Abstract
The DNA damage response pathway controlled by the BRCA and Fanconi Anemia (FA) genes can be disrupted by genetic or epigenetic mechanisms in breast cancer. Defects in this pathway may render the affected tumors hypersensitive to DNA damaging agents. The identification of these defects poses a challenge because of the large number of genes involved in the FA/BRCA pathway. Many pathway components form subnuclear repair protein foci upon exposure to ionizing radiation in-vitro, but it was unknown whether foci can be detected in live cancer tissues. Thus, the goal of this pilot study was to identify pathway defects by using a novel ex-vivo foci biomarker assay on tumor biopsies. Fresh pretreatment biopsy specimens from patients with locally advanced sporadic breast cancer were irradiated or mock-treated in the laboratory (ex-vivo). Foci formation of DNA repair proteins BRCA1, FANCD2, and RAD51 was detected by immunofluorescence microscopy. Three out of seven tumors showed intact radiation-induced foci formation while the other four tumors exhibited a defective foci response. Notably, three of the foci-defective tumors were ER/PR/HER2 (triple) negative, a phenotype that has been associated with BRCA1 deficiency. In conclusion, in this pilot study, we report the successful detection of BRCA1, FANCD2 and RAD51 foci in breast cancer biopsies irradiated exvivo. Our approach represents a potentially powerful biomarker assay for the detection of pre-existing and functionally important defects within the complex FA/BRCA pathway, which may ultimately allow us to tailor cancer treatment to the DNA repair profile of individual tumors.
Introduction
The tumor suppressor gene, BRCA1, is mutated in up to 50% of cases of familial early-onset breast cancer and in most families with hereditary breast and ovarian cancer. The vast majority of cancers arising in BRCA1 germline mutation carriers are of the basal-like subtype(1-3), a genetically distinct group of tumors characterized by markers expressed in normal basal/myoepithelial cells(4). Basal-like cancers only infrequently express estrogen (ER) or progesterone receptor (PR) and rarely express HER2(4), which has been referred to as a triple-negative phenotype. On a cellular level, loss of BRCA1 function leads to multiple abnormalities, including a defect in the homologous recombination (HR) pathway of DNA repair (reviewed in Ref. (5-7)). The HR defect is associated with a hypersensitivity to many agents that cause DNA double-double strand breaks (DSB) or interfere with DNA replication, such ionizing radiation (IR) or cisplatin.
Fanconi's Anemia (FA) is a rare hereditary childhood disorder characterized by many abnormalities including chromosomal instability and cancer predisposition (reviewed in Ref. (8, 9)). FA has autosomal or X-linked inheritance caused by mutation in any of 13 known genes (FANCA to N). The pathways controlled by the FA and BRCA1 proteins are connected on multiple levels including a functional BRCA1-FANCD2 interaction(10). Thus, these proteins are regarded as forming a common “FA/BRCA” pathway. The exact mechanisms by which the FA proteins are involved in the repair of damaged DNA and maintenance of chromosomal stability remain to be elucidated. Accumulating evidence suggests that the FA proteins participate in the maintenance of DNA replication forks, thereby cooperating with BRCA1 to promote HR(8). Recently, the breast cancer associated genes BRIP1 and PALB2 have been identified as FA genes FANCJ and FANCN, respectively(11, 12), further strengthening the importance of the FA/BRCA pathway for breast carcinogenesis.
BRCA1 mutations are rarely found in sporadic breast cancer (reviewed in Ref. (13)), but reduced BRCA1 expression has been reported in a subset of these cancers, particularly in patients with triple- negative tumors(14-17). However, whether reduced BRCA1 protein expression has functional consequences remains unknown. In addition, given the network-like complexity of the FA/BRCA pathway, assessing the expression of individual known proteins is unlikely to reveal the overall incidence of defects that can occur anywhere in the pathway(9).
One important aspect that determines the effectiveness of the cellular response to cytotoxic therapies is the recruitment of pathway components to nuclear regions containing the damaged DNA(8, 9, 18-20). The cytological manifestation of this nuclear redistribution is the formation of protein foci, which can be visualized by immunofluorescence (IF) microscopy. The repair activity of the FA/BRCA network is less dependent on protein expression levels than on the ability to localize these proteins into foci, in order to coordinate and execute repair(10, 21-23). Thus, the ability of cells to form repair foci may serve as a functional biomarker of the integrity of the FA/BRCA pathway and associated resistance to chemotherapeutics(7, 9). An important feature of the FA/BRCA pathway is that its activity is frequently revealed only when cells are exposed to DNA damaging agents. In untreated repair-proficient cells, BRCA1 and RAD51 foci may be visible in S-phase even in the presence of pathway defects, and the fraction of cells with foci and the number of foci per cells increases post-irradiation(22, 24, 25). In contrast, BRCA1-deficient cells have an impaired ability to mount a FANCD2 and RAD51 foci response(10, 22).
Therefore, the purpose of this pilot study was to monitor the activation of the FA/BRCA pathway in sporadic human breast cancers by subjecting biopsy specimens to IR ex-vivo. We report the successful detection of RAD51, BRCA1, and FANCD2 IR-induced foci (IRIF) in these tissues. The ability of individual tumor cells to form IRIF allowed the classification of tumors as having functional versus non-functional BRCA1 status. Our approach represents a potentially powerful functional biomarker assay that may enable us to detect pre-existing and clinically relevant defects within the complex FA/BRCA DNA damage response pathway.
Results
Detection of IRIF in breast cancer cells ex-vivo
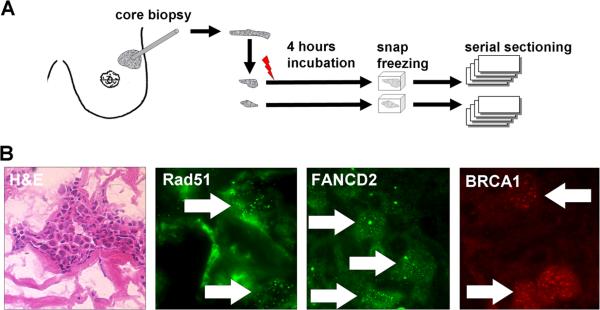
For visualization of BRCA1, FANCD2 and RAD51 foci in live tumor tissues, core biopsies from previously untreated breast cancers were subjected to X-irradiation with 8 Gy or mock-treatment (Fig. 1a). Samples were incubated in a cell culture incubator for 4 hours to allow for foci formation and then snap-frozen for later analysis. Viable tumor was identified on serial sections by H&E staining (Fig. 1b). Repair protein foci could be readily visualized in individual irradiated cells (Fig. 1b), but were often difficult to detect in the non-irradiated sample from the same tumor (see below). The observed foci are morphologically similar to IRIF seen in human cell lines (22).
FIGURE 1.
Detection of repair protein foci in fresh frozen breast cancer tissues. A. Illustration of the experimental approach. B. Representative images illustrating the detection of areas containing viable tumor as well as subnuclear RAD51, FANCD2, and BRCA1 foci.
IR induces RAD51 foci formation irrespective of sectioning
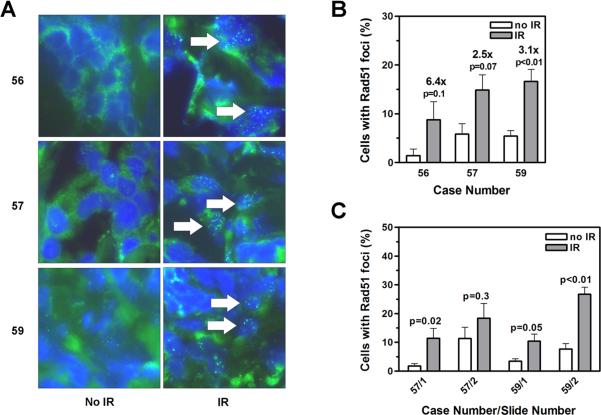
Three out of 7 irradiated tumor tissues clearly demonstrated an increased percentage of cell nuclei containing RAD51 foci when compared to the non-irradiated controls (Fig. 2a). Nine to 17% of cells contained RAD51 foci while the fraction of positive cells was significantly lower in the nonirradiated samples (range, 1-6%) (Fig. 2b). To assess the impact of potential intra-tumoral heterogeneity, we also compared sections from more distant locations within each biopsy, which is illustrated in Fig. 2c. For example, in tumor #57, there was some heterogeneity as the induction of foci was less pronounced in section #2 (1.7-fold) compared to section #1 (5.5-fold). This was due to a higher background of RAD51 foci in the non-irradiated sample. Specifically, in one of the captured high-power images the percentage of positive cells was high at 28%, likely reflecting a particularly proliferative group of tumor cells in that location, as spontaneous RAD51 foci are known to form in S-phase. In another tumor (#59), there was less variation in the non-irradiated sample, with the percentage RAD51-positive cells ranging from 0% to 8% (mean of 3.5%, Section #1) and 0% to 11% (7.1%, #2) for each 100X image. The corresponding induction of foci was approximately 3- to 3.5-fold. Together, these data indicate that IR-induced RAD51 foci formation can be reliably detected and semiquantified using 1-2 sections from tumor tissues irradiated ex-vivo.
FIGURE 2.
A. Representative images illustrating the induction of RAD51 foci in three tumors (cases 56, 57, 59) following exposure of samples to ionizing radiation (IR) ex-vivo. B. Quantification of cells with RAD51 foci. Bars represent means with upper standard error. Relative fold induction with IR is given. P-value, unpaired t-test, two sided. C. RAD51 foci analysis analogous to panel B. Numbers indicate slide numbers per case.
Correlation of RAD51 IRIF with BRCA1 and FANCD2 IRIF
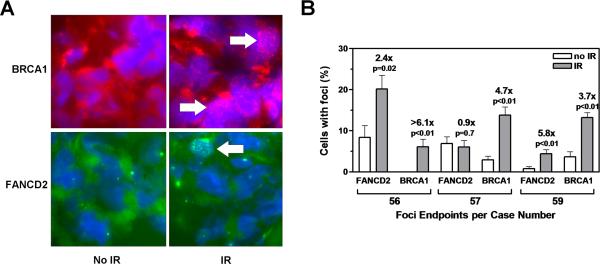
The ability of cells to form RAD51 IRIF is dependent upon intact BRCA1 function(22). We, therefore, expected that the same tumors that demonstrated RAD51 IRIF would also show IR-inducible BRCA1 foci formation. In the three tumors with intact RAD51 foci response, BRCA1 foci were largely absent in non-irradiated samples (0-3.7%) (Fig. 3a) but could be detected in the treated samples at frequencies of 6-14% (Fig. 3b). The relative induction of BRCA1 foci by IR was of similar magnitude as for RAD51. In addition, the formation of FANCD2 IRIF paralleled the induction of BRCA1 and RAD51 foci in tumors #56 and #59. However, in tumor #57, no FANCD2 IRIF were observed as the percentage of cells with foci was almost identical in the untreated and irradiated samples (~6-7%). The underlying mechanism was not known since the formation of RAD51 IRIF indicated normal BRCA1 function and the presence of spontaneous FANCD2 foci suggested that the upstream nuclear FANC core complex was intact.
FIGURE 3.
A. Representative images as in Figure 2A. B. Quantitative analysis of foci as in Figure 2B
Concordant lack of RAD51, BRCA1 and FANCD2 IRIF
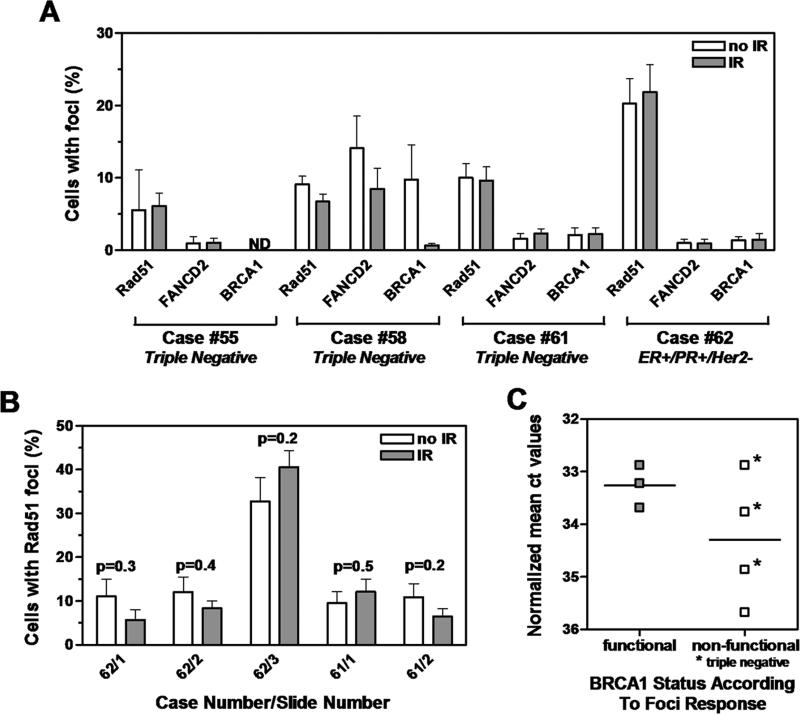
In the remaining 4 tumors, no RAD51 IRIF could be observed (Fig. 4a). Of note was the variation in the mean fraction of RAD51-positive cells, ranging from 6% to 22% among the 4 samples, which likely is a reflection of tumor heterogeneity. We, therefore, analyzed additional sections to exclude the possibility that heterogeneity in spontaneous foci formation could have masked the effects of IR (Fig. 4b). However, no significant IR-mediated induction of foci formation was revealed on any of the sections, with relative foci induction compared to untreated samples fluctuating around 1 (range, 0.5 - 1.2). Defective RAD51 foci formation suggested a defect in BRCA1 function, which should also lead to a failure of damage-induced but not spontaneous FANCD2 foci formation. Indeed, in all 4 cases, IR was insufficient to trigger the formation of additional FANCD2 or BRCA1 foci above background frequencies (Fig. 4a). BRCA1 expression is often impaired in breast cancers, which could have functional consequences. Thus, when we analyzed BRCA1 gene expression by RT-PCR, 2 of the 4 tumors classified as potentially BRCA1 dysfunctional in light of the abrogated foci response (#58, 62) also showed much lower expression levels than the 3 tumors classified as having functional BRCA1 status (#56, 57, 59) (Fig. 4c). The other 2 tumors with defective foci response (#55, 61) demonstrated BRCA1 expression levels overlapping with those seen in the IRIF-proficient tumors. Importantly, 3 of the 4 tumors classified as BRCA1-deficient (#55, 58, 61) but none of the BRCA1-proficient tumors were triple-negative (Fig. 4c).
FIGURE 4.
Identification of BRCA1-deficient tumors. A. Quantification of RAD51, FANCD2 and BRCA1 foci as in Figure 2B. Indicated is also the hormone receptor status for each of the four tumors. B. Quantification of RAD51 foci over several slides as in Figure 2C. C. Relative BRCA1 expression by quantitative RT-PCR is plotted against tumor classification based on foci analysis. Lines represent the logarithmic mean.
Discussion
We report here fore the first time the detection of BRCA1, FANCD2, and RAD51 foci in human breast cancer tissues. Strikingly, foci formation could be induced by exposing fresh biopsy samples to IR ex-vivo. The extent of foci induction, i.e., < 30% of cells scored positive, was less than observed for tumor cell lines(21, 22). This is likely related to the more homogeneous nature of cell populations that have been passaged numerous times. Further, the development of replication-associated repair protein foci is related to the growth fraction within the population of tumor cells, which is lower in tumor tissues than in cell lines.
Importantly, based on our foci analysis, we find a putative BRCA1 defect in 4 out of 7 tumors. This defect was characterized not only by an inability to form BRCA1 IRIF but also by an impairment of the downstream components FANCD2 and RAD51, as was predicted from cell line studies(10, 22). In a fifth case (#57), BRCA1 and RAD51 foci formation was intact and FANCD2 foci were present at baseline, but FANCD2 foci did not increase in number further with IR. The underlying genetic or epigenetic defect in this tumor remains to be identified. While a statistical analysis of our small study population is naturally limited, it is assuring to note that 3 of the 4 BRCA1-deficient tumors are triple negative, as would be predicted. These data also suggest that functional BRCA1 deficiency may occur in non-triple-negative tumors as well, a notion that is supported by preliminary results from ongoing large-scale cell line screens in our laboratories (Willers et al., Powell et al., unpublished data). Conversely, not all triple-negative sporadic breast cancers will be BRCA1 defective(14). It remains to be established whether BRCA1 defects, as defined by our foci assay, are predictors of clinical sensitivity to chemotherapeutics such as platinum compounds or topoisomerase II inhibitors(26, 27). While our pilot study is too small to address this question, the observed radiographic responses to neoadjuvant doxorubicin in patients with BRCA1 dysfunctional cancers are entirely consistent with this hypothesis (Taghian et al. unpublished data).
Characterizing the activity of the cellular DNA damage response to cytotoxic agents poses a challenge due to the spatiotemporal complexity of the response network(18-21). Identifying functional nodal points, such as BRCA1, FANCD2, or RAD51, offers an attractive approach to characterize the activity of the network. The advantage of using foci as biomarkers is that they can detect repair defects due to various mechanisms such as epigenetic events or gene mutations. Moreover, they provide a global measurement of network function without needing to know the identities of all the components, many of which are still unknown(9). As such, measuring BRCA1 foci formation should be more informative than monitoring BRCA1 mRNA or protein expression levels. While in Fig. 4 two tumors with relatively low BRCA1 expression levels also had a foci formation defect, two other tumors revealed DNA repair defects based on foci analysis despite having BRCA1 expression levels that were similar to the BRCA1-proficient tumors.
In conclusion, our study suggests that DNA repair foci can be utilized to detect functionally relevant BRCA1 defects in sporadic human breast cancers. Confirmatory studies with larger patient populations and automated foci scoring are required. Whether BRCA1 dysfunction translates into chemo- or radiosensitivity of the affected tumor is an important topic of ongoing and future investigations.
Material and Methods
Study population
We obtained pretreatment biopsies from eight breast cancer patients (cases 55-62), consecutively enrolled onto Dana-Farber/Harvard Cancer Center Protocol 99-278, Neoadjuvant Chemotherapy in Palpable Breast Cancer: Evaluation of Physiologic, Radiological and Molecular Markers in Predicting Response (AGT, principal investigator). The study was approved by the Institutional Review Board and written informed consent was obtained from every patient.
Ex-vivo treatment
Fresh core biopsies were transported in chilled complete cell culture medium to the laboratory and processed within 1-1.5 hours post-biopsy. Biopsy tissues were arbitrarily divided into two samples and subjected to 8 Gy X-irradiation (Siemens Stabilipan 2 X-ray generator operated at 250 kVp and 12 mA, dose rate of 2.08 Gy/min) or mock-treatment. Samples were incubated in humidified cell culture incubators at 37°C and 5% CO2 and snap frozen in OCT (Sigma-Aldrich) after 4 hours. Viable tumor was identified on serial cryosections by H&E staining. One specimen (#60) was of poor quality and not analyzed further.
Immunofluorescence (IF) microscopy
Adjacent cryoslides were analyzed for subnuclear foci formation by IF microscopy (Olympus BX51). Slides were fixed in 100% methanol at –20°C for 10 minutes and immersed in acetone for 30 seconds, followed by permeabilization with 0.5% Triton-X-100 in PBS for 15 minutes. After blocking in 3% BSA and 0.2% Triton-X-100 in PBS, rabbit polyclonal anti-RAD51 (Oncogene), mouse monoclonal anti-BRCA1 (Oncogene) and rabbit polyclonal anti-FANCD2 (Novus) were used at appropriate dilutions of 1:150, 1:50, and 1:200-400, respectively. Incubation with the primary antibodies was carried out at 37°C for at least 2 hours. Slides were then washed with PBS and incubated with Alexa-488 or Alexa-568 (Molecular Probes) or FITC-conjugated (Pierce) secondary antibodies at 37°C for 50 minutes. Slides were washed and counterstained with 1 ug/ml DAPI (Sigma-Aldrich). Slides were mounted in antifade reagent (Biorad). The specificity of the antibodies was confirmed by monitoring increased foci formation in irradiated versus untreated control cell lines (not shown). In order to semiquantify foci formation, each tumor sample was analyzed by capturing at least 8 random 100X images on 1-3 sections. On each high power image, typically 20-100 nuclei were scored. Only nuclei that could be clearly discerned were counted. A nucleus was scored as positive if containing at least 2 foci. Tumor nuclei were identified by DAPI staining (not shown). If DAPI was unable to distinguish between individual tumor nuclei due to the underlying tissue architecture, these cells were censored and not included in the count.
RT-PCR
Slides were subjected to laser capture microdissection and real-time quantitative PCR(28). TaqMan™ real-time PCR was performed using an ABI 7900HT (Applied Biosystems) as previously described(29). Each gene was analyzed in triplicate and the relative standard curve method was used for linear regression analysis of unknown samples in which β-actin levels served as an internal normalizing control. The sequences of the PCR primer pair and the fluorogenic TAMRA and MGB probes (5’ to 3’), for BRCA1 and b-actin, respectively, were as follows: FP: GGCTATGCAAGGGTCCCTTAA, RP: TGAATCAGCATCTTGCTCAATTG, Probe: VICTCTCCCTTGGAAATCTGCCATGAGCA-TAMRA; FP: CTTCCTGGGCATGGAGTCC, RP: ACGTCACACTTCATGATGGAGTT, Probe: VIC-ATCCACGAAACTAC.
Acknowledgements
Supported in part by an Avon-NCI Progress for Patients Award on the Dana-Farber/Harvard Cancer Center SPORE in Breast Cancer CA089393 (to AGT).
References
- 1.Foulkes WD, Stefansson IM, Chappuis PO, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 2.Lakhani SR, Reis-Filho JS, Fulford L, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11(14):5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 3.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25(43):5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 5.Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22(37):5784–5791. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25(43):5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 7.Powell SN, Kachnic LA. Therapeutic exploitation of tumor cell defects in homologous recombination. Anticancer Agents Med Chem. 2008;8(4):448–460. doi: 10.2174/187152008784220267. [DOI] [PubMed] [Google Scholar]
- 8.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 11.Levitus M, Waisfisz Q, Godthelp BC, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J. Nat Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 12.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 13.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 14.Turner NC, Reis-Filho JS, Russell AM, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26(14):2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 16.Staff S, Isola J, Tanner M. Haplo-insufficiency of BRCA1 in sporadic breast cancer. Cancer Res. 2003;63(16):4978–4983. [PubMed] [Google Scholar]
- 17.Rice JC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis. 2000;21(9):1761–1765. doi: 10.1093/carcin/21.9.1761. [DOI] [PubMed] [Google Scholar]
- 18.Essers J, Houtsmuller AB, van Veelen L, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. Embo J. 2002;21(8):2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118(6):699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10(15):886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 21.Shi W, Ma Z, Willers H, et al. Disassembly of MDC1 foci is controlled by ubiquitinproteasome-dependent degradation. J Biol Chem. 2008;283(46):31608–31616. doi: 10.1074/jbc.M801082200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Molecular & Cellular Biology. 2004;24(2):708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24(13):5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22(8):1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 25.Scully R, Chen J, Ochs RL, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90(3):425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275(31):23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 27.Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, Powell SN. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007;67(15):7078–7081. doi: 10.1158/0008-5472.CAN-07-0601. [DOI] [PubMed] [Google Scholar]
- 28.Elkahloun AG, Gaudet J, Robinson GS, Sgroi DC. In situ gene expression analysis of cancer using laser capture microdissection, microarrays and real time quantitative PCR. Cancer Biol Ther. 2002;1(4):354–358. [PubMed] [Google Scholar]
- 29.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100(10):5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]