Abstract
Membrane protein spectroscopic studies are challenging due to the difficulty introduced in preparing homogenous and functional hydrophobic proteins incorporated into a lipid bilayer system. Traditional membrane mimics such as micelles or liposomes have proved to be powerful in solubilizing membrane proteins for biophysical studies, however, several drawbacks have limited their applications. Recently, a nanosized complex termed lipodisq nanoparticles was utilized as an alternative membrane mimic to overcome these caveats by providing a homogeneous lipid bilayer environment. Despite all the benefits that lipodisq nanoparticles could provide to enhance the biophysical studies of membrane proteins, structural characterization in different lipid compositions that closely mimic the native membrane environment is still lacking. In this study, the formation of lipodisq nanoparticles using different weight ratios of POPC/POPG lipids to SMA polymers was characterized via solid-state nuclear magnetic resonance (SSNMR) spectroscopy and dynamic light scattering (DLS). A critical weight ratio of (1/1.25) for the complete solubilization of POPC/POPG vesicles has been observed and POPC/POPG vesicles turned clear instantaneously upon the addition of the SMA polymer. The size of lipodisq nanoparticles formed from POPC/POPG lipids at this weight ratio of (1/1.25) was found to be about 30 nm. We also showed that upon the complete solubilization of POPC/POPG vesicles by SMA polymers, the average size of the lipodisq nanoparticles is weight ratio dependent, when more SMA polymers were introduced, smaller lipodisq nanoparticles were obtained. The results of this study will be helpful for a variety of biophysical experiments when specific size of lipid disc is required. Further, this study will provide a proper path for researchers working on membrane proteins to obtain pertinent structure and dynamic information in a physiologically relevant membrane mimetic environment.
Keywords: lipodisq nanoparticle, POPC/POPG vesicle, 3:1 SMA polymer, dynamic light scattering, SSNMR spectroscopy
1. INTRODUCTION
Magnetic resonance spectroscopic studies of membrane proteins remain highly challenging due to the requirement of a membrane-mimicking environment that maintains the integrity and stability of membrane proteins outside their native cellular environment 1. The most commonly applied method to solubilize membrane proteins in aqueous solution is detergent-formed micelles 2, which are widely used to solve high resolution three-dimensional structures of membrane proteins 2,3. However, the lack of a lipid bilayer and the limitation of the size of micelles may not preserve the membrane proteins’ structural and dynamic integrities under physiological conditions 1,3,4,5.
To better maintain the structural integrity of membrane proteins, several classes of membrane mimics (e.g. liposomes, bicelles, or nanodiscs) comprising lipid bilayer have been developed previously 6,7,8,9. Despite the advantages, several drawbacks have limited their applications for spectroscopic studies. In the case of liposomes, homogenous liposome samples are not readily obtainable, especially when membrane proteins are to be incorporated. Also, the inaccessibility of liposomal interior raises challenges for the cytoplasmic domain studies of membrane proteins 10. Furthermore, it is not easy to incorporate large amounts of proteins into liposomes which is problematic given that sometimes a higher protein to lipid molar ratio is required for biophysical studies 11,12. A second alternative bicelles, are artificial lipid bilayer discs formed by the mixture of long-chain phospholipids (e.g. DMPC) and short-chain phospholipids (e.g. DHPC). Bicelles are favorable for the study of interactions within membrane proteins that are not retained in micelles 13,14. Bicelles are able to provide accessibility for the interaction study of both extracellular and cytoplasmic domains of membrane protein 10. However, the specific types of lipids amenable to bicelle formation limit its applications as the lipid compositions in the membrane have been shown to influence the function of antimicrobial peptides and amyloid peptides 10,15,16. The introduction of nanodiscs has provided a great tool to study membrane proteins in a native-like membrane environment 17,18. Nanodiscs consist of lipids surrounded by a membrane scaffold protein such as apolipoprotein to form a discoidal bilayer. Nanodiscs can be formed with different types of lipids, which gives nanodiscs great advantages over bicelles since many membrane proteins require specific types of lipids for structural folding and functional reconstitution 19,20,21. Also, it has been reported that membrane scaffold proteins helped to improve the stability of nanodiscs compared to other membrane-mimicking systmes 22. The drawbacks of using nanodiscs are that it requires detergent for protein incorporation and the absorbance properties of the membrane scaffold protein may interfere with the membrane protein of interest 9,17.
An alternative membrane mimic is highly desirable for the proper functional and structural characterization of membrane proteins. In this study, we characterize the recently developed lipodisq nanoparticle system (Figure S1 A) as a potential membrane mimic system 23,24,25. Unlike nanodiscs, lipodisq nanoparticles are formed from lipids solubilized by polymers instead of membrane scaffold proteins, thus they should not interfere with the absorbance properties of the membrane proteins of interest 26,27,23. Also, no detergent is needed for protein incorporation into the lipodisq nanoparticles. The polymer for solubilizing lipids consists of styrene and maleic acid (SMA) at a molar ratio of 3:1 (Figure S1 B). Though lipodisq nanoparticles show a high potential to become a good membrane mimetic to enhance biophysical studies of membrane proteins, its structural characterization in a native-membrane mimetic environment is still lacking. Here, we use POPC/POPG lipids at a molar ratio of 9/1 and SMA polymers to form lipodisq nanoparticles. POPC is the most common phospholipid found in mammalian cell membranes, and POPG was chosen to mimic the 10 – 20 molar percent anionic phospholipids typically found in mammalian membranes 28,29. The weight ratio of lipid to polymer was varied and different lipodisq nanoparticle samples were characterized by solid-state NMR (SSNMR) and dynamic light scattering (DLS). Transmission electron microscopy (TEM) was also used to confirm the size and homogeneity of vesicles and lipodisq nanoparticle samples.
2. MATERIALS AND METHODS
2.1 Materials
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (POPG), 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) (sodium salt) (LMPG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC) were purchased from Avanti Polar Lipids (Alabaster, Alabama, USA). 3:1 SMA polymer was purchased from Malvern Cosmeceutics (Tewkesbury, Gloucestershire, UK). N-[2-Hydroxyethyl]piperazine-N’-2-ethanesulfonic acid (HEPES) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Sodium chloride (NaCl) was purchased from Fisher Scientific (Pittsburgh, PA, USA).
2.2 Preparation of POPC/POPG vesicles and Lipodisq nanoparticles
POPC/POPG vesicles were composed of POPC and POPG with molar ratio of [POPC] / [POPG] = 9 / 1. POPC and POPG powdered lipids were suspended in the buffer (100 mM NaCl, 20 mM HEPES, pH 7.0) to a final concentration of 25 mM. Lipid slurry was vortexed vigorously to mix completely and vesicles were formed spontaneously after ten freeze/sonication cycles. Liposomes were than subject to extrusion through a 400 nm filter membrane followed by a 200 nm filter membrane. SMA polymers were dissolved in the buffer (100 mM NaCl, 20 mM HEPES, pH 7.0) to a final concentration of 5% (m/v) followed by 1 min of water-bath sonication to a complete dissolution. Lipodisq nanoparticles with different weight ratio of lipid to polymer (1/0.25, 1/0.5, 1/0.75, 1/1.25, 1/1.75, and 1/2.25) were formed by adding SMA polymers dropwise to the POPC/POPG vesicles. Samples were then mixed well by vortexing and equilibrated through two freeze/sonication cycles. Samples were allowed to equilibrate at room temperature overnight. For solid state NMR experiment, sample was concentrated to around 100 ul at 14,000 g using Amicon concentrator at cutoff of 3 kD according to manufacturer’s instructions.
2.3 Preparation of LMPG micelles and DMPC/DHPC bicelles
LMPG micelles were formed by dissolving LMPG in buffer (100 mM NaCl, 5 mM HEPES, pH 7.0) to a final concentration of 42 mM (20 mg/ml). DMPC/DHPC bicelles were composed of DMPC and DHPC with molar ratio of q = [DMPC] / [DHPC] = 3.2. DMPC and DHPC powdered lipids were dissolved in 5 mM HEPES buffer (pH 7.0) to a final concentration of 12% (m/v). Three cycles of the following: heating at 40 °C for 15 min, vortexing for 1min, cooling on ice for 15 min and again vortexing for 1 min, were performed until the lipid suspension was clear.
2.4 Dynamic light scattering (DLS)
DLS measurements were performed on a Zetasizer nano series (Malvern Instruments) at 25 °C in disposable 40 µl micro cuvettes. Data were collected for 20 sec and averaged for 10 scans. The distance distribution is shown on a log scale using Igor Pro (WaveMetrics).
2.5 Solid-state NMR spectroscopy
A 500 MHz WB Bruker Avance solid-state NMR spectrometer and a Bruker 4mm triple resonance NMR probe (Billerica, MA, USA) were used to collect the 31P solid-state NMR spectra. 31P NMR spectra were recorded with 1H decoupling using a 7 µs π/2 pulse for 31P and a 3 s recycle delay, 2 K scans were averaged, and the free induction decay was processed using 100 Hz of line broadening. The spectral width was set to 500 ppm. All of the data were collected at 25 °C except for DMPC/DHPC bicelle, which was collected at 42 °C. 31P NMR spectra were plotted using the software Igor Pro (WaveMetrics).
2.6 Transmission electron microscopy (TEM)
One drop of control POPC/POPG vesicle and lipodisq nanoparticle (1/1.25) samples were adsorbed to 200 mesh copper carbon-coated grids for 10 s for fully absorbance. Grids were stained with two drops of 1.5% ammonium molybdate. Images were recorded using a Joel-1200 electron microscope operated at 120 kV.
3. RESULTS AND DISCUSSIONS
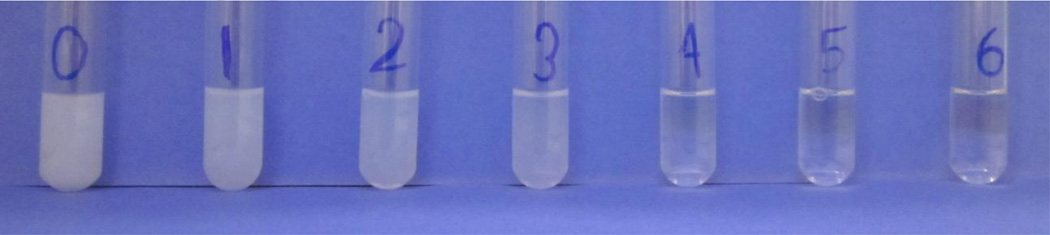
Figure 1 shows the visible observation of the transparency of POPC/POPG vesicles titrated with different amounts of SMA polymers. From left to right, the weight ratio of POPC/POPG lipid to SMA polymer gradually increased from (1/0) (tube 0) to (1/2.25) (tube 6). At a weight ratio of (1/0), the sample was opaque and cloudy. As the weight ratio increased, the sample appeared less cloudy. When the critical weight ratio of (1/1.25) (tube 4) was reached, the sample turned to clear, which indicated the formation of a homogenous sample. The addition of the SMA polymer instantaneously turned the sample clear, which indicates the potential of using lipodisq nanoparticles for efficient sample preparation. These results revealed a qualitative observation of the solubilization of POPC/POPG vesicles and demonstrated the optimal amount of SMA polymers needed to fully dissolve POPC/POPG vesicles at a weight ratio of (1/1.25). After the formation of liposomes, SMA lipodisq nanoparticles are quick and easy to prepare.
Figure 1.
Visible observation of lipodisq nanoparticles formation through the combination of POPC/POPG vesicles and SMA polymers at different lipid to polymer weight ratios and normal POPC/POPG vesicles as a control: (0) control POPC/POPG vesicles, (1) lipodisq nanoparticles (1/0.25), (2) lipodisq nanoparticles (1/0.5), (3) lipodisq nanoparticles (1/0.75), (4) lipodisq nanoparticles (1/1.25), (5) lipodisq nanoparticles (1/1.75), (6) lipodisq nanoparticles (1/2.25).
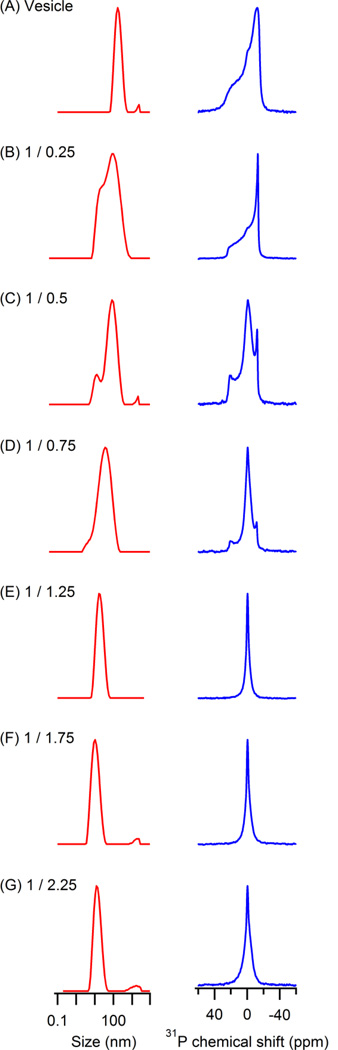
31P SSNMR and DLS experiments were performed to investigate the formation of lipodisq nanoparticles from POPC/POPG vesicles and SMA polymers. 31P SSNMR spectra provided unique structural and dynamic information on the phospholipid head groups, and DLS measured the size distribution of each sample. Both experiments were performed and the results were compared side by side. Figure 2 shows DLS data (left, red) and the corresponding 31P SSNMR data (right, blue) for control POPC/POPG vesicles and lipodisq nanoparticles formed with different weight ratios of lipid to polymer. Both DLS and 31P SSNMR results clearly indicate a change in the size of POPC/POPG vesicles from larger to smaller upon adding SMA polymers. As observed in Figure 2 A through E, there was a gradual shift of average size from about 250 nm to about 30 nm (see Table 1). Furthermore, a broader size distribution was observed when the weight ratio of lipid to polymer was greater than (1/1.25), which indicated an incomplete dissolution of POPC/POPG vesicles at these weight ratios and the existence of heterogeneity in these samples (Figure 2 B – D, The right most small peak may be due to small amounts of sample inhomogeneity which is not significant when compared to the main peak). This transition was supported by the corresponding 31P SSNMR results. An obvious 31P powder pattern shape was observed in Figure 2A, and this powder pattern gradually decreased from Figure 2B through 2D, accompanied by a gradual increasing 31P isotropic peak. When the weight ratio of (1/1.25) was reached, as shown in Figure 2E, the 31P powder pattern disappeared and only the isotropic peak was left, suggesting the formation of small lipodisq nanoparticles corresponding to fast isotropic motion. This formation of an isotropic peak when the weight ratio of lipid to polymer was (1/1.25) corresponded to the complete dissolution of the lipids as observed in Figure 1. Furthermore, inspection of the DLS and SSNMR results from samples with weight ratios of (1/0.75) to (1/1.25), revealed a change in particle size from about 90nm to about 30nm on average and a change in chemical shift width from 45 ppm to 6 ppm (see Table 1), clearly indicating the formation of small and homogenous lipodisq nanoparticles at the specific weight ratio of (1/1.25). These results indicated that the weight ratio was crucial in the formation of the lipodisq nanoparticles, and a threshold of weight ratio has to be reached in order to make a homogenous lipodisq nanoparticle suspension. We observed that the samples with weight ratios smaller than (1/1.25) (Figure 2 F and G), the uniformity of the lipodisq nanoparticle sample was unchanged, though the size of the particle was smaller (see Table 1). This result suggested that by modifying the weight ratio at the critical dissolution point, the lipodisq nanoparticles with a specific size can be formed. The DLS and 31P NMR studies are useful to help characterize lipodisq nanoparticles and for studying lipid systems when a specific size is required.
Figure 2.
DLS data (left, red) and 31P SSNMR data (right, blue) for control POPC/POPG vesicles and lipodisq nanoparticles generated from different lipid to polymer weight ratios: (A) control POPC/POPG vesicles, (B) lipodisq nanoparticles (1/0.25), (C) lipodisq nanoparticles (1/0.5), (D) lipodisq nanoparticles (1/0.75), (E) lipodisq nanoparticles (1/1.25), (F) lipodisq nanoparticles (1.75), (G) lipodisq nanoparticles (1/2.25).
Table 1.
Particle size in nm and 31P NMR spectrum line shape width in ppm of each sample.
| Sample names | Vesicles | Lipodisq nanoparticles | Bicelles | Micelles | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1/0.25 | 1/0.5 | 1/0.75 | 1/1.25 | 1/1.75 | 1/2.25 | ||||
| Particle size (nm) | 250 | 190 | 40–180 | 90 | 31 | 19 | 15 | 9 | 3 |
| 31P Line shape width (ppm) | 42 | 38 | 35 | 34 | 6 | 8 | 10 | 1 | 1.5 |
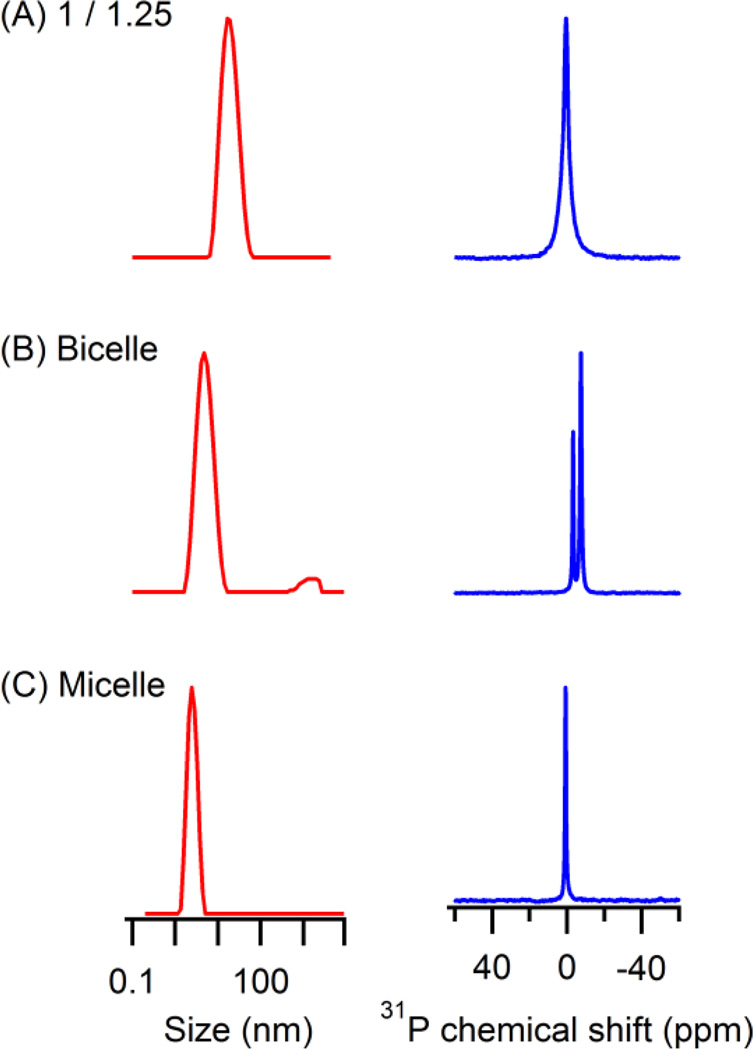
In order to further characterize the lipodisq nanoparticles as a model membrane mimic, 31P powder spectrum and size distribution of the lipodisq nanoparticles at the weight ratio of (1/1.25) were compared with other model membrane mimics (LMPG micelles and DMPC/DHPC bicelles). Figure 3 shows DLS data (left, red) and 31P SSNMR data (right, blue) of these three membrane mimics. DLS analysis of the particles demonstrated that the lipodisq nanoparticles had a narrow size distribution, which is comparable to micelles and bicelles. Under these sample conditions, the lipodisq nanoparticles are larger in size than both the micelle and bicelle samples. This is advantageous for incorporating larger integral membrane proteins. The isotropic 31P NMR linewidths are broader for the lipodisq nanoparticle sample when compared to the micelle and bicelle samples. This is because of the larger size and the corresponding slower motion of the lipodisq nanoparticles in solution. Also, the 31P NMR spectrum of the lipodisq nanoparticles is comparable to another commonly utilized powerful membrane mimic system called nanodiscs that have been reported previously in the literature 30. This result indicated that the homogeneity of the lipodisq nanoparticles has strong potential to serve as an alternative membrane mimic. Most importantly, the larger size and lipid bilayer properties of the lipodisq nanoparticles shows great promise over micelles and bicelles as a membrane mimic.
Figure 3.
Comparison of DLS data (left, red) and SSNMR data (right, blue) of lipodisq nanoparticles (1/1.25) at 25 °C, DMPC/DHPC bicelles at 42 °C, and LMPG micelles at 25 °C.
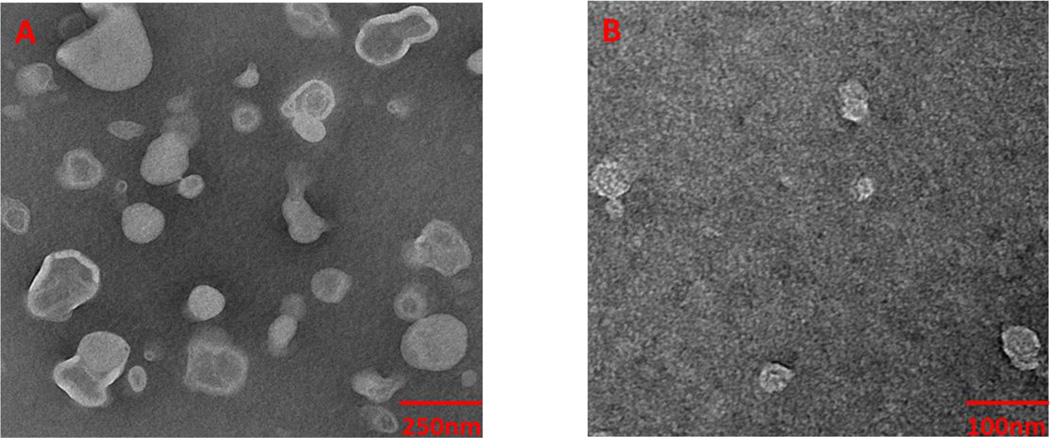
To confirm the size of the lipodisq nanoparticles obtained from the DLS measurements, transmission electron microscopy (TEM) experiments were conducted. Figure 4 shows TEM micrographs of uranyl acetate stained POPC/POPG vesicles and lipodisq nanoparticles (1/1.25). An average size of about 240 nm for POPC/POPG vesicles and about 35 nm for lipodisq nanoparticles (1/1.25) were obtained, which agreed with the analysis of the particles by DLS (Figures 2 and 3). Also, TEM results indicated that the lipodisq nanoparticle (1/1.25) sample was more homogenous in size when compared to POPC/POPG vesicles, which is a significant improvement in membrane mimetic systems, making the lipodisq nanoparticle system very attractive as an alternative method for membrane mimicking systems.
Figure 4.
TEM micrographs of uranyl acetate stained (A) control POPC/POPG vesicles and (B) lipodisq nanoparticles (1/1.25).
In conclusion, the formation of lipodisq nanoparticles from POPC/POPG lipids and SMA polymers was successfully demonstrated. A critical weight ratio (lipid to polymer) of (1/1.25) or smaller was required for the complete dissolution of POPC/POPG vesicles to form homogenous lipodisq nanoparticles. Also, it was found that the lipid composition played an important role in making lipodisq nanoparticle samples. Furthermore, lipodisq nanoparticle samples of specific size can be formed by modifying weight ratio, which is useful for biophysical studies of membrane proteins when different sizes of membrane mimic are needed. Future studies will focus on the characterization of membrane proteins in the lipodisq nanoparticle system, especially on the study of protein-protein complexs. Overall, lipodisq nanoparticles are a new membrane mimic system that shows great potential to advance biophysical studies of membrane proteins.
Supplementary Material
Highlights.
POPC/POPG vesicles are titrated with SMA polymers to form lipodisqs.
The more SMA were added, the smaller lipodisqs were formed.
Lipodisqs became clear at the weight ratio of (1/1.25).
ACKNOWLEDGMENT
This work was generously supported by National Institutes of Health Grants R01 GM108026. Funding was also provided by National Science Foundation (NSF) Grant CHE - 1305664.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
REFERENCE
- 1.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 2.Prive GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41(4):388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Tate CG. Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol Biol. 2010;601:187–203. doi: 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- 4.van den Brink-van der Laan E, Chupin V, Killian JA, de Kruijff B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry. 2004;43(14):4240–4250. doi: 10.1021/bi036129d. [DOI] [PubMed] [Google Scholar]
- 5.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys J. 1999;76(5):2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666(1–2):105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 7.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J Am Chem Soc. 2004;126(47):15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis AA, Opella SJ. Bicelle samples for solid-state NMR of membrane proteins. Nat Protoc. 2007;2(10):2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- 9.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12(11):2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20(4):471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geertsma ER, Nik Mahmood NA, Schuurman-Wolters GK, Poolman B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc. 2008;3(2):256–266. doi: 10.1038/nprot.2007.519. [DOI] [PubMed] [Google Scholar]
- 12.Fang G, Friesen R, Lanfermeijer F, Hagting A, Poolman B, Konings WN. Manipulation of activity and orientation of membrane-reconstituted di-tripeptide transport protein DtpT of Lactococcus lactis. Mol Membr Biol. 1999;16(4):297–304. doi: 10.1080/096876899294517. [DOI] [PubMed] [Google Scholar]
- 13.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. Embo j. 2009;28(9):1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee D, Walter KF, Bruckner AK, Hilty C, Becker S, Griesinger C. Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130(42):13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 15.Vold RR, Prosser RS. Magnetically Oriented Phospholipid Bilayered Micelles for Structural Studies of Polypeptides. Does the Ideal Bicelle Exist? Journal of Magnetic Resonance, Series B. 1996;113(3):267–271. [Google Scholar]
- 16.Dürr UHN, Gildenberg M, Ramamoorthy A. The Magic of Bicelles Lights Up Membrane Protein Structure. 2012 doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390(8):805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 19.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. 2013 doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126(11):3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 23.Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Grobner G, Watts A. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew Chem Int Ed Engl. 2012;51(19):4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 24.Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett. 2012;12(9):4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 25.Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith HJ, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA. DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry. 2013;52(38):6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 2009;131(22):7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 27.Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas OR, Overduin M, Dafforn TR. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem Soc Trans. 2011;39(3):813–818. doi: 10.1042/BST0390813. [DOI] [PubMed] [Google Scholar]
- 28.Gennis RB. Biomembranes: Molecular Structure and Function. New York: Springe-Verlag; 1989. [Google Scholar]
- 29.Coey AT, Sahu ID, Gunasekera TS, Troxel KR, Hawn JM, Swartz MS, Wickenheiser MR, Reid RJ, Welch RC, Vanoye CG, Kang C, Sanders CR, Lorigan GA. Reconstitution of KCNE1 into lipid bilayers: comparing the structural, dynamic, and activity differences in micelle and vesicle environments. Biochemistry. 2011;50(50):10851–10859. doi: 10.1021/bi2009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ. Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry. 2011;50(42):8983–8985. doi: 10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.