Abstract
Thirty years ago it was shown that the non-enzymatic, template-directed polymerization of activated mononucleotides proceeds readily in a homochiral system, but is severely inhibited by the presence of the opposing enantiomer.1 This finding poses a severe challenge for the spontaneous emergence of RNA-based life, and has led to the suggestion that either RNA was preceded by some other genetic polymer that is not subject to chiral inhibition2 or chiral symmetry was broken through chemical processes prior to the origin of RNA-based life.3,4 Once an RNA enzyme arose that could catalyze the polymerization of RNA, it would have been possible to distinguish among the two enantiomers, enabling RNA replication and RNA-based evolution to occur. It is commonly thought that the earliest RNA polymerase and its substrates would have been of the same handedness, but this is not necessarily the case. Replicating D-and L-RNA molecules may have emerged together, based on the ability of structured RNAs of one handedness to catalyze the templated polymerization of activated mononucleotides of the opposite handedness. Such a cross-chiral RNA polymerase has now been developed using in vitro evolution. The D-RNA enzyme, consisting of 83 nucleotides, catalyzes the joining of L-mono- or oligonucleotide substrates on a complementary L-RNA template, and similarly for the L-enzyme with D-substrates and a D-template. Chiral inhibition is avoided because the 106-fold rate acceleration of the enzyme only pertains to cross-chiral substrates. The enzyme's activity is sufficient to generate full-length copies of its enantiomer through the templated joining of 11 component oligonucleotides.
A potential advantage of a cross-chiral polymerase is that it offers a new mode of recognition between enzyme and substrates that avoids Watson-Crick pairing and therefore may provide greater sequence generality. Opposing enantiomers of RNA are unable to form contiguous base pairs5,6 and must instead recognize each other through tertiary interactions.7 Similar to the way a protein polymerase recognizes nucleic acids, a cross-chiral RNA polymerase might recognize the shape of the RNA duplex while being largely indifferent to the identity of the bases. Considerable progress has been made in developing D-RNA enzymes that polymerize D-RNA substrates,8,9 but these enzymes have strong sequence preferences10 that currently preclude the RNA-catalyzed replication of RNA, a defining function of RNA-based life.
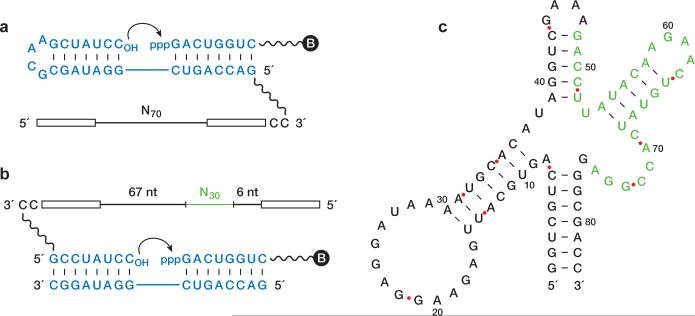
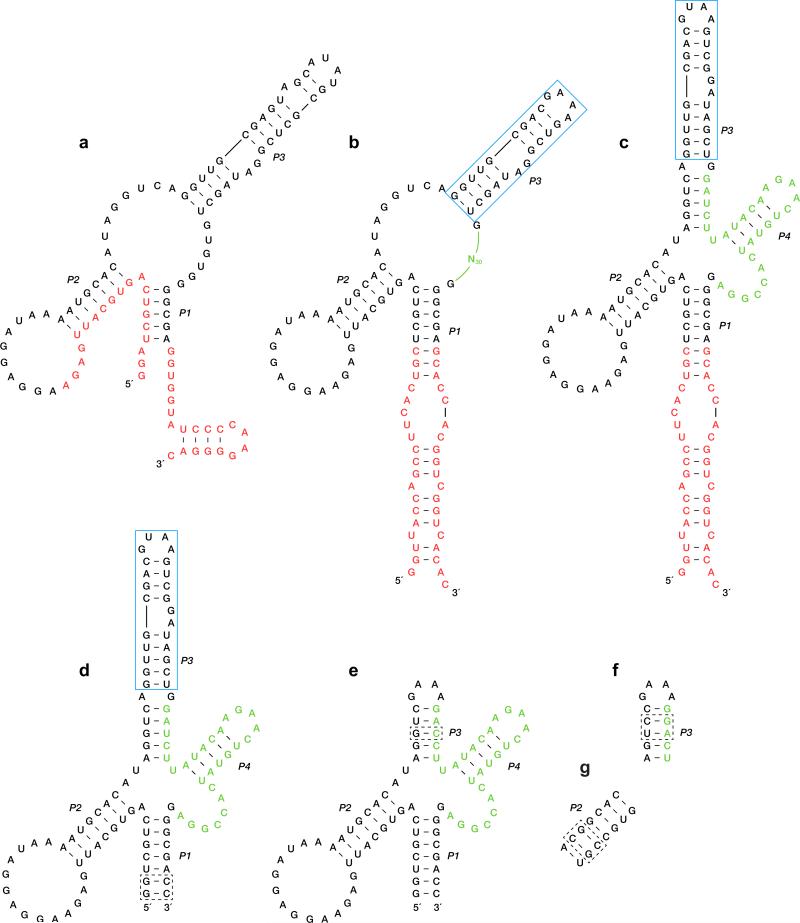
The search for a cross-chiral RNA polymerase began with a population of 1015 random-sequence D-RNAs that were tethered via a flexible linker to the template strand of a template-primer complex composed of L-RNA (Fig. 1a). A separate 5′-triphosphorylated, 3′-biotinylated L-oligonucleotide substrate was provided that could bind to the template adjacent to the primer. D-RNA molecules that catalyzed ligation of the substrate and primer were captured using streptavidin and selectively amplified. Following ten rounds of this procedure, a catalytic motif was identified and trimmed of extraneous nucleotides (Extended Data Figs 1a and 2a). This motif consists of a central core supported by three stem regions.
Figure 1. Evolution of a cross-chiral RNA ligase.
a, Reaction format during the first ten rounds of selective amplification, with the D-enzyme tethered to the L-template-primer complex, and with the 5′-triphosphorylated (ppp), 3′-biotinylated (B) L-substrate provided separately. L-nucleotides are shown in blue. The starting population contained 70 random-sequence nucleotides (N70), flanked by fixed primer-binding sites (open rectangles). Curved arrow indicates the site of ligation. b, Reaction format during rounds 11–16 of selective amplification, with the D-enzyme tethered to the L-primer, and with both the L-template and L-substrate provided separately. An additional 30 random-sequence nucleotides (N30, green) were inserted prior to round 11. c, Sequence and secondary structure of the final evolved enzyme. Nucleotides that derived from the N30 insert are shown in green.
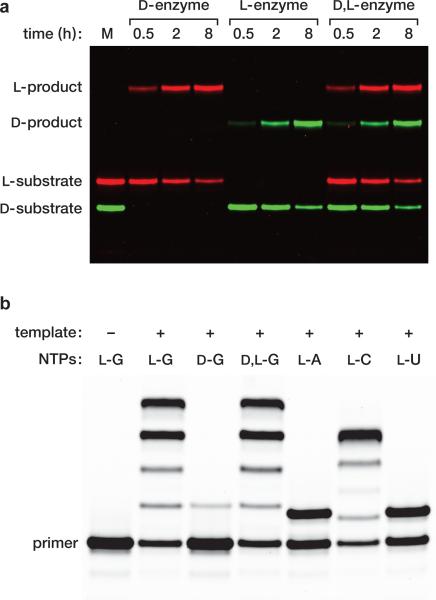
Figure 2. Cross-chiral ligation and polymerization.
a, Template-directed ligation of two oligonucleotides catalyzed by an RNA enzyme of the opposite handedness. The sequences of the substrates and template are as shown in Fig. 1b, but with the enzyme detached from the primer. The reactions employed 10 μM enzyme, 0.5 μM fluorescently-labeled upstream substrate, 4 μM downstream substrate, 2 μM template, 250 mM MgCl2, and 250 mM NaCl, which were incubated at pH 8.5 and 23 °C for 0.5, 2, or 8 h. The marker lane (M) contains the D and L upstream substrates alone, labeled with either fluorescein (green) or boron-dipyrromethene (red), respectively. (b) Template-directed polymerization of L-NTPs catalyzed by a D-RNA enzyme. The L-primer was tethered to the D-enzyme as shown in Fig. 1b and the L-template was provided separately. All templates had the primer-binding sequence shown in Fig. 1b, followed by 3′-CCCCAGUA-5′ for GTP addition, 3′-UUUUAGUA-5′ for ATP addition, 3′-GGGGAGUA-5′ for CTP addition, or 3′-AAAAAGUA-5′ for UTP addition. The reactions employed 0.5 μM enzyme-primer, 1 μM template, and 4 mM of the appropriate NTP, under conditions as above, except at 17 °C for 24 h. The reaction products were photocleaved to detach the extended primer prior to analysis by polyacrylamide gel electrophoresis.
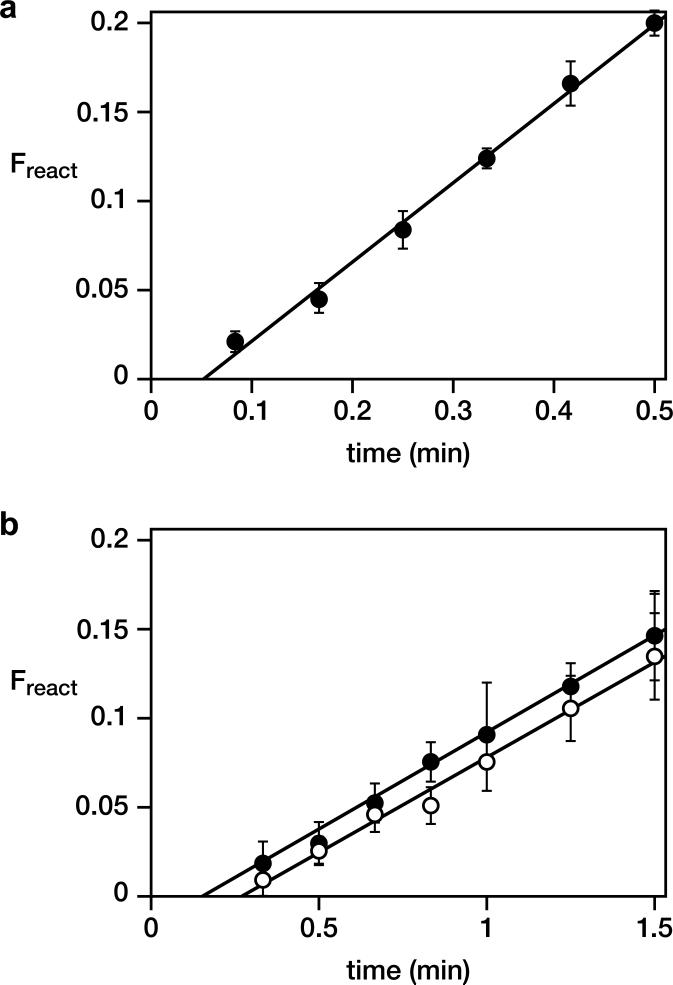
Next, four unpaired nucleotides within the central core were replaced by 30 random-sequence nucleotides (Extended Data Fig. 2b) and six additional rounds of selective amplification were carried out. For these additional rounds, the population of D-RNAs were tethered to the primer and both the template and substrate were provided as separate molecules (Fig. 1b). This was done to encourage the development of catalysts that are not beholden to a particular reaction format. An optimized D-enzyme was identified from the final evolved population (Extended Data Fig. 1b), again trimmed of extraneous nucleotides (Extended Data Fig. 2c–e), and resulting in an 83-nucleotide motif that catalyzes the ligation of L-RNA oligonucleotides on a L-RNA template (Fig. 1c). The rate of this reaction is 0.45 min–1 (Extended Data Fig. 3a), which is approximately 106-fold faster than the uncatalyzed rate of reaction.11
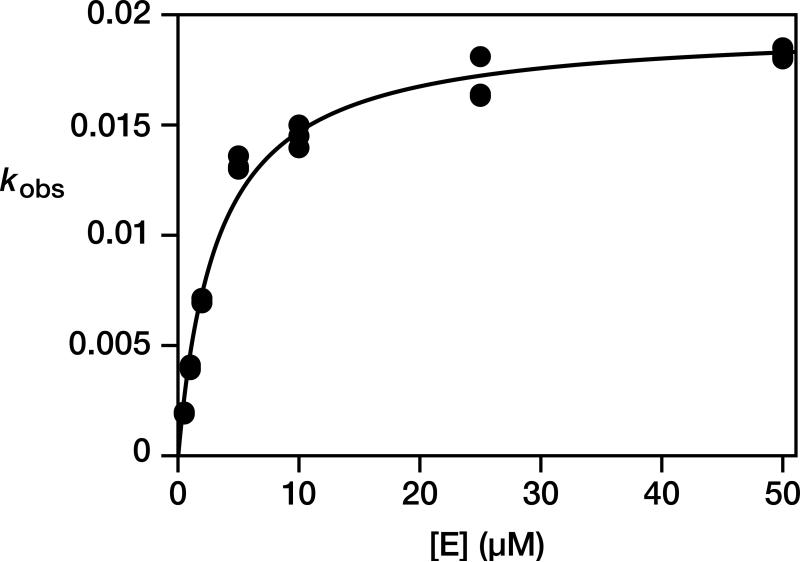
The RNA enzyme can operate on a separate template-substrate complex, recognizing that complex through tertiary interactions. The D-enzyme catalyzes the ligation of two L-RNA substrates on a L-RNA template, and the mirror-image L-enzyme behaves similarly with D-RNA substrates and a D-RNA template (Fig. 2a). Furthermore, the two enzymes can operate in a common mixture that contains both the L- and D-versions of the substrates and template. The D- and L-enzymes cannot interact through Watson-Crick pairing and do not appear to interact significantly through cross-chiral contacts. The intermolecular reaction exhibits saturation kinetics, with a kcat of 0.019 min–1 and Km of 3.3 μM (Extended Data Fig. 4). There is no detectable reaction when the template-substrate complex is of the same handedness as the enzyme, even at 50 μM concentration.
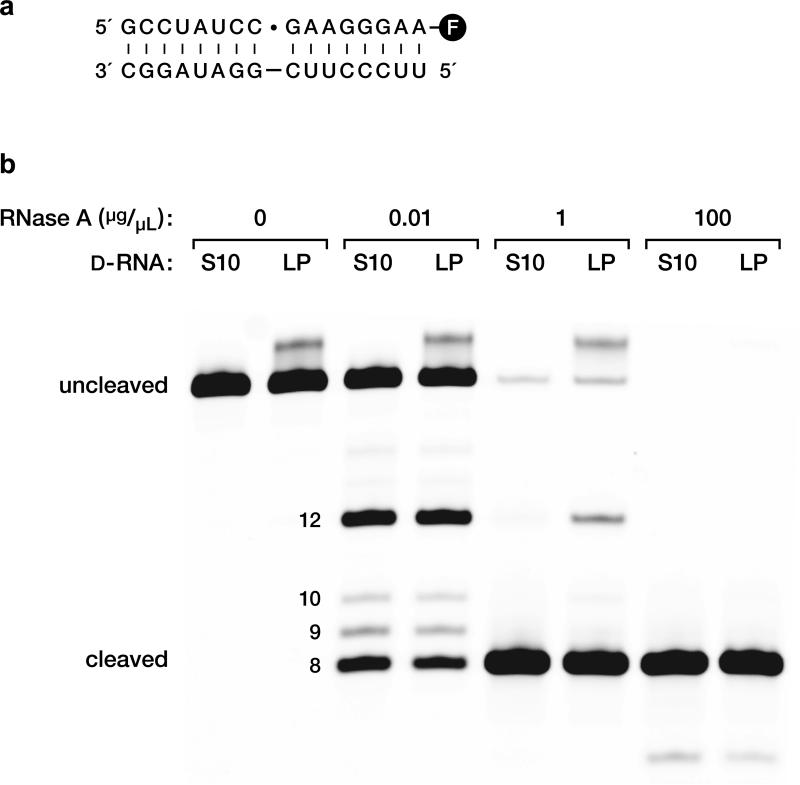
The products of the ligation of two D-RNA substrates were gel purified, then subject to cleavage by ribonuclease A, which cleaves 3′,5′- but not 2′,5′-phosphodiester linkages. Cleavage at the ligation junction was complete, demonstrating that the enzyme forms the “natural” 3′,5′-linkage (Extended Data Fig. 5).
Although the enzyme was selected on the basis of templated ligation activity, this reaction is mechanistically similar to the templated polymerization of NTPs. Other selected ligases have shown at least some polymerization activity,12,13 which is the case here too. The four L-NTPs were prepared by chemical synthesis and tested in various primer extension reactions with the D-RNA enzyme and a separate L-RNA template. Employing a template with the sequence 3′-CCCCAGUA-5ímmediately downstream from the primer-binding site, and supplying 4 mM L-GTP, the D-RNA enzyme catalyzes four successive GTP additions (Fig. 2b). When instead provided with D-GTP there is only a very low level of single-nucleotide addition. When provided with a racemic mixture of D,L-GTP the results are nearly identical to the reaction with L-GTP alone, with an observed rate of 0.11 min–1 in both cases (Extended Data Fig. 3b). Thus there is no chiral inhibition in the RNA-catalyzed polymerization reaction, unlike the situation with the non-enzymatic template-directed polymerization of activated mononucleotides.1
Other template-primer combinations were used to demonstrate the ability of the D-RNA enzyme to add each of the four L-NTPs on a complementary template (Fig. 2b). These experiments revealed that the enzyme does have sequence preferences, with addition to a 3′-terminal C or G residue being most efficient and addition to a 3′-terminal A or U residue being poor. Addition of GTP to a 3′-terminal C is especially efficient and mimics the ligation junction that was used during in vitro evolution. No attempt has yet been made to select directly for NTP addition or with different sequences surrounding the reaction site. Nonetheless, the current sequence tolerance of the enzyme is sufficient to enable the assembly of a variety of enantiomeric RNA products.
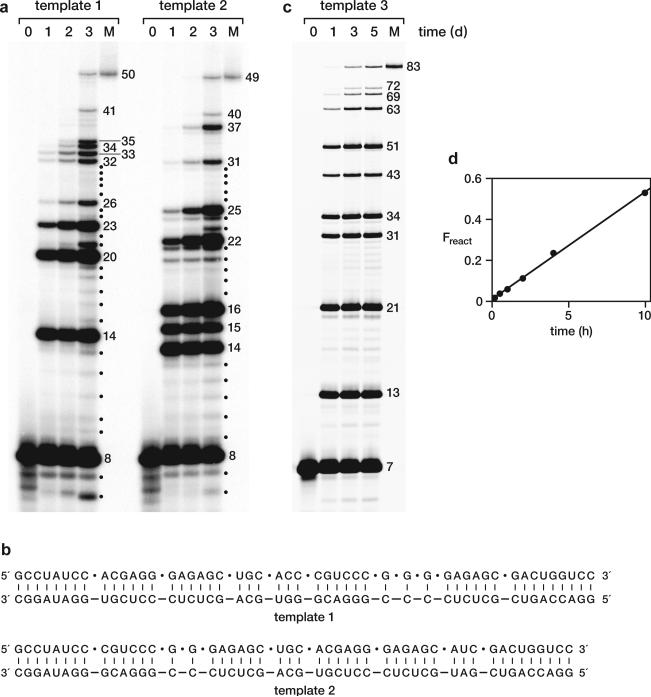
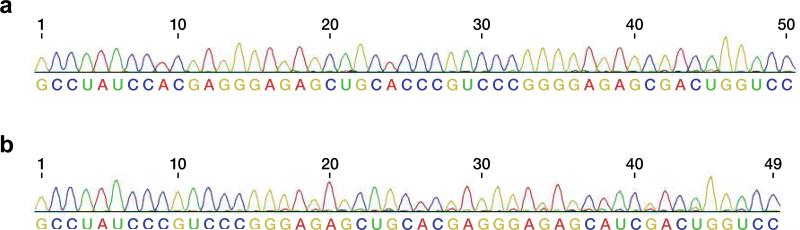
The RNA enzyme appears to be indifferent to the length of the substrates, so long as they are bound to a complementary template. As a demonstration of this property, a mixture of D-mono- and oligonucleotides were assembled on two different long D-RNA templates (Fig. 3a, b). The first required seven ligations and three NTP additions; the second required seven ligations and two NTP additions; and both resulted in the synthesis of full-length products. The ladder of 5′-labeled materials demonstrates that some additions are more efficient than others, likely reflecting a mixture of sequence preference, structural context, and competition among substrates. However, there is a clear progression of successive additions, culminating in the full-length product. The accurate assembly of the full-length materials was confirmed by sequence analysis (Extended Data Fig. 6).
Figure 3. Cross-chiral assembly of long RNAs.
a, Assembly of 50mer and 49mer D-RNAs on complementary D-RNA templates through multiple ligation and polymerization events, catalyzed by the L-RNA enzyme. The reaction mixtures were sampled at 0, 1, 2, and 3 d and the 5′-labeled products were analyzed by polyacrylamide gel electrophoresis in comparison to authentic full-length material (M). Numbers at the right indicate the size of successively assembled components. Dots indicate intermediate-length materials resulting from degradation of longer products. See Online Only Methods for reaction conditions. b, Sequences of substrates and templates used to assemble the two RNAs shown in a. Dots indicate the junctions for assembly. c, Assembly of the 83-nucleotide L-RNA enzyme on a complementary L-RNA template, catalyzed by the D-RNA enzyme of the same sequence. The reaction mixture was sampled at 0, 1, 3, and 5 d and the products were analyzed as above. Red dots in Fig. 1c indicate the junctions for assembly, with sequence modifications at positions 13, 14, 31, and 32, as shown in Extended Data Fig. 2g. d, Catalytic activity of the L-RNA enzyme that had been assembled by the D-RNA enzyme. The reaction conditions are as in Fig. 2a, but with 0.5 μM enzyme, 0.2 μM upstream substrate, 1 μM downstream substrate, and 0.5 μM template.
As a final test of the ability of the enzyme to synthesize enantiomeric products, the D-RNA enzyme was used to assemble 11 L-oligonucleotides to form a mirror copy of itself. The ten ligation junctions had either a C or G residue at the 3′-terminus and an A, U, or G residue at the 5′-terminus (Fig. 1b). The ladder of 5′-labeled materials again demonstrates successive additions culminating in the full-length product (Fig. 3c). This full-length material was gel purified and tested for enzymatic activity in a ligation reaction with two D-RNA substrates and a D-RNA template, confirming that it is fully functional (Fig. 3d). This is the first demonstration of an enzyme being synthesized by its enantiomer.
Biology is overwhelmingly homochiral, with only sparse examples of L-sugars and D-amino acids, such as L-arabinose in plant hemicellulose and D-alanine in bacterial peptidoglycan. There is no known example of a biopolymer containing subunits entirely of the “wrong” handedness. This is because the stereochemical handshake between biopolymers would seem to demand chiral uniformity. Yet macromolecules of opposite handedness can interact in their own fashion, including to bring about chemical transformations. The advantages of a cross-chiral polymerase for RNA-based life are twofold: first, both enantiomers are utilized, so polymerization does not deplete the supply of the “correct” enantiomer; and second, the interaction between D- and L-RNA does not allow consecutive Watson-Crick pairs that can contribute to sequence bias.
The question remains as to how a chirally pure RNA enzyme would arise in the first place, and moreover how there might be both D- and L- versions of such an enzyme. One possibility is that RNA-based life was preceded by a genetic system based on an achiral polymer,2,14 which then evolved the ability to synthesize RNA polymers. An achiral catalyst would generate both D- and L-RNA, but could distinguish between the homo- and heterochiral addition of monomers to the growing chain. A second possibility is that life began with the non-enzymatic replication of either D- or L-RNA,15,16 and subsequently evolved the ability to catalyze the cross-chiral polymerization of RNA. The products of cross-chiral polymerization could do so similarly, ultimately displacing the chemical replication process.
The cross-chiral polymerase is still a young enzyme, only 16 rounds of selective amplification away from random sequence. However, it has auspicious properties that likely can be improved through further in vitro evolution. It will be especially important to increase the catalytic rate of the enzyme and to enhance its ability to extend 3′-termini that end in either an A or U residue. The ultimate aim is to achieve cross-chiral RNA replication, which would require the enzyme to generate both strands of an RNA duplex, that is, both the enantiomeric enzyme and its complement. Cross-chiral replication does not require the D- and L-enzymes to have the same sequence, and even if initiated with enzymes of the same sequence, the two likely would soon drift apart. If early life did entail the cross-chiral polymerization of RNA, then there would have been an era when both sides of the mirror were indispensable. Subsequently, however, a key evolutionary innovation may have arisen on one side of the mirror, for example, the invention of instructed L-polypeptide synthesis by D-RNA. Then the other side of the mirror could go dark, leaving biology to follow a homochiral path.
Online-only Methods
Materials
Oligonucleotides were either purchased from IDT (San Diego, CA) or prepared by solid-phase synthesis using an Expedite 8909 DNA/RNA synthesizer with reagents and nucleoside phosphoramidites purchased from Glen Research (Sterling, VA), except L-2′-tbutyldimethylsilyl phosphoramidites and L-2′-triisopropylsilyloxymethyl phosphoramidites, which were from ChemGenes (Wilmington, MA). For the coupling of degenerate nucleotides (N), the concentration ratios of the four phosphoramidites were 3.0 : 2.0 : 2.3 : 2.5 for A : T : G : C, respectively, to achieve equal coupling efficiencies. Chemical triphosphorylation of synthetic oligoribonucleotides was carried out as described previously.17 All oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (PAGE) and desalted by ethanol precipitation. See Supplementary Table 1 for all oligonucleotide sequences and Extended Data Fig. 7 for chemical structure of synthetic linkers.
Histidine-tagged T7 RNA polymerase was purified from E. coli strain BL21 containing plasmid pBH161 (provided by William McAllister, State University of New York, Brooklyn). Thermus aquaticus (Taq) DNA polymerase was cloned from total genomic DNA and prepared as described previously.18 Superscript II RNase H– reverse transcriptase, Turbo DNase, and streptavidin-coated magnetic beads (MyOne C1 Dynabeads) were from Life Technologies (Carlsbad, CA). Bacteriophage T4 RNA ligase was cloned from total genomic DNA and prepared as described previously.19 RNase A and tRNA were purchased from Roche Applied Science (Indianapolis, IN) and passed through a 0.2 nm filter prior to use. Chemical reagents, including NTPs and dNTPs, were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. [γ-32P]ATP was from PerkinElmer (Waltham, MA).
Preparation of L-NTPs
Based on modification of previously described methods,20 the unprotected L-nucleoside (~0.7 mmol) was dissolved in 2 mL of either DMF (for C and U) or 1:1 DMF:DMSO (for A and G) in a flask containing activated molecular sieves, then stirred at 23 °C for 2 h. In a separate flask, also containing activated molecular sieves, 3 mL DMF, 0.14 mmol tributylammonium pyrophosphate, 0.85 mmol 2-chloro-4-H-1,2,3-benzodioxaphosphorin-4-one, and 0.5 mL tributylamine were added sequentially and stirred at 23 °C for 2 h. The pyrophosphate solution then was added slowly to the nucleoside solution at 4 °C and the reaction was allowed to proceed for 3 h before adding ~10 mL of 0.1 M iodine in THF/water/pyridine until a persistent brown color appeared. After 1 h, 10 mL of water was added to the mixture, which was stirred at 23 °C for 1 h. The NTP was recovered by ethanol precipitation, then purified by HPLC on a 250 × 10 mm Luna C18 column (Phenomenex, Torrance, CA) using a linear gradient of 0–10% acetonitrile in 20 mM triethylammonium acetate (pH 7.0), with UV detection at 254 nm.
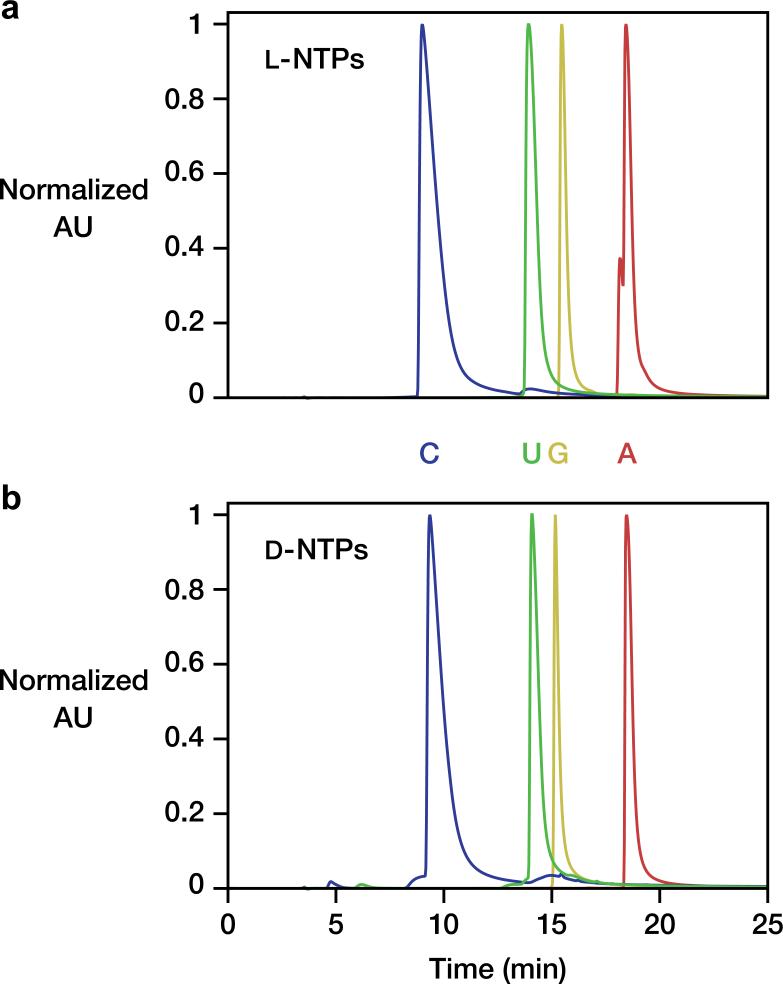
L-ATP was obtained in 52% crude yield. UV/vis: λmax = 261 nm; 1H-NMR (400 MHz, D2O): δ 8.56 ppm (s, 1H), 8.27 (s, 1H), 6.15 (d, J = 4.0 Hz, 1H), 4.84 (t, J = 8.0 Hz, 1H), 4.66–4.62 (m, 1H), 4.43–4.18 (m, 3H); 31P-NMR (162 MHz, D2O): δ –6.40 ppm (d, J = 9.7 Hz), –11.07 (d, J = 19.4 Hz), –22.05 (t, J = 21.1 Hz); HRMS (m/z): [M+H]– calc'd for C10H15N5O13P3 505.9885, found 505.9902. L-CTP was obtained in 13% crude yield. UV/vis: λmax = 270 nm; 1H-NMR (400 MHz, D2O): δ 8.03 (m, 1H), 6.17 (m, 1H), 6.04 (m, 1H), 4.46–4.42 (m, 1H), 4.37–4.24 (m, 3H); 31P NMR (162 MHz, D2O): δ –6.93 (m), –11.20 (d, J = 17.8 Hz), –22.28 (t, J = 19.4 Hz); HRMS (m/z): [M+H]– calc’d for C9H15N3O14P3 481.9772, found 481.9750. L-GTP was obtained in 33% crude yield. UV/vis: λmax = 251 nm; 1H-NMR (400 MHz, D2O): δ 8.12 (s, 1H), 5.94 (d, J = 8.0 Hz, 1H), 4.82 (t, J = 8.0 Hz, 1H), 4.61 (m, 1H), 4.36 (m, 1H), 4.29 (m, 2H); 31P NMR (162 MHz, D2O): δ –9.05 (m), –11.20 (d, J = 22.6 Hz), –22.72 (t, J = 19.4 Hz); HRMS (m/z): [M+H]– calc’d for C10H15N5O14P3 521.9834, found 521.9838. L-UTP was obtained in 27% crude yield. UV/vis: λmax = 257 nm; 1H-NMR (400 MHz, D2O): δ 8.01 (d, J = 8.0 Hz, 1H), 6.00–5.96 (m, 2H), 4.49– 4.38 (m, 2H), 4.31–4.20 (m, 2H); 31P NMR (162 MHz, D2O): δ –6.36 (m), –11.20 (d, J = 21.6 Hz), –22.12 (t, J = 19.4 Hz); HRMS (m/z): [M+H]– calc’d for C9H14N2O15P3 482.9613, found 482.9593. The purity of the four L-NTPs was confirmed by HPLC analysis in comparison to the four authentic D-NTPs (Extended Data Fig. 8).
In vitro evolution
The library of dsDNA templates was generated by extending 8 nmol Rev1 on 6 nmol Lib1 in a 200-μL reaction mixture containing 200 U/μL Superscript II reverse transcriptase, 0.25 mM each dNTP, 3 mM MgCl2, 75 mM KCl, and 50 mM Tris (pH 8.3), which was incubated at 42 °C for 1 h. The products were added directly to a 5-mL in vitro transcription mixture containing 300 U/μL T7 RNA polymerase, 5 mM each NTP, 25 mM MgCl2, 10 mM DTT, 2 mM spermidine, and 40 mM Tris (pH 7.9), which was incubated at 37 °C for 2.5 h. Then 2 U/μL DNase I was added, the mixture was incubated at 37 °C for 1 h, and the RNA was ethanol precipitated and purified by PAGE.
The pool of D-RNAs (40 nmol in round 1, 1–2 nmol in subsequent rounds) were ligated to the L-RNA primer (Tem1) in a reaction mixture containing 20 U/μL T4 RNA ligase, 10 μM pool RNA, 10 μM Rev2, 7 μM Tem1, 10 mM MgCl2, 1 mM DTT, 50 mM Tris (pH 7.5), and 10% DMSO, which was incubated at 16 °C overnight. The ligated products were ethanol precipitated, purified by PAGE, concentrated using an Amicon YM-10 spin filter (Millipore, Billerica, MA), and ethanol precipitated.
A portion of the ligated pool RNA (10 nmol in round 1, 0.2–0.5 nmol in subsequent rounds) was annealed with Sub1 in a mixture containing 2 μM pool RNA, 5 μM Sub1, 250 mM NaCl, and 50 mM Tris (pH 7.6), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Each round was initiated by adding to this mixture an equal volume of a solution containing 100 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 7.6), and 0.1% TWEEN-20. The RNA-catalyzed reaction was carried out at 23 °C for 16 h during rounds 1–6, 2 h during round 7, and 5 min during rounds 8–10, then quenched by adding an excess of EDTA relative to the concentration of Mg2+.
During round 1, the quenched reaction mixture was applied to a 400-μL bed volume streptavidin agarose column (Thermo Scientific, Rockford, IL), which was washed sequentially with 2 mL Buffer A (50 mM NaCl, 1 mM EDTA, 25 mM Tris (pH 7.6), and 0.1% TWEEN-20), 4 × 2 mL Buffer B (8 M urea, 50 mM NaCl, 1 mM EDTA, 25 mM Tris (pH 7.6), and 0.1% TWEEN-20), 2 × 0.5 mL Buffer C (25 mM NaOH and 1 mM EDTA), and 2 mL water. The captured RNA was reverse transcribed directly on the resin in a 400-μL reaction mixture containing 10 U/μL Superscript II reverse transcriptase, 1 μM Rev1, 0.25 mM each dNTP, 3 mM MgCl2, 75 mM KCl, and 50 mM Tris (pH 8.3), which was incubated at 42 °C for 2 h. The resin then was washed with 2 × 1 mL water and the cDNA was eluted with 2 × 0.5 mL 25 mM NaOH, immediately neutralized with 50 μL 1 M Tris (pH 7.6), and desalted by ethanol precipitation.
During rounds 2–10, biotinylated molecules were captured by incubating the quenched reaction mixture with 1–5 mg streptavidin-coated magnetic beads at 23 °C for 1 h. The beads were washed sequentially with 0.5 mL Buffer A, 4 × 0.5 mL Buffer B, and 2 × 0.5 mL Buffer C, then suspended in 500 μL buffer A and blocked with Buffer A containing 5 μg tRNA. Following a final wash with 2 × 0.5 mL Buffer A, the captured RNA was released from the beads by photolysis (350 nm) at 4 °C for 30 min. The eluted material was ethanol precipitated, then reverse transcribed in a 50-μL reaction mixture under the same conditions as above. The reverse transcriptase was inactivated by heating at 70 °C for 5 min.
The cDNA from each round was PCR amplified using primers Fwd1 and Rev1. Error-prone PCR21 was conducted following round 6. The PCR products were used to transcribe the subsequent pool of RNAs, as described above. Following the 10th round, the amplified DNA was cloned into E. coli using the TOPO TA Cloning Kit (Life Technologies). Bacteria were grown at 37 °C for 16 h on LB agar plates containing 50 μg/mL carbenicillin. DNA from individual colonies was PCR amplified and sequenced by Genewiz (La Jolla, CA).
Rounds 11–16 were conducted as above, except for the following modifications. The library of dsDNA templates was prepared by extending 8 nmol Rev3 on 6 nmol Lib2. The sequence of Lib2 was based on clone 10.2, with 30 random-sequence nucleotides inserted between the P1 and P3 stems (Extended Data Fig. 2a,b). This site was chosen because of its proximity to the presumed catalytic center of the 10.2 enzyme. Sub3 was used as the L-RNA primer. RNA-catalyzed ligation employed a separate template (Tem2) and different connectivity between the enzyme and upstream substrate (Fig. 1b). cDNA from each round was PCR amplified using primers Fwd2 and Rev3. Error-prone PCR was conducted following rounds 11–15. The stringency of selection was increased by progressively reducing the reaction time: 16 h in rounds 11–13, 20 min in round 14, 5 min in round 15, and 2 min in round 16. The amplified dsDNA from round 16 was cloned and sequenced, as described above.
Preparation of individual RNA enzymes
D-RNA enzymes were prepared by in vitro transcription of dsDNA templates, which were generated by cross-extending 0.5 μM each of S1 and S2 (for D-16.12t), S3 and S4 (for D-16.12tx), or S5 and S6 (for D-16.12ts), under the same conditions used to prepare the starting library. The extended products were transcribed and purified as described above.
The L-16.12tx enzyme was prepared by templated ligation of two shorter L-RNAs, using the D-16.12t enzyme as the catalyst. A mixture was prepared containing 2 μM D-16.12t, 35 μM S7, 50 μM S8, 50 μM S9, 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. The ligation reaction was initiated by adding an equal volume of a solution containing 200 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 8.5), and 0.1% TWEEN-20, which was incubated at 23 °C for 16 h, then quenched with EDTA. The products were ethanol precipitated, dissolved in 2 mL 50 mM Tris (pH 7.6), and incubated with 500 μg RNase A for 2 h at 37 °C to digest the D-16.12t enzyme. The RNase was extracted with phenol/chloroform, then the L-RNA products were ethanol precipitated, purified by PAGE, concentrated using an Amicon YM-10 spin filter, and again ethanol precipitated.
Determination of regiospecificity of ligation
3′-labeled ligated products, containing a cytosine residue immediately upstream from the ligation junction, were subject to cleavage by RNase A. A mixture was prepared containing 2 μM L-16.12tx, 20 μM Sub6, 40 μM Sub7, 40 μM Tem4, 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Ligation was carried out as described above, then the ligated products were purified by PAGE, desalted by ethanol precipitation, and dissolved in water. RNase A digestion was carried out in a mixture containing 2 μM ligated RNA, various amounts of RNase A, and 50 mM Tris (pH 7.6), which was incubated at 23 °C for 1 min, then quenched by adding 20 μg/μL tRNA and immediately analyzed by PAGE.
Analysis of ligation activity
For cis-ligation reactions, the D-RNA enzyme first was joined to Sub3 in a reaction mixture containing 4 U/μL T4 RNA ligase, 10 μM RNA enzyme, 7 μM Sub3, 10 mM MgCl2, 1 mM DTT, 50 mM Tris (pH 7.5), and 10% DMSO, which was incubated at 16 °C overnight. The ligated products were ethanol precipitated, purified by PAGE, concentrated using an Amicon YM-10 spin filter, and desalted by ethanol precipitation. The components for cross-chiral ligation were assembled in a mixture containing 0.1 μM enzyme-Sub3, 2 μM Sub1, 1 μM Tem2, 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Ligation was initiated by adding an equal volume of a solution containing 500 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 8.5), and 0.1% TWEEN-20, incubating at 23 °C for various times, then quenching with EDTA. The reaction products were photolyzed (350 nm) at 4 °C for 10 min, then analyzed by PAGE.
For trans-ligation reactions, 20 μM D- or L-16.12tx enzyme was mixed with 1 μM 5′-labeled upstream substrate (L-Sub4 or D-Sub5, respectively), 8 μM downstream substrate (L-Sub1 or D-Sub2, respectively), 4 μM template (L-Tem2 or D-Tem3, respectively), 250 mM NaCl, and 50 mM Tris (pH 8.5), heated at 70 °C for 3 min, then slowly cooled to 23 °C. Ligation was carried out as above and the products were analyzed by PAGE.
Analysis of polymerization activity
The D-16.12t enzyme first was joined to Sub3, as described above. The components for cross-chiral polymerization were assembled in a mixture containing 1 μM enzyme-Sub3, 2 μM various L-RNA templates (Tem5–8), 8 mM of the appropriate NTP, 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Polymerization was initiated by adding an equal volume of a solution containing 500 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 8.5), and 0.1% TWEEN-20, incubating at 17 °C for various times, then quenching with EDTA. The reaction products were photolyzed and analyzed by PAGE, as described above.
Cross-chiral synthesis of long RNAs
The reaction components were assembled in a mixture containing 80 μM L-16.12tx, 10 μM RNA template (Tem9 or Tem10), 4 μM [5′-32P]-labeled Sub7, 20 μM each hexanucleotide substrate (Sub8–10), 50 μM each trinucleotide substrate (Sub12–14), 20 μM Sub11, 10 mM D-GTP, 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Cross-chiral synthesis was initiated by adding an equal volume of a solution containing 500 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 8.5), and 0.1% TWEEN-20, incubating at 17 °C for various times, then quenching with EDTA. The biotinylated template was removed by adding 0.5 mg magnetic beads, shaking at 23 °C for 1 h, washing with 2 × 0.5 mL Buffer A, eluting the ligated products with 2 × 200 μL 25 mM NaOH, and immediately neutralizing with 1 M Tris (pH 7.6). The products were ethanol precipitated and analyzed by PAGE in comparison to authentic full-length materials (Std1 for the 50mer RNA; Std2 for the 49mer RNA).
The full-length RNA was excised from the gel, eluted overnight, concentrated using an Amicon YM-10 spin filter, and reverse transcribed as described above. The resulting cDNA was purified by PAGE, PCR amplified using primers Fwd3 and Rev4, and cloned and sequenced as described above, confirming the accurate synthesis of full-length RNA (Extended Data Fig. 6).
For cross-chiral synthesis of the L-16.12ts enzyme, an 83-nucleotide L-RNA template was prepared by cross-chiral ligation of Tem11 and Tem12, using S11 as a splint and D-16.12ts as the catalyst, under the same conditions used to prepare L-16.12tx. The components for synthesis of L-16.12ts were assembled in a mixture containing 80 μM D-16.12ts, 10 μM template, 50 μM each trinucleotide substrate (Sub15 and Sub16), 20 μM each of the other substrates (Sub17–25), 250 mM NaCl, and 50 mM Tris (pH 8.5), which was heated at 70 °C for 3 min, then slowly cooled to 23 °C. Cross-chiral synthesis was initiated by adding an equal volume of a solution containing 500 mM MgCl2, 250 mM NaCl, 50 mM Tris (pH 7.6), and 0.1% TWEEN-20, incubating at 17 °C for 5 d, then quenching with EDTA. The full-length products were purified by PAGE, then tested for activity in a trans-ligation reaction, as described above.
Extended Data
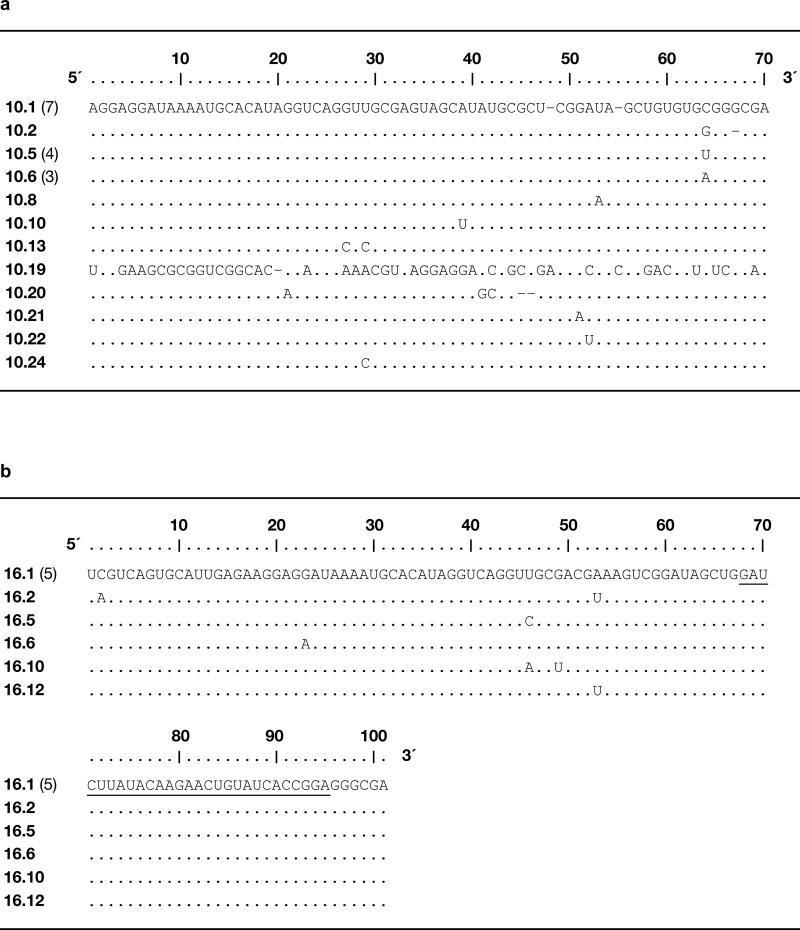
Extended Data Figure 1.
Sequences of individual clones isolated after each phase of the in vitro evolution process. a, 23 clones isolated after round 10. b, 10 clones isolated after round 16, with the underlined sequence derived from the 30 random-sequence nucleotides that were inserted following round 10. Clone numbers are shown at the left, with numbers in parentheses indicating duplicate sequences among the clones that were analyzed. Nucleotides within the fixed primer-binding sites are not shown.
Extended Data Figure 2.
Sequence and secondary structure of evolved and engineered variants of the cross-chiral RNA enzyme. a, Clone 10.2, isolated after round 10. Nucleotides within the primer-binding sites are shown in red. b, Lib2, based on clone 10.2, with 30 random-sequence nucleotides (N30, green) inserted between the P1 and P3 stems. c, Clone 16.12, isolated after round 16. d, Truncated version of clone 16.12, replacing the primer-binding sites with two G-C pairs (dashed box). e, The 16.12t enzyme, removing the extended portion of the P3 stem (blue box) and replacing a G-U pair within the P3 stem by a G-C pair (dashed box). f, Variant 16.12tx, replacing two base pairs compared to 16.12t (dashed box) to facilitate assembly by RNA-catalyzed ligation of two shorter RNAs. g, Variant 16.12ts, replacing two additional base pairs compared to 16.12tx (dashed box) to facilitate assembly catalyzed by the enantiomeric enzyme.
Extended Data Figure 3.
Catalytic activity of the D-16.12t enzyme. a, Ligation of two L-oligonucleotides on a L-RNA template, according to the reaction format shown in Fig. 1b. Reaction conditions: 0.05 μM enzyme-primer, 1 μM downstream substrate, 0.5 μM template, 250 mM MgCl2, 250 mM NaCl, pH 8.5, 23 °C. b, Polymerization of L-GTP by extension of a L-oligonucleotide primer on a complementary L-RNA template (see Fig. 2b). Reaction conditions as above, but with no downstream substrate and with either 4 mM L-GTP (solid circles) or 4 mM each D- and L-GTP (open circles).
Extended Data Figure 4.
Kinetic analysis of the reaction of the D-16.12t enzyme with a separate L-template/primer/substrate complex. The reactions were carried out as described in the Online Only Methods, except that the concentration of enzyme was varied, always in at least 10-fold excess over the concentration of template/primer/substrate complex. Values for kobs were obtained for each concentration of enzyme based on the initial rate of reaction, then fit to the Michaelis-Menten equation: kobs = kcat [E] / (Km + [E]). This gave values for kcat of 0.019 ± 0.001 min–1 and for Km of 3.3 ± 0.3 μM. Reaction conditions: 0.5–50 μM enzyme, 0.1 μM template/primer (Tem13), 0.2 μM downstream substrate (Sub1), 250 mM MgCl2, 250 mM NaCl, pH 8.5, 23 °C
Extended Data Figure 5.
Analysis of the regiospecificity of ligation. a, D-RNA substrates and template for ligation, catalyzed by the L-16.12tx enzyme. Dot indicates the ligation junction, which is also the site for RNase A cleavage that is closest to the 3′ end of the ligated product. The downstream substrate is labeled at the 3′ end with fluorescein (circled F). b, RNase A digestion of the ligated products (LP) in comparison to authentic all-3′,5′-linked RNA of the same sequence (S10). Reaction conditions: 0–100 μg/μL RNase A, 2 μM RNA, 50 mM Tris (pH 7.6), 23 °C, 1 min.
Extended Data Figure 6.
Sequence analysis of long RNAs obtained by cross-chiral synthesis. a, Full-length 50mer D-RNA assembled through seven ligations and three NTP additions. b, Full-length 49mer D-RNA assembled through seven ligations and two NTP additions. See Fig. 3a, b for substrate sequences and reaction products. See Online Only Methods for reaction conditions and sequencing procedure.
Extended Data Figure 7.
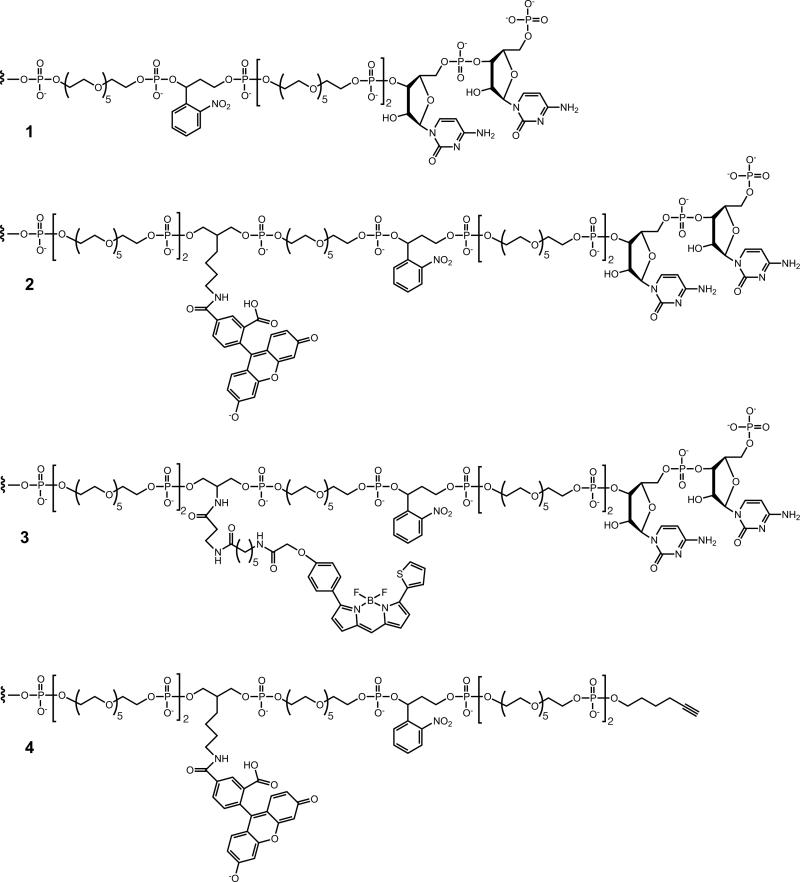
Chemical structure of linkers used to prepare various enzyme and enzyme-primer molecules (see Supplementary Table 1). 1, CpC dinucleotide tethered via a photocleavable linker to the 5′ end of Tem1, enabling joining by T4 RNA ligase to the pool of RNAs in rounds 1–10, as shown in Fig. 1a. 2, CpC dinucleotide tethered via a photocleavable, fluorescein-labeled linker to the 5′ end of Sub2, enabling joining by T4 RNA ligase to the pool of RNAs in rounds 11–16, as shown in Fig. 1b. 3, CpC dinucleotide tethered via a photocleavable, boron-dipyrromethene-labeled linker to the 5′ end of Sub4, used in the D-RNA- catalyzed ligation of L-RNA (Fig. 2a, red). 4, CpC dinucleotide tethered via a photocleavable, fluorescein-labeled linker to the 5′ end of Sub5, used in the L-RNA-catalyzed ligation of D-RNA (Fig. 2a, green).
Extended Data Figure 8.
HPLC analysis of synthetic L-NTPs. a, Elution of the four L-NTPs. b, Elution of the four authentic D-NTPs. HPLC conditions: C18 column, linear gradient of 0– 10% acetonitrile in 20 mM triethylammonium acetate (pH 7.0), UV detection at 254 nm.
Supplementary Material
Acknowledgments
This work was supported by grant NNX10AQ91G from NASA and by grant 287624 from the Simons Foundation. J.T.S. was supported by Ruth L. Kirschstein National Research Service Award No. F32 GM101741 from the National Institutes of Health.
Footnotes
Author Contributions J.T.S. and G.F.J. conceived the project, designed the experiments, and wrote the paper. J.T.S. carried out the experiments.
The authors declare no competing financial interests.
Supplementary Information is available in the online version of the paper.
References
- 1.Joyce GF, et al. Chiral selection in poly(C)-directed synthesis of oligo(G). Nature. 1984;310:602–604. doi: 10.1038/310602a0. [DOI] [PubMed] [Google Scholar]
- 2.Joyce GF, Schwartz AW, Miller SL, Orgel LE. The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc. Natl. Acad. Sci. USA. 1987;84:4398–4402. doi: 10.1073/pnas.84.13.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klussmann M, et al. Thermodynamic control of asymmetric amplification in amino acid catalysis. Nature. 2006;441:621–623. doi: 10.1038/nature04780. [DOI] [PubMed] [Google Scholar]
- 4.Hein JE, Tse E, Blackmond DG. A route to enantiopure RNA precursors from nearly racemic starting materials. Nat. Chem. 2011;3:704–706. doi: 10.1038/nchem.1108. [DOI] [PubMed] [Google Scholar]
- 5.Ashley GW. Modeling, synthesis, and hybridization properties of (L)-ribonucleic acid. J. Am. Chem. Soc. 1992;114:9731–9736. 1992. [Google Scholar]
- 6.Garbesi A, et al. L-DNAs as potent antimessenger oligonucleotides: a reassessment. Nucleic Acids Res. 1993;21:4159–4165. doi: 10.1093/nar/21.18.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sczepanski JT, Joyce GF. Binding of a structured D-RNA molecule by an L-RNA aptamer. J. Am. Chem. Soc. 2013;135:13290–13293. doi: 10.1021/ja406634g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 9.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 10.Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohatgi R, Bartel DP, Szostak JW. Kinetic and mechanistic analysis of nonenzymatic, template-directed oligoribonucleotide ligation. J. Am. Chem. Soc. 1996;118:3332–3339. doi: 10.1021/ja953712b. [DOI] [PubMed] [Google Scholar]
- 12.Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 13.McGinness KE, Joyce GF. RNA-catalyzed RNA ligation on an external RNA template. Chem. Biol. 2002;9:297–307. doi: 10.1016/s1074-5521(02)00110-2. [DOI] [PubMed] [Google Scholar]
- 14.Böhler C, Nielsen PE, Orgel LE. Template switching between PNA and RNA oligonucleotides. Nature. 1995;376:578–581. doi: 10.1038/376578a0. [DOI] [PubMed] [Google Scholar]
- 15.Inoue T, Orgel LE. A nonenzymatic RNA polymerase model. Science. 1983;219:859–862. doi: 10.1126/science.6186026. [DOI] [PubMed] [Google Scholar]
- 16.Adamala K, Szostak JW. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul N, Springsteen G, Joyce GF. Conversion of a ribozyme to a deoxyribozyme through in vitro evolution. Chem. Biol. 2006;13:329–338. doi: 10.1016/j.chembiol.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Pluthero FG. Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Res. 1993;21:4850–4851. doi: 10.1093/nar/21.20.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LK, Ho CK, Pei Y, Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1: Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 20.Caton-Williams J, Hoxhaj R, Fiaz B, Huang Z. Use of a novel 5′-regioselective phosphitylating reagent for one-pot synthesis of nucleoside 5′-triphosphates from unprotected nucleosides. Curr. Protocols Nucleic Acid Chem. 2013;52:1.30.1–21. doi: 10.1002/0471142700.nc0130s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.