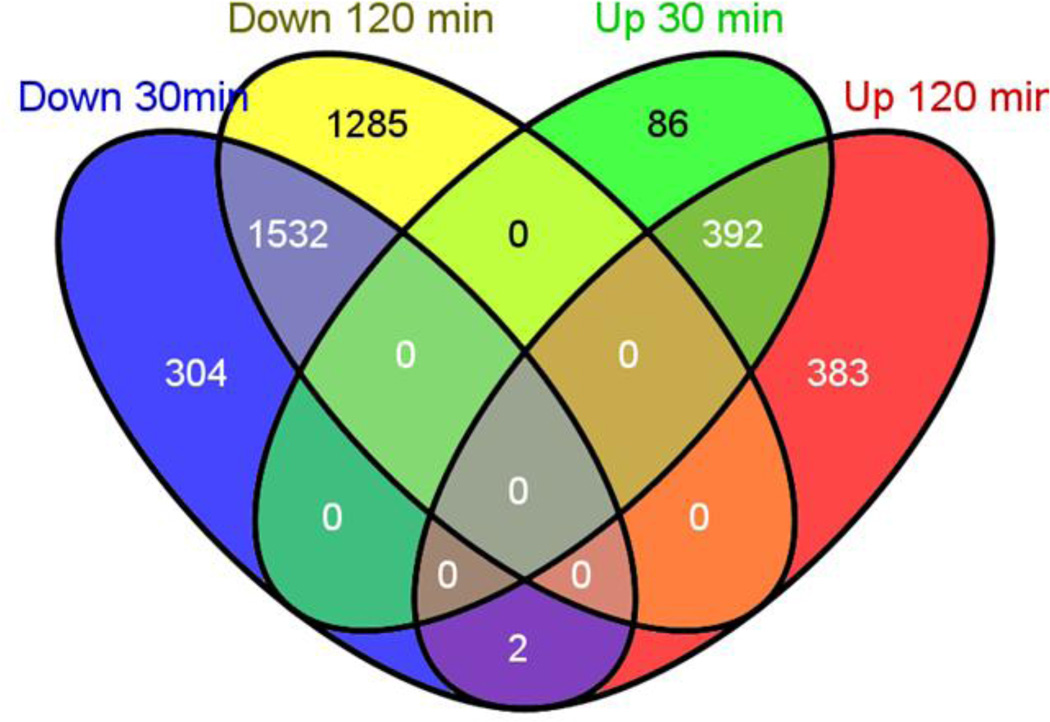
Figure 1. Differentially expressed genes in A. fumigatus in hypoxia are similar at 30 and 120 minutes.
Venn diagram showing the intersection of up- or down-regulated transcripts at the different time points. More genes were differentially regulated at 120 minutes. Most genes were regulated in the same way at 30 or 120 minutes, few genes were uniquely differentially expressed at a single time point.
Table 1.
Gene Ontology enrichment of differentially expressed genes in hypoxia.
| GO Term1 | Fold2 difference 30 min |
Fold2 difference 120 min |
Description | p-value3 |
|---|---|---|---|---|
| GO:0006396 | 0.14 | 0.05 | RNA processing | 1.90 × 10−9 |
| GO:0006412 | 0.23 | 0.14 | Translation | 3.61 × 10−3 |
| GO:0006520 | 0.35 | 0.27 | Cellular amino acid metabolic process | 2.70 × 10−5 |
| GO:0006725 | 0.17 | 0.12 | Cellular aromatic compound metabolic process | 7.10 × 10−8 |
| GO:0006807 | 0.17 | 0.12 | Nitrogen compound metabolic process | 7.22 × 10−9 |
| GO:0008033 | 0.14 | 0.07 | tRNA processing | 6.31 × 10−3 |
| GO:0030515 | 0.21 | 0.12 | snoRNA binding | 2.51 × 10−4 |
| GO:0034062 | 0.17 | 0.10 | RNA polymerase activity | 7.13 × 10−6 |
| GO:0034660 | 0.17 | 0.08 | ncRNA metabolic process | 3.98 × 10−3 |
| GO:0044452 | 0.21 | 0.14 | Nucleolar part | 7.74 × 10−3 |
The 10 most representative GO terms with the highest level ontology are represented.
Average fold differences of transcripts associated with each GO term as compared to normoxia. Values below zero signify down-regulation in hypoxia.
p-values as calculated by hypergeometric distributions in the GOEAST server [22].
The magnitude of the change in transcript levels for many genes (268 up-regulated and 285 down-regulated) was significantly different between 30 and 120 minutes suggesting that responses at 30 minutes are amplified at 120 minutes (Table S4). For 126 genes, the response was delayed until 120 minutes, because the genes were not significant at 30 minutes. Seventeen known or putative transcriptional regulators were differentially regulated between 30 and 120 minutes (Table 2), including some involved in stress response and cellular morphology and division (e.g. StuA). These transcriptional regulators might be responsible for propagating early responses to hypoxia for prolonged periods.
Table 2.
Transcription factors differentially regulated at later (120 minutes) compared to early (30 minutes) exposure to hypoxia.
| A1163 gene id |
AF293 gene id |
Description | Fold1 difference |
|---|---|---|---|
| AFUB_015380 | Afu1g15850 | C6 transcription factor, putative | 17.55 |
| AFUB_015560 | Afu1g16220 | C6 transcription factor, putative | 13.33 |
| AFUB_037000 | Afu8g06010 | C6 transcription factor, putative | 8.93 |
| AFUB_004520 | Afu8g01150 | C6 transcription factor, putative | 7.89 |
| AFUB_001060 | Afu6g13680 | APSES transcription factor Xbp1, putative | 7.38 |
| AFUB_039500 | Afu3g09670 | C6 transcription factor OefC | 6.07 |
| AFUB_066180 | Afu4g09080 | C2H2 transcription factor (Seb1), putative, putative | 6.02 |
| AFUB_023920 | Afu2g07900 | APSES transcription factor StuA | 5.01 |
| AFUB_036440 | Afu3g12730 | Transcription factor TFIIIB complex subunit Brf1, putative | 4.27 |
| AFUB_015020 | Afu1g15470 | C6 transcription factor (UaY), putative | 3.70 |
| AFUB_009640 | Afu1g10230 | C2H2 transcription factor (Egr2), putative | 3.69 |
| AFUB_016540 | Afu1g17150 | C6 transcription factor, putative | 3.66 |
| AFUB_022410 | Afu2g05380 | C6 transcription factor, putative | 3.33 |
| AFUB_080790 | Afu8g02640 | C6 transcription factor, putative | 0.22 |
| AFUB_013000 | Afu1g13510 | C6 transcription factor FacB/Cat8 | 0.20 |
| AFUB_080380 | Afu8g07360 | C6 transcription factor, putative | 0.18 |
| AFUB_077130 | Afu6g11110 | C6 transcription factor, putative | 0.06 |
Values below above and below one signify up or down-regulation at 120 minutes compared to 30 minutes, respectively.
Comparison with microarrays and later time points
RNAseq provides finer detail on the genomic and transcriptomic adaptations to varying physiological conditions compared to our previous microarrays conducted at 2 h, 6, 12 h, and 24 h [9]. For example, a much lower limit of detection is obtained and identification of non-coding regions is possible. Due to the differences in sensitivity and dynamic range between the two platforms, neither expression levels nor fold differences between conditions are directly comparable. Instead, we compared the results from gene ontology and pathway enrichment. The GO and pathways increased and reduced in response to hypoxia were similar between early and late time points (Tables 1 and S3). Namely, the reduced transcript levels of ribosome biosynthesis, purine and pyrimidine biosynthesis, and proteasome activity were also observed in early time points. RNAseq data, however, did not reveal a reduction in transcript levels of oxidative phosphorylation genes at 30 minutes, only at 120 minutes. We observed a significant down-regulation at 120 minutes of glycan and glycerolipid synthesis. Transcript levels of ergosterol biosynthetic genes were also similarly regulated for all timepoints (Table S5). Both srbA and srbB transcripts were significantly increased at 30 and 120 minutes. erg3A, erg3B, erg7, erg24, and erg25A were all significantly increased under hypoxia at 30 and 120 minutes. We also observed an increase in erg11A (>64 fold increase), which was not included in the microarray design. Erg3C, erg4B, and hmgA were significantly decreased for all timpoints. We detected the increase of erg10A (2 – 7 fold increase), but a significant decrease of erg10B (3.5 – 11 fold decrease). The data here support our initial observations and hypothesis that transcript levels for sterol and cell wall biosynthetic enzymes are changed as a consequence of lowered precursor availability potentially leading to cell envelop structures better suited for lower oxygen concentrations. Using RNAseq we did not detect any significant up-regulation of histone deacetylases as we had observed using microarrays, which might indicate that those genes are not affected until longer exposure to hypoxic conditions.
Hypoxia-specific genes
We found thirty-two genes consistently expressed only in hypoxia at both 30 and 120 minute (>20 RPKM, Table 3), which are likely to be specifically responsive to oxygen deprivation. These include a sensor histidine kinase response regulator (AFUB_101210) and a C6 transcriptional factor (AFUB_088390). The receiver domain of AFUB_101210 is 34% identical to the Saccharomyces cerevisiae and Candida albicans SSK1, while the histidine kinase domain is 27% identical to C. albicans NIK1. These proteins form part of a two-component response regulator system involved in hyphal development and virulence regulation. An AFUB_101210 null mutant did not have a significant growth defect or morphology change in hypoixia. Interestingly, the null mutant had a significant altered morphology when ethanol was used as the sole carbon source in hypoxia (data not shown). The data suggest that this histidine kinase might be specifically involved in ethanol utilization in hypoxia. In C. albicans, SSK1 is required for adaptation to oxidative stress [24] and in A. fumigatus NIK1 was associated with development, osmotic stress, and antifungal resistance [25]. AFUB_088390 is related to NIT4, a nitrate utilization transcription factor that also is involved hyphal and spore formation in Neurospora crassa [26,27]. Interestingly, neither gene was identified among the differentially expressed genes using microarrays at later time points. These results suggest that specific transcriptional regulators are activated early during hypoxia and may activate signaling cascades that amplify and extend the response to hypoxia.
Table 3.
Identification of hypoxia-specific genes.
| Gene Id | Description | 30 min1 | 120 min1 |
|---|---|---|---|
| AFUB_001360 | Conserved hypothetical protein | 17.99 | 118.99 |
| AFUB_001370 | Conserved hypothetical protein | 36.97 | 47.26 |
| AFUB_004010 | Exo-beta-1,3-glucanase (Exg1), putative | 10.52 | 12.98 |
| AFUB_017290 | Conserved hypothetical protein | 12.06 | 22.28 |
| AFUB_017390 | Beta-alanine synthase, putative | 13.12 | 14.98 |
| AFUB_029260 | Conserved hypothetical protein | 22.35 | 5.87 |
| AFUB_031180 | Conserved hypothetical protein | 16.03 | 26.82 |
| AFUB_036240 | FAD binding domain protein | 6.87 | 7.75 |
| AFUB_036410 | Conserved hypothetical protein | 42.71 | 26.78 |
| AFUB_039440 | Hypothetical protein | 7.04 | 20.07 |
| AFUB_044650 | 67 kDa myosin-cross-reactive antigen family protein | 37.05 | 126.13 |
| AFUB_047120 | Acetyltransferase, GNAT family family | 19.98 | 16.56 |
| AFUB_047130 | Short-chain dehydrogenase/reductase family protein, putative | 16.90 | 18.53 |
| AFUB_047950 | Integral membrane protein | 34.26 | 117.72 |
| AFUB_049300 | Integral membrane protein | 9.85 | 20.08 |
| AFUB_054380 | Hypothetical protein | 40.62 | 26.44 |
| AFUB_062190 | Beta-lactamase, putative | 6.94 | 13.84 |
| AFUB_062200 | Conserved hypothetical protein | 20.33 | 30.13 |
| AFUB_062380 | Conserved hypothetical protein | 20.81 | 33.93 |
| AFUB_068810 | Conserved hypothetical protein | 25.32 | 8.58 |
| AFUB_081050 | UDP-glucose,sterol transferase, putative | 35.99 | 36.32 |
| AFUB_084110 | Conserved hypothetical protein | 26.44 | 9.22 |
| AFUB_084920 | Hypothetical protein | 19.99 | 27.19 |
| AFUB_088390 | C6 transcription factor, putative | 20.80 | 19.13 |
| AFUB_090060 | Conserved hypothetical protein | 44.29 | 17.93 |
| AFUB_090930 | Stearoyl-CoA desaturase | 30.42 | 61.33 |
| AFUB_091830 | Conserved hypothetical protein | 14.78 | 24.77 |
| AFUB_092360 | Hypothetical protein | 37.37 | 19.08 |
| AFUB_094970 | MFS multidrug transporter, putative | 12.29 | 7.41 |
| AFUB_100020 | MFS monosaccharide transporter, putative | 12.07 | 7.23 |
| AFUB_101210 | Sensor histidine kinase/response regulator, putative | 48.25 | 21.65 |
Fold difference compared to normoxia conditions. Hypoxia specific genes were identified as having < 4 RPKM in normoxia and >20 RPKM in hypoxia.
Interestingly, we found evidence for a non-coding RNA (Afu-182) whose expression was high under normoxic conditions and ~40 times lower in hypoxia (Figure 2). The 216 bp element found on chromosome 4 is conserved in A. nidulans, A. oryzae and A. niger [15]. It is located within the 3’-UTR of the putative INSIG homolog (AFUB_064770). In mammals, INSIG regulates sterol homeostasis through HMG-CoA reductase degradation under sterol replete conditions and sterol mediated retention of the Scap-SREBP complex on the ER membrane [28]. In yeast, the INSIG homolog Ins1 does not participate in regulation of the yeast SREBP homolog Sre1 but regulates sterol homeostasis through interactions with HMG-CoA reductase [29]. The A. fumigatus Ins1 homolog (insA) transcript had a minor 2-fold reduction in hypoxia, suggesting that the ncRNA was specifically decreased compared to the protein-coding gene. Many non-coding RNAs function as anti-sense RNAs targeting mRNA to regulate transcription and/or translation. A motif analysis of this element identified two putative microRNAs within the locus. Predicted secondary structure revealed typical microRNA organization, approximately 65 nucleotides each with a stem loop arrangement [30] (Figure 3). The candidate microRNA sequences were used to identify 5’-seed and 3’pairing sequences and compared to newly identified 3’- UTRs for potential binding targets. Twelve genes had motifs that matched the first candidate microRNA (Table 4), including erg5, and hmgX. Nine of those genes were significantly differentially regulated in hypoxia, 3 up-regulated and 9 down-regulated. Five 3’-UTRs had potential targets for the second microRNA, however, these genes were poorly annotated and not significantly regulated under hypoxia (data not shown). The role of Afu-128 as an oxygen-sensing mechanism merits further investigation.
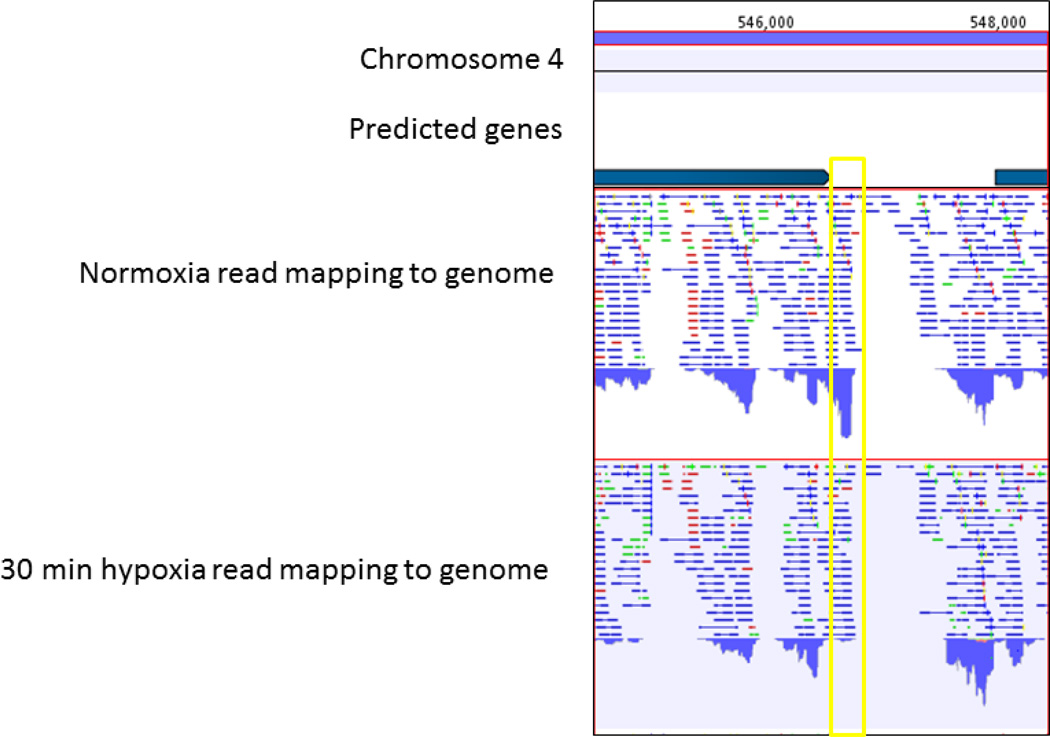
Figure 2. Hypoxia repression of the ncRNA Afu-182 detected by RNAseq.
RNAseq reads were mapped to A1163 genome and expression of non-coding elements >200bp was inspected visually. The genes on chromosome 4 are depicted as dark blue rectangles with arrow demonstrating the coding strand; coordinates refer to location of chromosome 4. Paired-reads (blue), forward (green), and reverse reads were mapped and accumulated if > than 40 read depth as bar graphs (below read mapping). The figure highlights the coding region and 3’UTR of AFUB_064770, Afu182, intergenic region, and the 5’UTR and beginning of AFUB_064780.
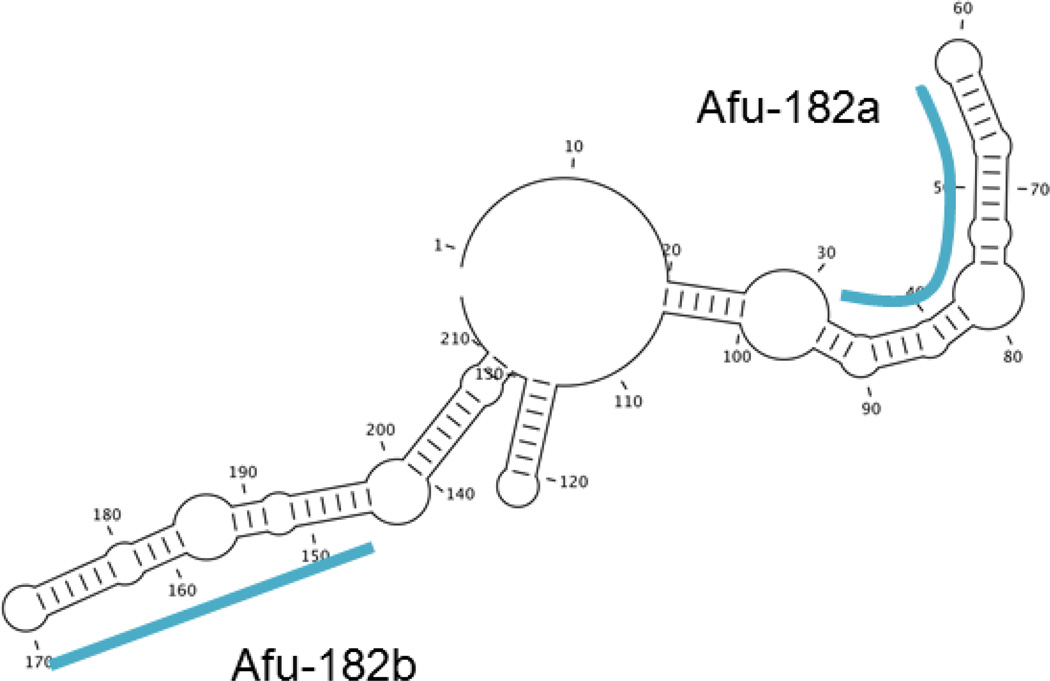
Figure 3. Predicted secondary structure of Afu182 suggests presence of microRNAs.
The secondary structure was predicted using PFOLD demanding the lowest free energy. Two candidate microRNA motifs (blue) were detected and used for identification of potential 5’ seed and gene targets in the genome.
Table 4.
Target sequence and expression of candidate Afu-182 ncRNA regulated genes.
| Sequence | 5' seed match |
3' pairing sequence |
A1163 annotation | Comments1 | Fold2 change |
|---|---|---|---|---|---|
| AFUB_057650 | TTCCTAC | CTATCCTCC | Cytochrome b5 reductase, putative | Cbr1 homolog | 5.28 |
| AFUB_025410 | TTCCTAC | ATATCCTCC | Cytochrome P450 family protein, putative | Erg5/Cyp52g1 | 4.85 |
| AFUB_082079 | TTCCTAC | TTATCCTTC | Hypothetical protein | 3.70 | |
| AFUB_021280 | TTCCTAC | TTATCTTCC | Conserved hypothetical protein | HmgX homolog | 1.47 |
| AFUB_005310 | TTCCTAC | ATATCCTCC | Small monomeric GTPase SarA, putative | 0.96 | |
| AFUB_005320 | TTCCTAC | ATATCCTCC | Serine/threonine protein phosphatase PP1, putative | 0.89 | |
| AFUB_046020 | TTCCTAC | TTATCCTTC | Pyrroline-5-carboxylate reductase | SrbA-regulated | 0.78 |
| AFUB_064770 | TTCCTAC | TTATCCTCC | Conserved hypothetical protein | INSIG-homolog | 0.55 |
| AFUB_025420 | TTCCTAC | ATATCCTCC | Biotin apo-protein ligase, putative | 0.35 | |
| AFUB_057110 | TTCCTAC | TTATCATCC | Ankyrin repeat protein (Yar1), putative | Unfolded protein binding | 0.15 |
| AFUB_046380 | TTCCTAC | TTGTCCTCC | Conserved hypothetical protein | 0.15 | |
| AFUB_061230 | TTCCTAC | TTCTCCTCC | 3–5 exoribonuclease (Csl4), putative | 0.15 |
Additional information obtained from AF293 annotation [33].
Average fold difference at 30 and 120 minutes compared to normoxia. Values above and below zero signify up- and down-regulation in hypoxia, respectively.
DISCUSSION
In treating patients with IA, limiting fungal growth, and therefore damage to host tissues, is a major therapeutic goal. Thus, exploring and defining mechanisms of fungal growth in the context of the infection microenvironment is a significant goal expected to yield an urgently needed therapeutic advance. Hypoxia is a major component of the immediate microenvironment surrounding the fungus in the lungs during IA [2,3]. However, mechanisms of obligate aerobic fungal adaptation to hypoxic conditions are relatively unstudied. We had previously studied the long-term adaptation of A. fumigatus to hypoxia using microarrays [9]. Here, we used a more sensitive platform to characterize early A. fumigatus transcriptional responses to hypoxia. Most significantly, we found that a large fraction (>23%) of genes were responsive to hypoxia as early as 30 minutes. Several transcription factors were differentially expressed, and seventeen of those factors changed with longer exposures. We found a small set of genes whose expression was only detected in hypoxia, including a C6 transcriptional regulator and a sensor histidine kinase response regulator. Both of these proteins share homology with regulators known to be involved in oxidative stress and nitrate metabolism in A. fumigatus and other pathogenic fungi. It is likely that early responses to hypoxia include the activation of specific transcriptional regulators, which affect transcript levels of other regulators that amplify and propagate the hypoxic signal.
The potential regulation of the hypoxia response in A. fumigatus by an ncRNA is a novel and exciting finding. Our data show that Afu-182 levels are significantly reduced under hypoxic conditions. This element is associated with the putative A. fumigatus INSIG homolog, insA. In mammals, INSIG functions as a chaperone for proteins containing sterol-sensing domains and promotes the ubiquitinylation and degradation of the HMG-CoA reductase (HMGR) [31], thus controlling the available cholesterol precursors. When cholesterol or oxysterol concentrations within the mammalian cell increase, INSIG-driven ubiquitinylation and degradation of HMGR leads to a reduction in the available sterol precursors, and ultimately cholesterol itself [31]. In contrast, the S. cerevisiae INSIG homolog, Nsg1p, although also involved in regulation HMGR abundances, stabilizes the protein and protect it from proteosomal degradation [32]. The role of ncRNAs in lipid and sterol metabolism in mammals is beginning to be understood. Several microRNAs control posttranscriptional expression of SREBP, INSIG, and cholesterol transporters [13,14]. A decrease of Afu-182 could be acting to promote translation of the AFUB_064770 transcript, increasing the abundance of the INSIG homolog, which in turn would result in a higher turnover of HMGR and lower accumulation of potentially toxic sterol intermediates in the cell. In addition to INSIG, we found that out of eleven additional candidate microRNA target genes, nine were differentially regulated in response to hypoxia, including two that are probably involved in sterol homeostasis (erg5 and hmgX). The actual mechanism of Afu-182 regulation of lipid metabolism and the hypoxia response in A. fumigatus is an area of ongoing research.
RNAseq data improved the annotation of the A. fumigatus A1163 genome. The resulting information allows better prediction of regulatory elements including the target sites for microRNAs as described above. The improvements from this A1163 RNAseq data combined with the AF293 improvements recently published [33] provide an invaluable resource for studying gene function, regulation, and evolution in A. fumigatus. Taken together, these data have identified new candidate genes and ncRNAs to explore in the context of A. fumigatus in vivo growth and host damage that are expected to yield a novel treatment advance for increasingly common and refractory cases of IA.
Supplementary Material
ACKNOWLEDGEMENTS
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contracts number HHSN272200900007C, and grant number R01AI81838 (RAC). BMB was supported with funds from the Freeport-McMoRan Foundation.
References
- 1.Rhodes JC. Aspergillus fumigatus: growth and virulence. Med Mycol. 2006;44(Suppl 1):S77–S81. doi: 10.1080/13693780600779419. [DOI] [PubMed] [Google Scholar]
- 2.Grahl N, et al. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS pathogens. 2011;7(7):e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willger SD, et al. Aspergillus fumigatus metabolism: clues to mechanisms of in vivo fungal growth and virulence. Medical Mycology. 2009;47(S1):S72–S79. doi: 10.1080/13693780802455313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Synnott JM, et al. Regulation of the hypoxic response in Candida albicans. Eukaryotic cell. 2010;9(11):1734–1746. doi: 10.1128/EC.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun CD, et al. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS pathogens. 2007;3(2):e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willger SD, et al. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS pathogens. 2008;4(11):e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blatzer M, et al. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS genetics. 2011;7(12):e1002374. doi: 10.1371/journal.pgen.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker BM, et al. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC genomics. 2012;13(1):62. doi: 10.1186/1471-2164-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker BM, et al. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genomics. 2012;13:62. doi: 10.1186/1471-2164-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawson RB. Control of lipid metabolism by regulated intramembrane proteolysis of sterol regulatory element binding proteins (SREBPs) Biochem Soc Symp. 2003;(70):221–231. doi: 10.1042/bss0700221. [DOI] [PubMed] [Google Scholar]
- 11.McPherson RA, Gauthier A, et al. Molecular regulation of SREBP function: the Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochem Cell Biol. 2004;82(1):201–211. doi: 10.1139/o03-090. [DOI] [PubMed] [Google Scholar]
- 12.Dong XY, Tang SQ. Insulin-induced gene: a new regulator in lipid metabolism. Peptides. 2010;31(11):2145–2150. doi: 10.1016/j.peptides.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 13.Jeon T-I, et al. An SREBP-Responsive microRNA Operon Contributes to a Regulatory Loop for Intracellular Lipid Homeostasis. Cell Metabolism. 2013;18(1):51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science. 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jöchl C, et al. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic acids research. 2008;36(8):2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cramer RA, et al. Immune responses against Aspergillus fumigatus: what have we learned? Current opinion in infectious diseases. 2011;24(4):315. doi: 10.1097/QCO.0b013e328348b159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. PMCID:2672628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 22.Hulsegge I, et al. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. doi: 10.1186/1753-6561-3-S4-S10. PMCID:PMC2712740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chauhan N, et al. Candida albicans Response Regulator Gene SSK1 Regulates a Subset of Genes Whose Functions Are Associated with Cell Wall Biosynthesis and Adaptation to Oxidative Stress. Eukaryotic cell. 2003;2(5):1018–1024. doi: 10.1128/EC.2.5.1018-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara D, et al. NikA/TcsC Histidine Kinase Is Involved in Conidiation, Hyphal Morphology, and Responses to Osmotic Stress and Antifungal Chemicals in Aspergillus fumigatus. PLoS ONE. 2013;8(12):e80881. doi: 10.1371/journal.pone.0080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu YH, et al. Sequence-specific DNA binding by NIT4, the pathway-specific regulatory protein that mediates nitrate induction in Neurospora. Molecular Microbiology. 1995;15(5):935–942. doi: 10.1111/j.1365-2958.1995.tb02362.x. [DOI] [PubMed] [Google Scholar]
- 27.Colot HV, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proceedings of the National Academy of Sciences. 2006;103(27):10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein JL, et al. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Burg JS, et al. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8(6):522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espenshade PJ, Hughes AL. Regulation of Sterol Synthesis in Eukaryotes. Annual review of genetics. 2007;41(1):401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 32.Flury I, et al. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. The EMBO Journal. 2005;24(22):3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerqueira GC, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2013;42(Database issue):D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.