Abstract
Background
Leishmaniases are tropical zoonotic diseases, caused by parasites from the genus Leishmania. New World (NW) species are related to sylvatic cycles although urbanization processes have been reported in some South American Countries such as Colombia. This eco-epidemiological complexity imposes a challenge to the detection of circulating parasite species, not only related to human cases but also infecting vectors and reservoirs. Currently, no harmonized methods have been deployed to discriminate the NW Leishmania species.
Findings
Herein, we conducted a systematic and mechanistic High-Resolution Melting (HRM) assay targeted to HSP70 and ITS1. Specific primers were designed that coupled with a HRM analyses permitted to discriminate six NW Leishmania species. In order to validate the herein described algorithm, we included 35 natural isolates obtained from human cases, insect vectors and mammals. Our genotyping assay allowed the correct assignment of the six NW Leishmania species (L. mexicana, L. infantum (chagasi), L. amazonensis, L. panamensis, L. guyanensis and L. braziliensis) based on reference strains. When the algorithm was applied to a set of well-characterized strains by means of PCR-RFLP, MLEE and monoclonal antibodies (MA) we observed a tailored concordance between the HRM and PCR-RFLP/MLEE/MA (KI = 1.0). Additionally, we tested the limit of detection for the HRM method showing that this is able to detect at least 10 equivalent-parasites per mL.
Conclusions
This is a rapid and reliable method to conduct molecular epidemiology and host-parasite association studies in endemic areas.
Keywords: Leishmania, Genotyping, Real-Time PCR, High-resolution Melting
Findings
Leishmaniasis is a tropical infection caused by the kinetoplastid parasites of the genus Leishmania, and is naturally transmitted to humans and among reservoirs by sandfly bites [1]. With a worldwide distribution and a large number of clinical cases, Leishmaniasis affects around 12 million individuals and 350 million are at risk of contracting the disease representing a serious issue in public health [2]. Additionally, control strategies against the disease are a challenge due to its eco-epidemiological complexity, involving sylvatic and domestic multi host-pathogen interactions (humans as well as a wide range of mammalian hosts). Recently, armed conflicts, increased tourism, natural disasters, deforestation, environmental and climate changes have contributed with new challenges to disease prevention and control: vectors distributions are reaching higher altitudes and latitudes increasing population at risk and changing the paradigm of poverty related disease [3-5].
The Neotropical Leishmania species responsible for a diversity of clinical symptoms are grouped in the two subgenera: Viannia and Leishmania. Within the Viannia subgenus, the species Leishmania (Viannia) braziliensis, Leishmania (Viannia) panamensis and Leishmania (Viannia) guyanensis are the most important causal agents of the New World tegumentary Leishmaniasis. From the Leishmania subgenus, the species Leishmania (Leishmania) infantum (chagasi) (related to Visceral Leishmaniasis) and the species of Leishmania (Leishmania) amazonesis and Leishmania (Leishmania) mexicana are responsible for the mucocutaneous, cutaneous and diffuse manifestations [6,7]. These parasite species have a wide geographic distribution from Mexico to the north of Argentina [6], with a significant increase in the last years in the number of cases in countries such as Bolivia, Brazil, Colombia and Peru [6,8-10]. The epidemiological complexity can be accompanied by a clinical difficulty when some infections, such as the one caused by L. braziliensis, appear in a cutaneous form and after self-healing reappear years later in a mucocutaneous form. This kind of situations makes species identification a critical step in clinical diagnosis and management.
As in most parasitic diseases, the challenge remains in having the ability to perform a rapid diagnosis of the disease and the characterization and genotyping of the different isolates obtained from infected individuals [11]. So far, the techniques typically employed to characterize and discriminate Leishmania species are Multi Locus Enzyme Electrophoresis (MLEE) [12] and multiple methods based on Polymerase Chain Reaction (PCR) such as Multilocus Sequence Typing (MLST) [13], PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) [14], Multiplex PCR [15] and PCR followed by sequencing [16]. These methods usually use microsatellites genes as amplification targets such as: kinetoplastid DNA (kDNA), telomeric sequences, internal transcribed spacer (ITS1), Heat Shock Proteins such as HSP70 and HSP60, and genes involved in metabolic processes like Mannose Phosphate Isomerase (MPI) and 6-phosphogluconate dehydrogenase (6GPD) [11-13,17].
From this great variety of molecular targets, kDNA, ITS1, and HSP70 have been the widest employed markers due to their abundance in the number of copies across Leishmania genome which increases the sensitivity of the assays [18-20]. However, traditional methods have multiple disadvantages including long procedures that entail an increased risk of DNA contamination, interpretation and processing of complex data, low sensitivity, high cost and cross-reactivity [21]. To overcome these disadvantages, Real-Time PCR methods, particularly those that use SYBR Green, TaqMan probes or FRET [11-13,17,22], have become available in the last few years to ensure better reproducibility, specificity, sensitivity, and velocity during diagnosing and genotyping [23,24]. Some of these methods can distinguish between the two subgenera L. (Leishmania) and L (Viannia) at a complex level, but with many cross-reactions of human DNA and other mammals [11]. Albeit, other methods can only distinguish among the species of the L. (Viannia) complex [13-17].
From this perspective, it is mandatory to develop and implement a technique that allows specific, rapid, robust, and cost-effective identification of the Leishmania species circulating in the New World to be employed in both clinical and eco-epidemiological studies. High Resolution Melting (HRM) is a technique that has been widely used in the genotyping of bacteria, fungi, protozoan parasites and vertebrates [25-29] and could be used for identification and genotyping of these Leishmania species [30]. The objective of this study was to evaluate the potential of the HSP70 and ITS1 genes in genotyping six Leishmania species present in the New World through the use of High Resolution Melting (HRM).
Promastigotes of the reference strains L. panamensis (MHOM/PA/71/LS94), L. guyanensis (MHOM/BR/75/M4147), L. braziliensis (MHOM/BR/75/M2903), L. amazonensis (IFLA/BR/67/PH8), L. mexicana (MHOM/BZ/82/BEL21) and L. infantum (chagasi) (MHOM/TN/80/IPT1) were kindly provided by the Centro Internacional de Entrenamiento e Investigaciones Medicas (CIDEIM). Parasites were maintained in a biphasic medium of Liver Infusion (Tobie medium infused with human blood) and Triptone (LIT) supplemented with Bovine fetal serum inactivated at 20% and incubated at 25°C for 5 days to reach the late logarithmic growth phase. Aliquots of 200 μL of the LIT biphasic culture medium were obtained and then used for DNA extraction using the Roche High Pure PCR Template Preparation Kit™ following the respective instructions of the insert. The obtained DNA was quantified in a NanoDrop 2000 at a wavelength of 260 nm.
HSP70 and ITS1 genes were used given their good performance as reported by other authors [24-26]. For the HSP70 gene, primer design was carried out using the sequences available on GenBank, TriTrypDB and www.itg.be/leishmaniahsp70 of New World Leishmania spp. (L. mexicana (XM_003877072.1); L. amazonensis (L14605.1); L. braziliensis (AF291716.1); L.guyanensis (EU599093.1); L. infantum (chagasi) (XM_003392632.1); L. panamensis (FN395055.1)). Multiple alignments (1380 bp) were performed using the software Mega 5 [31]. Primers were designed using the software PRIMER BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) from the consensus sequence obtained by the multiple alignments, with an expected amplicon size of 337 bp for all the sequences. For the amplification of ITS1 gene, the primers reported by El Tai et. al (2000) were used and adapted to the conditions of Real-Time PCR of this study. The expected amplicon size in this case was 300–350 pb.
Real-Time PCR was coupled together with HRM analysis using the HSP70 and ITS1 genes as targets in a Real-Time PCR system 7500 (Applied Biosystems, Inc., CA, USA) with 21 μL amplification reactions. The reaction mix contained 1X of Master Mix MeltDoctor HRM (Applied Biosystems, Inc., CA, USA), a 5 μM solution of each primers HSP70F (5’ AGG TGA AGG CGA CGA ACG 3’) and HSP70R (5’ CGC TTG TCC ATC TTT GCG TC 3’) for the amplification of HSP70 and for ITS1, LITSR (5’ CTG GAT CAT TTT CCG ATG 3’) and L5.8S (5’ TGA TAC CAC TTA TCG CAC TT 3’) [32], 6.6 μL of water and 10 ng/μL of DNA template. Real-Time PCR cycle conditions were adjusted to the following protocol: 95°C for 10 minutes (1 cycle) followed by a 40-cycles amplification (95°C for 15 seconds (denaturation) and then 60°C for 1 minute). After the Real-Time PCR, a dissociation of the amplicon was performed and followed immediately by a fusion step. The thermal profile consisted in denaturation at 95°C for 10 seconds, annealing at 60°C for 1 minute, High Resolution Melting at 95°C for 30 seconds, and a final annealing stage at 60°C for 15 seconds. During this process, the amplicons obtained from PCR were denatured prior to the development of melting curves in the inflexion point where changes in fluorescence with respect to changes in temperature (dF/dT) were recorded with a ramp of 0.1°C/seg [25]. Each DNA sample was analyzed in duplicate. Finally, the profiles of the normalized and derived fusion curves were obtained. Analyses of the obtained melting values were conducted with the software Graphpad Prism 6 (GraphPah Software, San Diego California USA, www.graphpad.com) to determine mean and standard deviations. To verify the reproducibility of the genotyping assay, this process was performed on 15 different days with the same DNA aliquots under the same conditions. The contamination controls of the study always included two controls of reaction mix without DNA (controls without templates - NTC). To evaluate the cross-reactivity, uninfected human and mouse DNA were tested and compared to the known positive range of melting temperatures for New World Leishmania species. Serial dilutions (factor 10) from 106 to 1 parasite per mL were performed to establish the detection limit, which was defined as the minimum number of parasites necessary to generate a melting curve inside the reference values.
To determine the accuracy and reliability of the method herein reported, we analyzed 35 Leishmania strains obtained from humans, reservoirs and insect vectors from Colombia that were previously characterized by PCR-RFLP, monoclonal antibodies and MLEE. The HRM was applied in blind to this set of well-characterized strains and examined by duplicate. Based on this characterization, we determined the concordance between PCR-RFLP, MLEE and the HRM algorithm by means of the Kappa Index.
Primers for the HSP70 gene designed to obtain specific melting temperatures were able to discriminate the subgenera L. (Leishmania) and L. (Viannia) and the complexes L. mexicana from L. infantum (chagasi) - L. amazonensis and L. panamensis from L. braziliensis - L. guyanensis (Figure 1A). For the ITS1 gene, the obtained ranges allowed clear discrimination of the species L. infantum (chagasi) from L. amazonensis and L. braziliensis from L. guyanensis (Figure 1B). These results were used to construct the herein showed algorithm (Figure 2) which, following the amplification of HSP70 and then ITS1, can be used to discriminate six Leishmania species. No cross-reactivity was observed during the assays for HSP70 or ITS1 genes with uninfected human and mouse DNA. The detection limit for the HRM assay was determined on average in all examined species to be 10 parasites/mL for both markers. Based on the designed algorithm, we tested 35 well-characterized Leishmania strains. We were able to obtain reliable Tm curves for each strain that were in accordance with the ranges found in the reference strains (Table 1). We calculated the Kappa Index considering PCR-RFLP/MLEE as the gold standard and the results showed a High concordance between HRM and PCR-RFLP/MLEE (KI = 1.0).
Figure 1.
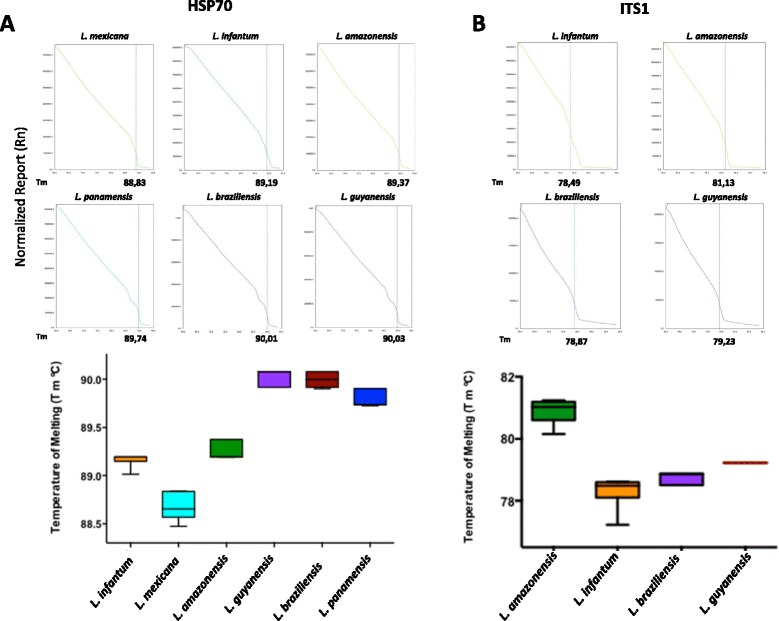
Melting Temperature profiles and scatter plots of the HRM genotyping assay. A. Representative normalized melting curves of each species of Leishmania for the genes HSP70 and ITS1. B. Box diagram showing the range of obtained temperatures of melting (Tm) for the curves of each species of Leishmania.
Figure 2.
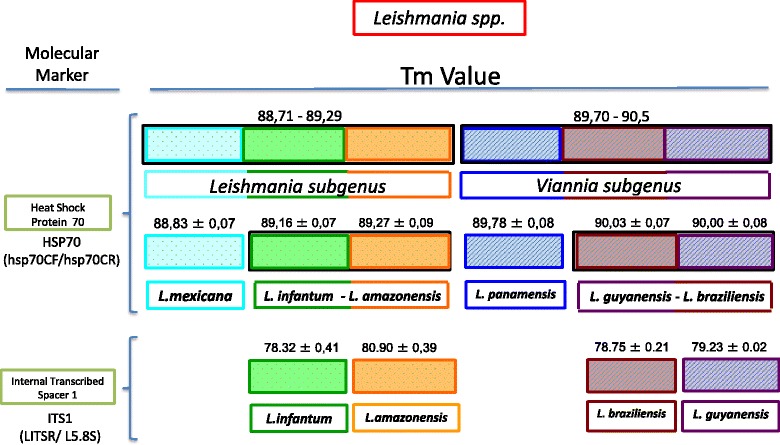
Algorithm that shows the use of HRM assays for genes HSP70 and ITS1 in differentiating species of New World Leishmania .
Table 1.
Comparison of HRM assay and PCR-RFLP/MLEE in the set of well-characterized Leishmania strains
| Sample name | Geographical origin | Host | PCR-RFLP/MLEE | HSP70 | Tm | ITS1 | Tm |
|---|---|---|---|---|---|---|---|
| 6 | Meta/Colombia | Homo sapiens | L. guyanensis | L. guyanensis/L. braziliensis | 90,08 | L. guyanensis | 79,22 |
| 7 | Santander/Colombia | Lutzomyia umbratilis | L. guyanensis | L. guyanensis/L. braziliensis | 90,07 | L. guyanensis | 79,21 |
| 71 | Boyaca/Colombia | Lutzomyia umbratilis | L. guyanensis | L. guyanensis/L. braziliensis | 90,07 | L. guyanensis | 79,23 |
| 875 | Santander/Colombia | Homo sapiens | L. infantum (chagasi) | L. guyanensis/L. braziliensis | 90,07 | L. chagasi | 79,21 |
| 924 | Santander/Colombia | Homo sapiens | L. panamensis | L. panamensis | 89,91 | L. panamensis | - |
| 113 | Meta/Colombia | Homo sapiens | L. braziliensis | L. guyanensis/L. braziliensis | 90,08 | L. braziliensis | 78,77 |
| 12 | Boyaca/Colombia | Homo sapiens | L. braziliensis | L. guyanensis/L. braziliensis | 90,09 | L. braziliensis | 78,55 |
| 13 | Boyaca/Colombia | Homo sapiens | L. guyanensis | L. guyanensis/L. braziliensis | 90,06 | L. guyanensis | 79,22 |
| 15 | Boyaca/Colombia | Didelphis marsupialis | L. panamensis | L. panamensis | 89,73 | L. panamensis | - |
| 19 | Meta/Colombia | Lutzomyia spinicrassa | L. braziliensis | L. guyanensis/L. braziliensis | 90,09 | L. braziliensis | 78,77 |
| 20 | Meta/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,73 | L. panamensis | - |
| 21 | Meta/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,83 | L. panamensis | - |
| 23 | Meta/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,73 | L. panamensis | - |
| 86 | Santander/Colombia | Homo sapiens | L. infantum (chagasi) | L. infantum/L. amazonensis | 89,14 | L. chagasi | 78,33 |
| 87 | Santander/Colombia | Homo sapiens | L. infantum (chagasi) | L. infantum/L. amazonensis | 89,17 | L. chagasi | 78,56 |
| 94 | Santander/Colombia | Homo sapiens | L. infantum (chagasi) | L. infantum/L. amazonensis | 89,19 | L. chagasi | 78,55 |
| 094b | Antioquia/Colombia | Didelphis marsupialis | L. mexicana | L. mexicana | 88,75 | L. mexicana | - |
| 245 | Antioquia/Colombia | Homo sapiens | L. amazonensis | L. infantum/L. amazonensis | 89,19 | L. amazonensis | 80,61 |
| 280 | Antioquia/Colombia | Homo sapiens | L. amazonensis | L. infantum/L. amazonensis | 89,19 | L. amazonensis | 80,77 |
| 916 | Antioquia/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,79 | L. panamensis | - |
| 916 | Antioquia/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,73 | L. panamensis | - |
| 984 | Antioquia/Colombia | Homo sapiens | L. braziliensis | L. guyanensis/L. braziliensis | 90,04 | L. braziliensis | 78,77 |
| 985 | Antioquia/Colombia | Lutzomyia spinicrassa | L. braziliensis | L. guyanensis/L. braziliensis | 90,08 | L. braziliensis | 78,89 |
| 986 | Meta/Colombia | Homo sapiens | L. braziliensis | L. guyanensis/L. braziliensis | 90,09 | L. braziliensis | 78,91 |
| 990 | Meta/Colombia | Homo sapiens | L. guyanensis | L. guyanensis/L. braziliensis | 90,04 | L. guyanensis | 79,22 |
| 094b | Meta/Colombia | Homo sapiens | L. infantum (chagasi) | L. infantum/L. amazonensis | 89,22 | L. chagasi | 78,67 |
| 094b | Meta/Colombia | Homo sapiens | L. infantum (chagasi) | L. infantum/L. amazonensis | 89,29 | L. chagasi | 78,66 |
| 101 | Boyaca/Colombia | Homo sapiens | L. amazonensis | L. infantum/L. amazonensis | 89,14 | L. amazonensis | 81,11 |
| 123 | Boyaca/Colombia | Homo sapiens | L. braziliensis | L. guyanensis/L. braziliensis | 90,02 | L. braziliensis | 78,66 |
| 157 | Santander/Colombia | Homo sapiens | L. guyanensis | L. guyanensis/L. braziliensis | 90,04 | L. guyanensis | 79,22 |
| 146 | Santander/Colombia | Homo sapiens | L. guyanensis | L. guyanensis/L. braziliensis | 90,01 | L. guyanensis | 79,22 |
| 147 | Santander/Colombia | Didelphis marsupialis | L. mexicana | L. mexicana | 88,37 | L. mexicana | - |
| 222 | Santander/Colombia | Homo sapiens | L. panamensis | L. panamensis | 89,82 | L. panamensis | - |
| 2698 | Santander/Colombia | Homo sapiens | L. panamensis | L. panamensis | 89,81 | L. panamensis | - |
| 1453 | Santander/Colombia | Lutzomyia trapidoi | L. panamensis | L. panamensis | 89,88 | L. panamensis | - |
Currently, the methods employed for the discrimination of New World Leishmania species are laborious, subjective and often lacking of specificity [21]. Therefore, it is crucial to count on a low-cost, effective and rapid tool that allows an accurate detection and identification of the Leishmania species present in the New World or at least the most prevalent. Herein, the proposed algorithm allowed identifying L. mexicana, L. infantum (chagasi), L. amazonensis, L. panamensis, L. braziliensis and L. guyanensis with a Real-Time PCR coupled with HRM analysis after the amplification of HSP70 gene and the subsequent amplification of ITS1 gene. The HRM methodology is considered as an excellent option to reduce some problems of the previously molecular techniques used for Leishmania genotyping such as the need of modified oligonucleotides, low accuracy, limited high-throughput applications, etc. [33] since this is a closed-tube assay (does not require additional precautions to prevent crossover of PCR products), avoids sequencing processes, laborious procedures and high costs as well as offering robust and high efficiency in its results [25,34]. On average, this type of test has been calculated to be three times faster and five times cheaper than other types of analysis such as MLST and RFLPs [17].
Additionally, our algorithm helps to differentiate between L. (L). mexicana and L. (L). amazonensis, two species that in previous studies could not be identified using HRM tests [11,17]. This differentiation is important, and although L. (L.) amazonensis infections usually exhibit a milder form of the disease, it can also cause diffuse forms that do not respond adequately to existing treatments [35]. On the other hand, our algorithm can also differentiate between L. braziliensis (the main etiologic cause of cutaneous Leishmaniasis) and L. guyanensis (with high prevalence in the Amazon region). This is of great importance due to its eco-epidemiological implications and the close genetic relationship of the members of the subgenus Viannia that has represented a limited number of techniques that can be used to correctly identify them [36]. As well, HSP70 and ITS1 showed a higher resolution in characterizing the members of the subgenera Viannia and Leishmania than other genes. For example, markers like kDNA can only discriminate between fewer members of these subgenera but are nonetheless used in many assays as a gold standard for diagnosis of Old and New World Leishmaniasis due to its high sensitivity [11,17].
The results of our study support the idea that genes such HSP70 and ITS1 are good candidates for genotyping neotropical Leishmania species, and that these genes allow detection of genetic variability in molecules associated with immunological processes (HSP70) and confer a high probability of being detected due to their high number of copies [35]. Similarly, regarding specificity, this method does not cause mammalian DNA cross-reactions like has been previously noted in studies using HRM [11]. This fact makes HRM a good candidate for identifying Leishmania in epidemiological studies. Also, the technique is reproducible, and this was retained even though the tests were duplicated and performed on different days, albeit under the same conditions of DNA and reactive concentrations.
We decided to apply this algorithm to a set of well-characterized strains by the gold standard method (PCR-RFLP/MLEE). The findings allowed observing that HRM was accurate enough to discriminate six Neotropical species and was 100% concordant with PCR-RFLP/MLEE which suggests its suitability in the application to a broad number of epidemiological questions. Another important feature of our assay besides its reproducibility was a tailored sensitivity obtained where the method is able to detect at least 10 parasites/mL and is considered a remarkable value of sensitivity. These advantages demonstrate that this method has a tremendous potential for the identification of New World Leishmania species and is a suitable choice for identification and characterization. Nonetheless, it is important to point out that the Leishmania strains employed in this implementation were isolated from Colombian hosts and unbiased range of Leishmania diversity was not considered. Therefore, we suggest that this novel method is implemented in strains from Meso-America in order to validate the feasibility of the technique.
Conclusions
In conclusion, the suggested HRM assay in this study is a reliable, robust and reproducible technique that allows the correct genotyping of six New World Leishmania species, in a rapid way (it does not require post-PCR treatments), little labor and low cost (around 7.5 USD) with a potential applicability in different areas of molecular epidemiology of Leishmaniasis such as host-parasite relationships, eco-epidemiology and infection dynamics in urban and wild areas. We encourage the scientific community from Leishmania endemic areas to test this novel approach in order to fully validate its reliability at a continental level.
Acknowledgements
Authors wish to thank CIDEIM for providing reference strains. This study was supported by the FAPA grant Vicerrectoría de Investigaciones, Universidad de Los Andes, Colciencias (Contrato 315 de ACA-INS). CA received a Colciencias Young Researcher award (Young Researcher 2012 - Agreement 0020–2012).
Footnotes
Carolina Hernández, Catalina Alvarez, and Camila González, are contributed equally to this work.
Competing interests
The author declared that they have no competing interests.
Authors’ contributions
JDR, CG, CA, CH designed the experiments. CA, CH, CML, MSA and JDR standardized the HRM assay and developed the implementation of the HRM assay across the 35 Leishmania isolates. JDR, CA, CH and CG wrote the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Carolina Hernández, Email: dcahernandezc@gmail.com.
Catalina Alvarez, Email: mc.alvarez68@uniandes.edu.co.
Camila González, Email: c.gonzalez2592@uniandes.edu.co.
Martha Stella Ayala, Email: mayala@ins.gov.co.
Cielo Maritza León, Email: cmlr8527@gmail.com.
Juan David Ramírez, Email: juand.ramirez@urosario.edu.co.
References
- 1.Weirather JL, Jeronimo SM, Gautam S, Sundar S, Kang M, Kurtz MA, Haque R, Schriefer A, Talhari S, Carvalho EM, Donelson JE, Wilson ME. Serial quantitative PCR assay for detection, species-discrimination and quantification of Leishmania spp. in human samples. J Clin Microbiol. 2011;49(11):3892–904. doi: 10.1128/JCM.r00764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27(5):305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez C, Wang O, Strutz SE, Gonzalez-Salazar C, Sanchez-Cordero V, Sarkar S. Climate change and risk of leishmaniasis in north america: predictions from ecological niche models of vector and reservoir species. PLoS Negl Trop Dis. 2010;4(1):e585. doi: 10.1371/journal.pntd.0000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw J. The leishmaniases–survival and expansion in a changing world. A mini-review. Mem Inst Oswaldo Cruz. 2007;102(5):541–547. doi: 10.1590/S0074-02762007000500001. [DOI] [PubMed] [Google Scholar]
- 5.Desjeux P. The increase in risk factors for leishmaniasis worldwide. Trans R Soc Trop Med Hyg. 2001;95(3):239–243. doi: 10.1016/S0035-9203(01)90223-8. [DOI] [PubMed] [Google Scholar]
- 6.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7(9):581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 7.Lainson R, Shaw JJ: New World Leishmaniasis. In Topley & Wilson’s Microbiology and Microbial Infections. John Wiley & Sons, Ltd; 2010. http://onlinelibrary.wiley.com/doi/10.1002/9780470688618.taw0182/abstract.
- 8.Lucas CM, Franke ED, Cachay MI, Tejada A, Cruz ME, Kreutzer RD, Barker DC, McCann SH, Watts DM. Geographic distribution and clinical description of leishmaniasis cases in Peru. Am J Trop Med Hyg. 1998;59(2):312–317. doi: 10.4269/ajtmh.1998.59.312. [DOI] [PubMed] [Google Scholar]
- 9.Velez ID, Carrillo LM, Lopez L, Rodriguez E, Robledo SM. An epidemic outbreak of canine cutaneous leishmaniasis in Colombia caused by Leishmania braziliensis and Leishmania panamensis. Am J Trop Med Hyg. 2012;86(5):807–811. doi: 10.4269/ajtmh.2012.11-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez E, Mollinedo S, Torrez M, Munoz M, Banuls AL, Le Pont F. Co-infection by Leishmania amazonensis and L. infantum/L. chagasi in a case of diffuse cutaneous leishmaniasis in Bolivia. Trans R Soc Trop Med Hyg. 2002;96(5):529–532. doi: 10.1016/S0035-9203(02)90428-1. [DOI] [PubMed] [Google Scholar]
- 11.Pita-Pereira D, Lins R, Oliveira MP, Lima RB, Pereira BA, Moreira OC, Brazil RP, Britto C. SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral leishmaniases. Parasit Vectors. 2012;5:15. doi: 10.1186/1756-3305-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arana M, Evans DA, Zolessi A, Cuentas AL, Arevalo J. Biochemical characterization of Leishmania (Viannia) braziliensis and Leishmania (Viannia) peruviana by isoenzyme electrophoresis. Trans R Soc Trop Med Hyg. 1990;84(4):526–529. doi: 10.1016/0035-9203(90)90025-A. [DOI] [PubMed] [Google Scholar]
- 13.Tsukayama P, Lucas C, Bacon DJ. Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol. 2009;39(3):355–362. doi: 10.1016/j.ijpara.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 14.de Andrade HM, Reis AB, dos Santos SL, Volpini AC, Marques MJ, Romanha AJ. Use of PCR-RFLP to identify Leishmania species in naturally-infected dogs. Vet Parasitol. 2006;140(3–4):231–238. doi: 10.1016/j.vetpar.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 15.Jorquera A, Gonzalez R, Marchan-Marcano E, Oviedo M, Matos M. Multiplex-PCR for detection of natural Leishmania infection in Lutzomyia spp. captured in an endemic region for cutaneous leishmaniasis in state of Sucre, Venezuela. Mem Inst Oswaldo Cruz. 2005;100(1):45–48. doi: 10.1590/S0074-02762005000100008. [DOI] [PubMed] [Google Scholar]
- 16.Schulz A, Mellenthin K, Schonian G, Fleischer B, Drosten C. Detection, differentiation, and quantitation of pathogenic leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J Clin Microbiol. 2003;41(4):1529–1535. doi: 10.1128/JCM.41.4.1529-1535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukayama P, Nunez JH, De Los Santos M, Soberon V, Lucas CM, Matlashewski G, Llanos-Cuentas A, Ore M, Baldeviano GC, Edgel KA, Lescano AG, Graf PC, Bacon DJ. A FRET-based real-time PCR assay to identify the main causal agents of New World tegumentary leishmaniasis. PLoS Negl Trop Dis. 2013;7(1):e1956. doi: 10.1371/journal.pntd.0001956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toz SO, Culha G, Zeyrek FY, Ertabaklar H, Alkan MZ, Vardarli AT, Gunduz C, Ozbel Y. A real-time ITS1-PCR based method in the diagnosis and species identification of leishmania parasite from human and dog clinical samples in Turkey. PLoS Negl Trop Dis. 2013;7(5):e2205. doi: 10.1371/journal.pntd.0002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arevalo J, Wilber Quispe Tintaya K, Dujardin JC. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol. 2004;42(5):2294–2297. doi: 10.1128/JCM.42.5.2294-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volpini AC, Passos VM, Oliveira GC, Romanha AJ. PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop. 2004;90(1):31–37. doi: 10.1016/j.actatropica.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 21.de Paiva Cavalcanti M, Dantas-Torres F, da Cunha Goncalves de Albuquerque S, Silva de Morais RC, de Brito ME, Otranto D, Brandao-Filho SP. Quantitative real time PCR assays for the detection of Leishmania (Viannia) braziliensis in animals and humans. Mol Cell Probes. 2013;27(3–4):122–128. doi: 10.1016/j.mcp.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 22.de Monbrison F, Mihoubi I, Picot S. Real-time PCR assay for the identification of cutaneous Leishmania parasite species in Constantine region of Algeria. Acta Trop. 2007;102(2):79–83. doi: 10.1016/j.actatropica.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Jara M, Adaui V, Valencia BM, Martinez D, Alba M, Castrillon C, Cruz M, Cruz I, Van der Auwera G, Llanos-Cuentas A, Dujardin JC, Arevalo J. Real-time PCR assay for detection and quantification of leishmania (viannia) organisms in skin and mucosal lesions: exploratory study of parasite load and clinical parameters. J Clin Microbiol. 2013;51(6):1826–1833. doi: 10.1128/JCM.00208-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wortmann G, Hochberg L, Houng HH, Sweeney C, Zapor M, Aronson N, Weina P, Ockenhouse CF. Rapid identification of Leishmania complexes by a real-time PCR assay. Am J Trop Med Hyg. 2005;73(6):999–1004. [PubMed] [Google Scholar]
- 25.Higuera SL, Guhl F, Ramirez JD. Identification of trypanosoma cruzi discrete typing units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasit Vectors. 2013;6(1):112. doi: 10.1186/1756-3305-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong SY, Xie S, Richardson LJ, Ballard SA, Dakh F, Grabsch EA, Grayson ML, Howden BP, Johnson PD, Giffard PM. High-resolution melting genotyping of Enterococcus faecium based on multilocus sequence typing derived single nucleotide polymorphisms. PLoS One. 2011;6(12):e29189. doi: 10.1371/journal.pone.0029189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldschmidt P, Degorge S, Benallaoua D, Semoun O, Borsali E, Le Bouter A, Batellier L, Borderie V, Laroche L, Chaumeil C. New strategy for rapid diagnosis and characterization of keratomycosis. Ophthalmology. 2012;119(5):945–950. doi: 10.1016/j.ophtha.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 28.Costa JM, Cabaret O, Moukoury S, Bretagne S. Genotyping of the protozoan pathogen Toxoplasma gondii using high-resolution melting analysis of the repeated B1 gene. J Microbiol Methods. 2011;86(3):357–363. doi: 10.1016/j.mimet.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen N, Ali RG, Ahmed JN, Arkell RM. High resolution melt analysis (HRMA); a viable alternative to agarose gel electrophoresis for mouse genotyping. PLoS One. 2012;7(9):e45252. doi: 10.1371/journal.pone.0045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schutz E, von Ahsen N. Influencing factors of dsDNA dye (high-resolution) melting curves and improved genotype call based on thermodynamic considerations. Anal Biochem. 2009;385(1):143–152. doi: 10.1016/j.ab.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.el Tai NO, Osman OF, el Fari M, Presber W, Schonian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg. 2000;94(5):575–579. doi: 10.1016/S0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- 33.Liew M, Nelson L, Margraf R, Mitchell S, Erali M, Mao R, Lyon E, Wittwer C. Genotyping of human platelet antigens 1 to 6 and 15 by high-resolution amplicon melting and conventional hybridization probes. J Mol Diagn. 2006;8(1):97–104. doi: 10.2353/jmoldx.2006.050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krypuy M, Newnham G, Thomas D, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6(1):295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva LA, de Sousa Cdos S, da Graca GC, Porrozzi R, Cupolillo E. Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol. 2010;10(1):77–83. doi: 10.1016/j.meegid.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, Marco JD, Mimori T, Gomez EA, Hashiguchi Y, Uezato H. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009;121(4):352–361. doi: 10.1016/j.exppara.2008.12.013. [DOI] [PubMed] [Google Scholar]


