Abstract
Background
Xylogen, a chimeric arabinogalactan protein containing a non-specific lipid transfer protein domain, can promote xylem cell differentiation. No comprehensive study has been carried out on the XYLP gene family in rice. As a first step in research on this gene family and as a useful strategy in general, a genome-wide analysis of the OsXYLP gene family is thus needed.
Results
In this study, we identified 21 XYLP genes from the rice genome and comprehensively analyzed their protein structures, phylogenetic relationships, chromosomal locations, and gene duplication status. Our results indicate that gene duplication has played major roles in the expansion of the OsXYLP gene family. We used expressed sequence tag, microarray, massively parallel signature sequencing, and quantitative real-time PCR data to analyze OsXYLP gene expression during various developmental stages and under abiotic stress conditions. We found that many OsXYLP genes are abundantly expressed in vascular tissues and seeds, with some genes regulated under hormonal or abiotic stresses. In addition, we identified knockout mutants of OsXYLP7 and OsXYLP16 and discovered that the mutant xylp7 has a defect in stem height.
Conclusions
We analyzed expression profiles of 21 XYLP genes and characterized the structures and evolutionary relationships of their proteins. Our results demonstrate that the rice XYLP gene family may play roles in plant vascular system development and hormone signaling. Among the 21 detected OsXYLPs, 19 are newly identified genes encoding arabinogalactan proteins. Our results provide comprehensive insights that will assist future research on the biological functions of the rice XYLP gene family.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-014-0299-y) contains supplementary material, which is available to authorized users.
Keywords: Rice, XYLPs, Arabinogalactan protein, Expression analysis, Non-specific lipid transfer protein
Background
Arabinogalactan proteins (AGPs) are a class of extracellular glycoproteins consisting of a core protein backbone and diverse type-II arabinogalactan (AG) polysaccharide chains made up of galactan and arabinose [1-4]. Typical AGP molecular weights range from 60 to 300 kDa. The protein backbones are usually rich in proline/hydroxyproline, alanine, serine, and threonine (PAST), with the hydroxyproline O-glycosylated by peripheral AG side chains that determine macromolecular heterogeneity [3,5]. AGPs are classified into several subclasses based on their core protein structures: classical AGPs, Lys-rich AGPs, AG peptides, non-classical AGPs, and chimeric AGPs [6-9]. According to their domain constitutions, chimeric AGPs can be further divided into three subclasses: fasciclin-like AGPs (FLAs) [7,10], xylogen-like proteins (XYLPs) [11,12], and phytocyanin-like AGPs (PLAs) [10,13,14]. Previous researchers have identified 98 AGPs in rice, including 11 classical AGPs, 15 AG peptides, 2 Lys-rich AGPs, 27 FLAs, 38 phytocyanin-like AGPs, and 3 non-classical AGPs [14-16]. AGPs can selectively bind to a synthetic dye, β-glucosyl Yariv reagent (β-GlcY). Although the precise underlying mechanism is unclear, this binding requires the presence of both the protein and AG chains. β-GlcY binding ability can thus be used as a distinguishing standard to identify AGPs [17,18]. Many studies on the biological function of AGPs have been performed using β-GlcY and polyclonal antibodies such as JIM8, JIM13, JIM14, LM2, and CCRC-M7 [19]. AGPs have been reported to be involved in various plant growth and developmental processes, such as cell expansion [20-22], cell proliferation [23-25], programmed cell death [26,27], cell wall plasticization [28], hormone response [29], salt tolerance [28,30], xylem differentiation [11], root growth and development [31], female and male gametogenesis [32-36], pollen tube growth [37,38], and zygotic division and embryo development [33,39-42].
Plant non-specific lipid-transfer proteins (nsLTPs), which are abundant small basic proteins that can transfer phospholipids between membranes, were first isolated from spinach leaves as phospholipid-binding proteins [43,44]. The lipid-binding properties of nsLTPs are derived from a unique structure: a region of eight strictly conserved cysteine residues. The eight cysteines bind to one another to form four disulfide bridges that give rise to a three-dimensional structure containing an internal hydrophobic cavityable to firmly bind lipids [44]. Xylogen, a 25–300-kDa glycoprotein, mediates local intercellular communication and is essential for tracheary element (TE) differentiation in in vitro Zinnia elegans xylogenic culture [44,45]. Xylogen is secreted from differentiating vascular cells and promotes the transformation of adjacent undifferentiated cells into TEs; it has a unique structure including AGP domains and an nsLTP domain, as typical structure of chimeric AGPs [11]. In a previous bioinformatic analysis of xylogen-type proteins in Arabidopsis [12], 13 AtXYLP (xylogen-like protein) genes with significant similarity to ZeXYP1 were identified and their expression profiles were analyzed.
Genome-wide analysis is a useful strategy for the elucidation of biological functions of the XYLP gene family. In this study, we identified 21 XYLP genes in the rice (Oryza sativa L.) genome and conducted a phylogenetic analysis. To obtain further information about OsXYLP gene expression patterns, we evaluated publicly available resources such as microarray and massively parallel signature sequencing (MPSS) databases. We then validated the digital expression data obtained for these genes through quantitative real-time PCR (qRT-PCR). In addition, we identified the knockout mutants of OsXYLP7 and OsXYLP16 and found that OsXYLP7 is involved in stem development. Our results provide a comprehensive understanding of OsXYLPs and may serve as a guide for research on the OsXYLP gene family.
Results
Identification of putative OsXYLPs
To identify xylogen-like proteins (XYLPs) in rice, we performed BLASTP searches across several rice protein databases using ZeXYP1, AtXYP1, and AtXYP2 protein sequences as queries [11]. After confirming the presence of nsLTP-like domains, AGP-like regions, and AG-type glycomodules and removing redundant sequences, we identified 21 OsXYLPs in rice (Table 1). To ensure the detection of all proteins in this family, we conducted additional BLASTP searches using protein sequences of the 21 identified OsXYLPs; these searches yielded no more XYLPs. Among the 21 OsXYLPs, we identified 19 new AGPs. The remaining 2 identified OsXYLPs, OsLTPL1 (OsLLA1) [16,46] and OsXYLP9 (OsLLA6) [46], were among 98 AGPs previously identified [14-16,46]. OsLTPL1 was first isolated as a β-GlcY-reactive arabinogalactan protein; and then OsLTPL1 and OsXYLP9 were identified as nsLTP-like AGPs.
Table 1.
The general information of rice XYLP genes
| Gene a | Subfamily b | RGAP locus c | RAP-DB locus d | Chromosome location e | Size(aa) f | Signal g | GPI h | FL-cDNA i | EST j | MI k | MPSS l |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OsLTPL1 | Clade A | LOC_Os03g26820 | Os03g0385400 | chr03:15307624..15309776 | 178 | √ | √ | √ | √ | √ | √ |
| OsXYLP2 | Clade A | LOC_Os03g26800 | Os03g0385100 | chr03:15300737..15301156 | 139 | √ | - | - | - | - | √ |
| OsXYLP3 | Clade A | LOC_Os07g30590 | Os07g0489000 | chr07:18102558..18103713 | 170 | √ | √ | √ | √ | √ | √ |
| OsXYLP4 | Clade A | LOC_Os07g43290 | Os07g0625800 | chr07:25920699..25922871 | 177 | √ | √ | √ | √ | √ | √ |
| OsXYLP5 | Clade A | LOC_Os03g09230 | Os03g0192600 | chr03:4820699..4822659 | 214 | √ | √ | √ | √ | √ | √ |
| OsXYLP6 | Clade B | LOC_Os03g20760 | Os03g0323900 | chr03:11751959..11753399 | 199 | √ | √ | √ | √ | √ | - |
| OsXYLP7 | Clade B | LOC_Os05g41030 | Os05g0489200 | chr05:24045555..24047844 | 210 | √ | √ | √ | √ | √ | √ |
| OsXYLP8 | Clade B | LOC_Os01g59870 | Os01g0814100 | chr01:34626255..34628601 | 187 | √ | √ | √ | √ | √ | √ |
| OsXYLP9 | Clade C | LOC_Os07g07790 | Os07g0174400 | chr07:3910661..3911056 | 188 | √ | √ | - | √ | √ | √ |
| OsXYLP10 | Clade C | LOC_Os07g07860 | Os07g0174900 | chr07:3947968..3949371 | 171 | √ | √ | - | √ | √ | √ |
| OsXYLP11 | Clade C | LOC_Os03g57990 | Os03g0794000 | chr03:33028009..33029105 | 189 | √ | √ | - | √ | √ | √ |
| OsXYLP12 | Clade C | LOC_Os07g07870 | Os07g0175000 | chr07:3950662..3951180 | 181 | √ | √ | - | - | √ | √ |
| OsXYLP13 | Clade C | LOC_Os03g57970 | Os03g0793800 | chr03:33023016..33023942 | 177 | √ | √ | √ | √ | √ | √ |
| OsXYLP14 | Clade C | LOC_Os07g07930 | Os07g0175600 | chr07:3980211..3981196 | 170 | √ | √ | √ | √ | √ | √ |
| OsXYLP15 | Clade C | LOC_Os04g38840 | Os04g0462200 | chr04:23093153..2309406 | 200 | √ | √ | √ | √ | √ | - |
| OsXYLP16 | Clade D | LOC_Os07g09970 | Os07g0198300 | chr07:5313828..5318057 | 207 | √ | √ | √ | √ | √ | √ |
| OsXYLP17 | Clade D | LOC_Os03g58940 | Os03g0804200 | chr03:33563702..33566613 | 195 | √ | √ | √ | √ | √ | √ |
| OsXYLP18 | Clade D | LOC_Os03g07100 | Os03g0167000 | chr03:3626392..3628578 | 187 | √ | √ | √ | √ | √ | √ |
| OsXYLP19 | Clade D | LOC_Os06g47200 | Os06g0686400 | chr06:28617447..28620310 | 150 | √ | √ | √ | √ | √ | √ |
| OsXYLP20 | Clade D | LOC_Os03g46150 | Os03g0664400 | chr03:26096042..26096992 | 243 | √ | √ | √ | √ | √ | √ |
| OsXYLP21 | Clade D | LOC_Os08g42040 | Os08g0532800 | chr08:26554159..26557021 | 179 | √ | √ | √ | √ | √ | √ |
aSystematic designation given to rice XYLPs.
bOsXYLPs are divided into four clades according to the sequence homology of their protein backbones.
cand dLocus numbers assigned by RGAP (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/) and RAP-DB (Rice Annotation Project Database, http://rapdb.dna.affrc.go.jp/), which can be converted by ID converter (http://rapdb.dna.affrc.go.jp/tools/converter/).
eChromosomal localization of rice XYLP genes.
fLength of the open reading frame in amino acids.
gN-terminal signal sequence predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
hGPI anchor signal predicted by big-PI (http://mendel.imp.ac.at/gpi/plant_server.html).
i ~ lFull-length cDNA; Expressed sequence tag profiles; microarray data; massively parallel signature sequencing.
√, exist; −, not exist.
We performed a multiple sequence alignment on the nsLTP-like domains of 21 OsXYLPs and 13 AtXYLPs to clarify the sequence characteristics of OsXYLPs (Additional file 1: Figure S1). It is noteworthy that the distribution of eight cysteine (Cys) residues is highly conserved, following an C-X-C-X-CC-X-CXC-X-C-X-C pattern, in both OsXYLPs and AtXYLPs. The hydrophobicity of the residue between Cys5 (C5) and Cys6 (C6) is also conserved, with the residue always leucine, isoleucine, or valine (Additional file 1: Figure S1). The conserved nature of the eight Cys residues and the hydrophobic residue, which in combination are involved in the formation of the three-dimensional structure that can firmly bind lipids, implies their important contribution to lipid-binding ability.
Protein structure and phylogenetic analysis
The OsXYLP protein sequences were submitted to several bioinformatic websites to predict the presence of signal peptides, glycosylphosphatidylinositol (GPI)-anchored signals, N-glycosylation sites, and AG glycomodules (Additional file 2: Table S1). All 21 OsXYLPs were expected to have an N-terminal signal peptide for targeting to the endoplasmic reticulum. All OsXYLPs except for OsXYLP2 were found to be GPI anchor proteins, indicating that these proteins might localize in the plasma membrane (Figure 1). In addition, putative AG glycomodules in all OsXYLPs were found to be distributed in the PAST-rich region before and/or after the nsLTP-like domain (Figure 1). Moreover, N-glycosylation sites in most of the OsXYLPs were located in the nsLTP-like domain and the PAST-rich region (Additional file 2: Table S1). The existence of signal peptides and AG glycomodules suggest that the 21 OsXYLPs may be chimeric AGPs.
Figure 1.
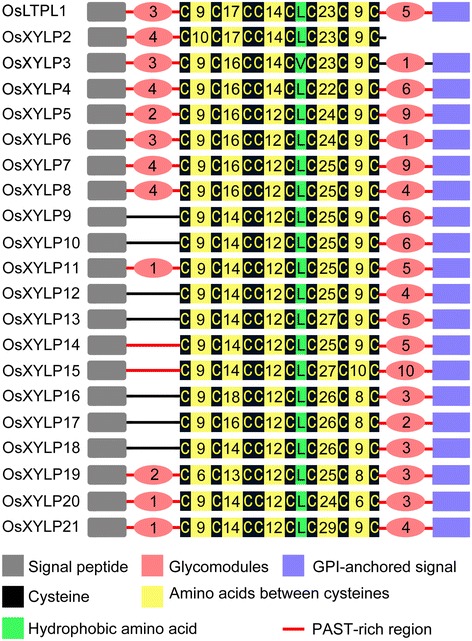
Protein structure of rice XYLPs. Gray boxes indicate the secretory signal sequence predicted by SignalP. The violet boxes indicate predicted the GPI-anchored signal. Dark red straights indicate glycoprotein-like Pro/Ala/Ser/Thr-rich regions (PAST > 35%). Light red circles with number indicate putative AG glycomodules and its number. Yellow and black boxes indicate nsLTP domains; black boxes indicate the eight conserved cysteine residues; the numbers in yellow boxes means the number of amino acid residues; the green boxes show the hydrophobic residues between C5 and C6.
Using the aligned full-length OsXYLP and AtXYLP protein sequences, we obtained an unrooted phylogenetic tree showing their phylogenetic relationships (Figure 2). With a few exceptions, all XYLPs in the tree are clustered according to their protein sequence homologies into four distinct, strongly supported clades (A–D). Family members with high sequence homology therefore cluster together in the tree. For instance, five XYLPs each from rice and Arabidopsis are placed in Clade A, with cysteine residues distributed following the conserved pattern: C-X9/10-C-X16/17-CC-X12/14-C-L/V/I-C-X22/23/24-C-X7/8/9-C (Figures 1 and 2). Clade B consists of five XYLPs, three OsXYLPs, and two AtXYLPs (Figure 2). The distribution of the eight cysteine residues in the 10 XYLPs in Clade C displays a highly conserved pattern: C-X9-C-X14-CC-X12-C-L/V-C-X25/27-C-X9/10-C (Figures 1 and 2). In addition, the putative AG glycomodules in all 10 XYLPs are located between the nsLTP-like domain and the GPI anchor signal. The major difference between clades A, B, and C vs. clade D is that OsXYLP19, OsXYLP20, and OsXYLP21 in the latter have low similarity to other XYLPs. Representatives of rice and Arabidopsis are present in each clade in the phylogenetic tree. Within each clade, species-specific XYLPs from rice and Arabidopsis are grouped separately, indicating that the evolutionary expansions of XYLPs in rice and Arabidopsis have occurred independently.
Figure 2.
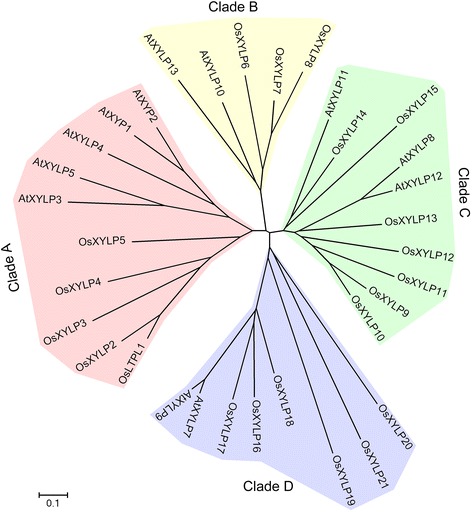
Phylogenetic relationship of XYLPs between rice and Arabidopsis . Four clades of XYLPs are show on different color backgrounds. Scale bar represent 0.1 amino acid substitution per site.
Chromosomal localization and gene duplication
We obtained the exact coordinates and orientations of OsXYLP genes from the Rice Genome Annotation Project (RGAP) database. The approximate locations of these genes are marked on the rice chromosome sketch shown in Figure 3. The OsXYLP genes are located on seven rice chromosomes: nine genes on chromosome 3, seven genes on chromosome 7, and one gene each on chromosomes 1, 4, 5, 6, and 8 (Figure 3). The OsXYLPs thus appear to be preferentially distributed.
Figure 3.
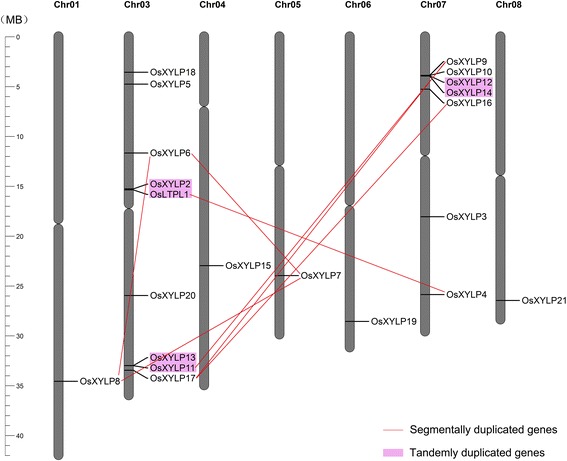
Chromosomal localization and gene duplication events of OsXYLP genes. Chromosome numbers are indicated at the top of each chromosome. The cleavages on the chromosomes indicate the position of centromeres. Genes present on duplicated segments of genome are connected by red lines, and tandem duplicated genes are marked with purple background.
We also investigated segmental and tandem duplications in the OsXYLP gene family. We found that nine OsXYLP genes (OsLTPL1 and OsXYLPs 4, 6, 7, 8, 9, 11, 16, and 17) located in the duplicated chromosomal segments of rice chromosomes mapped by RGAP with a maximal distance between collinear gene pairs of 500 kb (Figure 3). Additionally, six genes (OsLTPL1 and OsXYLPs 2, 11, 12, 13, and 14) are tandemly duplicated and separated by no more than five intervening genes. To summarize, 13 OsXYLP genes are associated with segmental and tandem duplications, indicating that evolution in this gene family has involved a large number of duplication events.
Expression patterns of OsXYLP gene
Expression patterns are important for analyzing the function of target genes. To investigate expression patterns of OsXYLP genes, we accordingly investigated three publicly available resources: expressed sequence tag (EST) profiles, MPSS tags, and microarray data.
We examined the availability of EST and full-length cDNA data by searching the Rice Annotation Project Database locus of OsXYLP genes in the UniGene database at NCBI (http://www.ncbi.nlm.nih.gov/unigene/) (Table 1). We discovered that 19 of 21 OsXYLP genes are represented by at least one full-length cDNA or EST. Both full-length cDNAs and ESTs are reported for 16 genes, whereas 3 genes are only represented by an EST. The data indicate that the OsXYLP genes, except for OsXYLP2, are expressed (Table 1). The EST data demonstrate that four genes are tissue-specifically expressed: OsLTPL1 in stems, OsXYLP13 and OsXYLP21 in shoot apical meristem (SAM), and OsXYLP18 in panicles (Additional file 3: Table S2).
MPSS is a sensitive quantitative method for gene expression analysis [47]. To analyze the expression pattern of the 21 OsXYLP genes, we obtained two 17-base and 20-base signatures in 10 different organs and tissues of rice from the MPSS database. MPSS signatures for 16 OsXYLP genes were available in at least one of the two libraries (Additional file 4: Table S3). Differential expression abundances, represented by the number of tags (transcripts per million [tpm]), were classified to indicate low (<50 tpm), moderate (50–500 tpm), and strong (>500 tpm) expression. Eight and seven genes displayed strong and moderate expression levels, respectively, and four genes were expressed at a low level (Additional file 4: Table S3). It is noteworthy that 10 genes showed abundant or specific expression in roots, leaves, stems, and panicles. The results of this analysis are consistent with the predicted roles of OsXYLP genes in vascular system development.
Microarrays provide a high-throughput approach for the analysis of gene expression patterns. Microarray data were obtained from a previous study of OsXYLP gene expression in various tissues, including young roots (YR), mature leaves (ML), young leaves (YL), shoot apical meristem (SAM), and various stages of panicle (P1–P6) and seed (S1–S5) development [48]. A hierarchical cluster analysis was performed by using the logarithmic signal values of OsXYLP genes (Additional file 5: Table S4) revealed that 20 of the 21 OsXYLPs genes are expressed in at least one vegetative or reproductive developmental stage (Figure 4). OsXYLP8 is abundantly expressed across the panicle development process (Figure 4A), while OsLTPL1 is expressed in all examined organs and tissues (Figure 4B). Five genes (OsXYLP4, OsXYLP11, OsXYLP13, OsXYLP14, and OsXYLP20) are mainly expressed in YR and P5 (Figure 4C). High expression levels were indicated for OsXYLP17 in P5 (Figure 4D), OsXYLP6 in YR and P4–P6 (Figure 4E), and OsXYLP15 in P3 (Figure 4F). OsXYLP5, OsXYLP9, and OsXYLP10 are highly expressed in YR (Figure 4G). The expression levels of OsXYLP3 and OsXYLP12 are relatively low in all examined organs and tissues (Figure 4H). OsXYLP7, OsXYLP18, and OsXYLP21 are highly expressed in panicles and seeds (Figure 4I), while the expression levels of OsXYLP16 and OsXYLP19 are high in all examined organs and tissues (Figure 4J).
Figure 4.
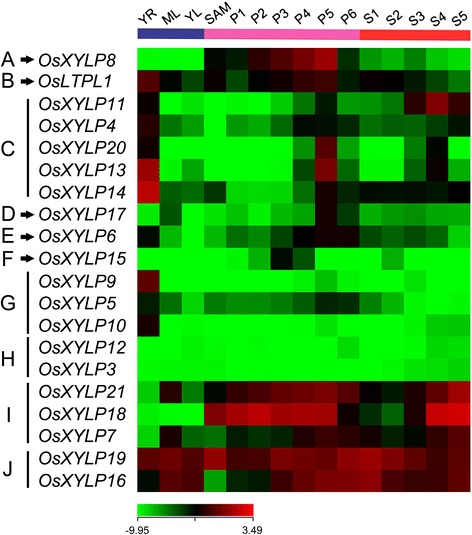
Expression profiles of OsXYLP genes in various organs and tissues. The microarray data (GSE6893) of OsXYLP genes expression are analyzed. A heat map representing hierarchical clustering of average log signal values of OsXYLP genes in various developmental stages are generated (samples are indicated at the top of each lane: YR, roots from 7-day-old seedlings; ML, mature leaves; YL, leaves from 7-day-old seedling, different stages of panicle development: SAM, up to 0.5 mm; P1, 0–3 cm; P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; P6, 22–30 cm and different stages of seed development: S1, 0–2 dap (days after pollination); S2, 3–4 dap; S3, 5–10 dap; S4, 11–20 dap; S5, 21–29 dap). Genes are divided into 10 groups: (A) SAM, P1-P6, S1-S5; (B) all examined organs and tissues; (C) YR, P4-P6; (D) ML, P5, P6; (E) YR, P4-P6; (F) P3; (G) YR; (H) low expression in all examined organs and tissues; (I) SAM, P1-P6, S3-S5; (J) all examined organs and tissues. The color scale (representing average log signal values) is shown at the bottom.
To validate the results of the digital expression analysis, we examined the expression levels of OsXYLP genes in five different tissues by qRT-PCR. The resulting gene expression patterns were in general agreement with the microarray and MPSS tag data (Figure 5). According to our PCR results, OsXYLP9, OsXYLP10, OsXYLP11, and OsXYLP14 are especially expressed in roots (R) (Figure 5A–D), OsLTPL8, OsXYLP15, and OsXYLP18 are predominantly expressed in P3 (Figure 5E–G), OsXYLP12 and OsXYLP17 are mainly expressed in P6 (Figure 5H and 5I), OsXYLP2 and OsXYLP20 are mainly expressed in roots and leaves (L) (Figure 5J and 5K), and OsXYLP6 is mainly expressed in leaves and P3 (Figure 5L). Four genes are mostly expressed in three tissues: OsXYLP13 in roots, leaves, and stems (Figure 5M), OsXYLP4 in roots, leaves, and P3 (Figure 5N), OsLTPL1 in roots, leaves, and P6 (Figure 5O), and OsXYLP7 in leaves, P3, and P6 (Figure 5P). In contrast, no obviously specific expressions were observed for OsXYLP5, OsXYLP16, OsXYLP19, and OsXYLP21 genes (Figure 5Q–T).
Figure 5.
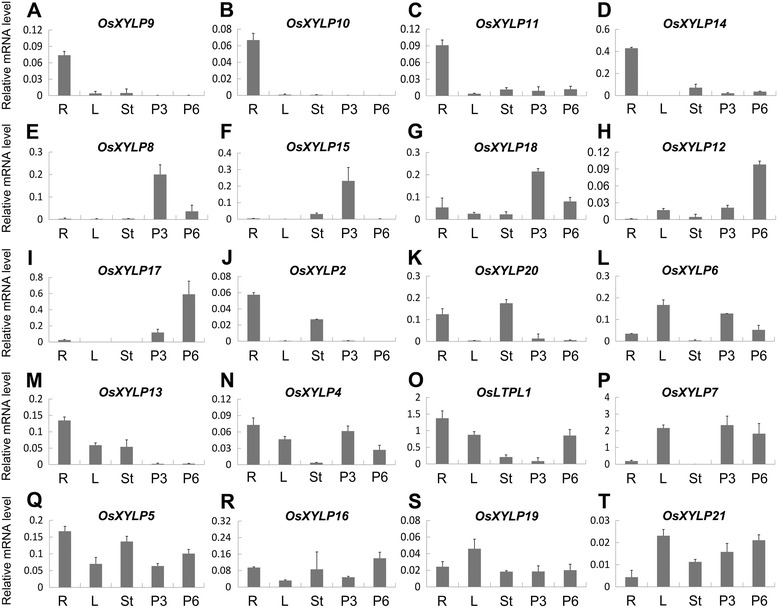
Real-time PCR analysis of representative OsXYLP genes in different developmental stages of vegetative and reproductive tissues and organs. The expression levels of OsXYLP genes in different tissues and organs (A-T). R, 7-day-old roots; L, 7-day-old leaves; St, 60-day-old stems; P3, 5–10 cm panicles; P6, 22–30 cm panicles. Error bars indicate standard deviations of independent biological replicates (n =2 or more).
Expression profiles of OsXYLP genes under abiotic stresses and hormone treatments
We analyzed the microarray data of 7-day-old seedlings under drought, salt, and cold stresses to investigate the abiotic stress response of OsXYLPs. Our results indicate that OsXYLP7 expression is up-regulated by drought stress, whereas OsXYLP8, OsXYLP13, and OsXYLP21 are down-regulated by drought and salt stresses (Figure 6). To verify the above results, we used qRT-PCR to detect the expression levels of these four genes in 7-day-old seedlings under three stress conditions for 3 hours (Figure 6B–E). The expression of OsXYLP7 was up-regulated under salt stress (Figure 6B), while OsXYLP8, OsXYLP13, and OsXYLP21 were significantly down-regulated by drought and salt stresses (Figure 6C–E). These results suggest that some OsXYLP genes may participate in abiotic stress pathways and play roles in the response to these stresses, especially drought and salt stresses.
Figure 6.
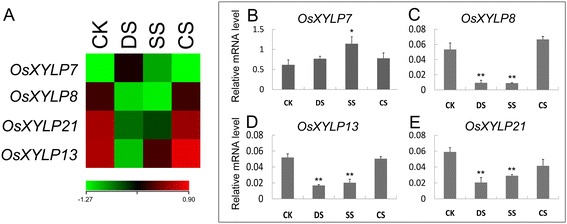
Differential expression profiles of OsXYLP genes under abiotic stresses. The microarray data (GSE6901) of gene expression under various abiotic stresses (CK, control; DS, drought stress; SS, salt stress; CS, cold stress) were used for cluster display. The average log signal values of OsXYLP genes are presented by a heat map. Under any of the given abiotic stress conditions, genes that exhibited ≥ 2-fold differential expression are shown (A). Real-time PCR were performed on these genes (B-E). The significance of difference between the controls and treatments are determined by using Origin 7.5, and are represented by two asterisks (**P < 0.01) and one asterisk (*0.01 < P < 0.05). The color scale (representing average log signal values) is shown at the bottom.
We used qRT-PCR to examine transcriptional levels of 12 representative OsXYLP genes under NAA, 6-BA, and GA treatments (Figure 7). Except for OsXYLP9 and OsXYLP19, the examined OsXYLP genes were up-regulated significantly in seedlings subject to NAA treatment (Figure 7). Only four genes (OsXYLP4, OsXYLP5, OsXYLP7, and OsXYLP16) displayed significant up-regulation under 6-BA treatment (Figure 7B,C,E, and K). Except for OsXYLP19, the expression levels of all examined genes were increased under GA treatment (Figure 7L). These results indicate that OsXYLPs may play roles in responses to these hormones.
Figure 7.
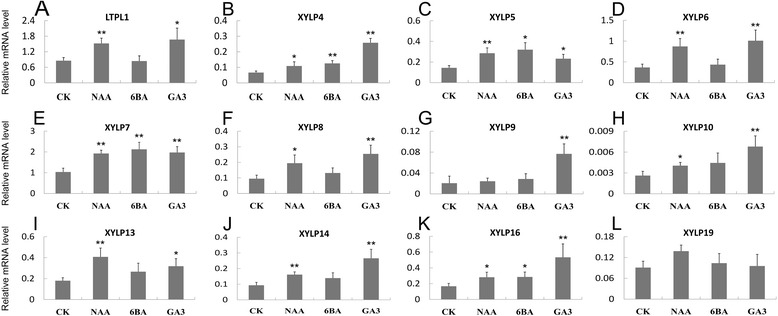
Real-time PCR analysis of OsXYLP genes under NAA, 6-BA and GA treatments. The expression levels of OsXYLP genes under different treatments (A-L). The significance of difference between the controls and treatments are determined by using Origin 7.5, and are represented by two asterisks (**P < 0.01) and one asterisk (*0.01 < P < 0.05). CK, control.
Comparative expression analysis of OsXYLP and AtXYLP genes
To provide more evidence for the deduced biological functions of XYLP genes, a comparative expression analysis of rice and Arabidopsis XYLP genes was performed using microarray and MPSS data from roots, leaves, inflorescences, pollen, and siliques/seeds and from plants under abiotic stresses (Figure 8; Additional file 4: Table S3; Additional file 5: Table S4). All OsXYLP and AtXYLP genes were found to be present in at least one of the databases, except for OsXYLP2 which was absent from the two data sets (Figure 8). Analysis of the integrated microarray and MPSS data revealed that 20 XYLP genes are expressed in at least two organs and tissues. Among the 20 genes, 6 XYLP genes showed specific expression patterns and 3 were entirely lowly expressed (Figure 8).
Figure 8.
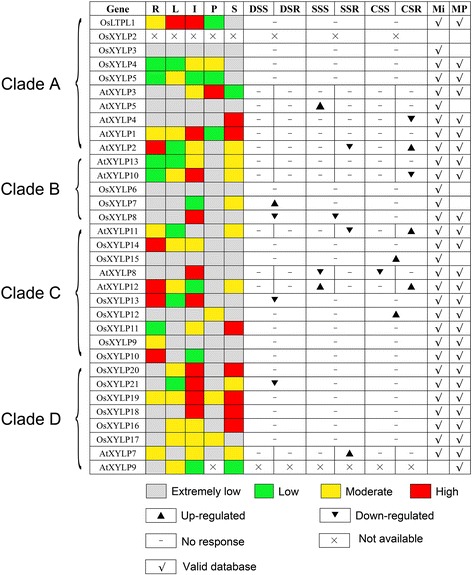
Comparison of expression levels between rice and Arabidopsis XYLP genes in different organs and under abiotic stresses. R, roots; L, leave; I, inflorescence; P, pollens; S, siliques or seeds; DSS and DSR; drought stressed shoots and roots; SSS and SSR, salt stressed shoots and roots; CSS and CSR, cold stressed shoots and roots; Mi, microarray data; MP, MPSS data.
The analysis furthermore revealed that some XYLP genes with close evolutionary relationships have similar expression patterns. For example, OsXYLP10, OsXYLP13, OsXYLP14, and AtXYLP12 are highly expressed in roots, as are OsXYLP18, OsXYLP19, and OsXYLP20 in inflorescences and seeds (Figure 8).
It is noteworthy that XYLP genes originating from gene duplication events, such as, segmental duplicated genes: OsXYLP6, OsXYLP7, and OsXYLP8; OsLTPL1 and OsXYLP4; tandem duplicated genes: OsXYLP11 and OsXYLP13, OsXYLP12 and OsXYLP14, do not show similar expression patterns and responses under abiotic stresses (Figure 8). These results are in accord with the conclusions of previous studies that the duplicated genes have frequently diverged from their ancestors, thus hinting that gene duplication has played an important evolutionary role by enriching biological functions of the XYLP gene family.
Identification of xylp7 and xylp16 mutants
To investigate the biological functions of OsXYLP genes in rice, we acquired four T-DNA insertion mutants from the Plant Functional Genomics Laboratory of Korea. Two mutants (xylp7 and xylp16) were successfully identified, and the expressions of OsXYLP7 and OsXYLP16 genes in their homozygous mutants were accordingly analyzed (Additional file 6: Figure S2).
We observed and measured stem and spike stalk lengths of mature xylp7 mutant plants. These lengths were found to be shorter in mutants than in the wild type, whereas no obvious distinction was observed in plant height (Figure 9A and B). The mutant xylp7 plants displayed a reduction in the length of internodes, except for the basal internode (Figure 9C). We examined the expression level of OsXYLP7 in different-aged stems by qRT-PCR. The results showed that OsXYLP7 is high expressed in 70–90 day old stems and low expressed in 60-day-old stems (Additional file 7: Figure S3). The xylp16 mutant plants showed no distinct phenotype compared with the wild type (data not shown).
Figure 9.
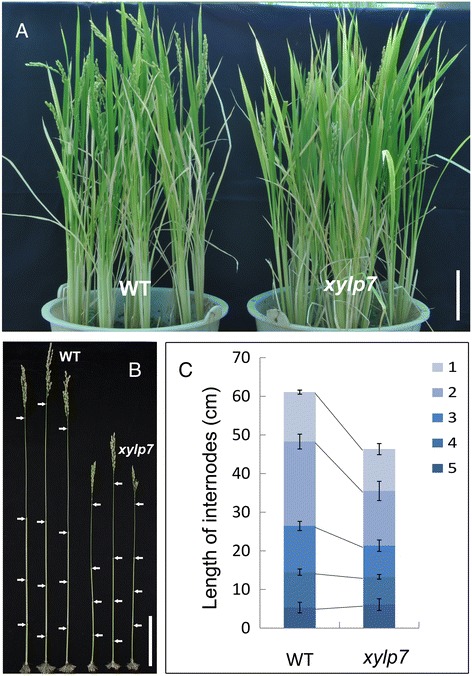
Phenotypes of wild type and xylp7 mutant plants. (A) Plants at the mature stage. Scale bar: 10 cm. (B) The stem of the wild-type and xylp7. Scale bar: 10 cm. (C) Comparison of the internode lengths between the wild-type and mutant xylp7. Error bars indicate standard deviations of independent biological replicates (n =5 or more).
Discussion
In this study, we used ZeXYP1, AtXYP1, and AtXYP2 protein sequences to search for xylogen-like proteins in the RGAP database (http://rice.plantbiology.msu.edu/). After confirming the presence of nsLTP and AGP domains, we identified 21 XYLP genes in rice. The XYLP proteins were found to have a unique structure: chimeric AGPs with a conserved nsLTP domain. We classified OsXYLP genes into four clades based on their phylogenetic relationships, arranged their genetic information, and inferred their expression patterns from three conventional and valid bioinformatic databases. Observations of xylp mutants hinted that rice XYLP genes may have a function in the development of organs with vascular systems.
Gene duplication, both tandem and segmental, plays important roles in genome evolution [49]. OsXYLP genes are located on seven rice chromosomes. Thirteen (61.90%) of the 21 OsXYLP genes are derived from gene duplications: 9 genes attributed to segmental duplication are localized on chromosomes 1, 3, 5, and 7; and 3 pairs of tandemly duplicated genes are distributed on chromosomes 3 and 7 (Figure 3). Most duplicated OsXYLP genes show diverse expression patterns, including those arising from tandem and segmental duplications, such as tandemly duplicated gene pairs: OsXYLP11 and OsXYLP13, OsXYLP12 and OsXYLP14; and segmentally duplicated gene pairs: OsXYLP6 and OsXYLP8, OsXYLP9 and OsXYLP17. Our analyses indicate that the duplication events not only contributed to the expansion of the OsXYLP gene family, but also created differences in expression between duplicated genes that may have given rise to genetic functional diversity over the course of evolution.
Analysis of EST, microarray, and MPSS signature data revealed that all OsXYLP genes are expressed (Table 1). The analysis also indicated that most OsXYLP genes have high expression levels in tissues with vascular system, such as roots, stems, leaves, and panicles. OsXYLP and AtXYLP protein sequences were aligned and divided into four clades, with OsLTPL1, OsXYLP2, 3, 4, and 5 sharing high sequence homology with AtXYP1 and AtXYP2. A double mutant of xyp1 and xyp2 in Arabidopsis, but neither single mutant, shows defects in vascular development, and AtXYP1 has been confirmed to have TE-inducing activity [11]. OsLTPL1 and OsXYLP5 are highly expressed in roots, panicles, and seeds, similar to AtXYP1, and OsXYLP4 and AtXYP2 have similar expression patterns in roots (Figure 5). Thus, it is possible that OsLTPL1, OsXYLP4, and OsXYLP5 function in the vascular system development of higher plants.
In previous studies, gibberellin plays roles in the control of cambial activity, differentiation of xylem fibers, and cell elongation of secondary xylem fibers [50]. Auxin and cytokinin coordinately post-transcriptionally regulate the accumulation of xylogen and are subsequently involved in the process of TE differentiation [11]. The hormones GA, auxin, and cytokinin have also been verified to affect secondary xylem development [51]. Studies indicate that hormones play important roles during plant vascular development [52]. In barley (Hordeum vulgare L.) aleurone protoplasts, β-GlcY inhibits GA-promoted induction of α-amylase, suggesting that AGPs are involved in GA function [53]. In our qRT-PCR analysis, all examined genes were up-regulated under GA treatment except for OsXYLP5, which showed little change. These results suggest that OsXYLP genes may participate in the hormone signaling pathway. Various genes have been reported to be involved in the GA signaling pathway and to have important roles in plant growth and development. For example, a rice mutant of the Dwarf1 gene has dark green leaves, compact panicles, and short, round grains [54], and a gibberellin-responsive gene, CsAGP1, is involved in stem elongation [29]. In our study, we identified a xylp7 mutant that has a significantly decreased stem height compared with the wild type, with every internode except for the basal internode observed to be shorter. These results suggest that OsXYLP7 may take part in the GA signaling pathway and is likely to have an important role in stem elongation.
Conclusions
We identified 21 XYLP genes from the rice genome and classified them into four clades according to their evolutionary relationships. We also elucidated their genomic characteristics, protein structures, duplication status, and expression patterns during different developmental stages as well as under abiotic stress treatments. Alterations in OsXYLP gene expression levels were observed under NAA, 6-BA, and GA treatments, indicating that OsXYLP genes may be involved in hormonal regulation. These data provide insights into the characteristics of OsXYLP genes. A mutant of OsXYLP7 showed defects in stem length, suggesting that OsXYLP7 has a function in the development of organs with vascular systems. In conclusion, this study has provided fundamental information on OsXYLP gene functions and is a first step in functional research of rice XYLPs. To our knowledge, this is the first report of xylogen-like proteins in Oryza sativa L., and 19 of the 21 identified OsXYLPs are new AGP genes.
Methods
Plant materials and treatment methods
Oryza sativa L. japonica cv. Nipponbare plants were cultivated in greenhouse at Wuhan University at 28°C with a 16 h light and 8 h dark cycle. Tissues and organs for expression analysis were: (i) 7-day-old roots (R, young root) and leaves (L, young leaves); (ii) 60-day-old stems (St, young stems); (iii) 5–10 cm panicles (P3) and (iv) 22–30 cm panicles (P6). For hormone treatments, the 7-day-old seedlings were transferred into deionized water contained 1 μM NAA (1-naphthylacetic acid), 5 μM 6-BA (6-Benzylaminopurine) or 5 μM GA (Gibberellin A3) for 3 h. For stress treatments, the 7-day-old seedling were transferred onto filter papers at 28°C as drought stress, placed in 400 mM NaCl solution at 28°C as salt stress, or stayed in sterile water at 4°C as cold stress for 3 h. The parallel control samples were kept the seedlings in sterile water at 28°C for 3 h. Stems of WT and xylp7 mutant were collected respectively, including stems of 60, 70, 80, and 90 days-old plants. All materials above were respectively collected and frozen immediately in liquid nitrogen, and stored in −80°C until RNA extraction.
Identification of OsXYLPs and bioinformatics analysis
Using the protein sequences of ZeXYP1, AtXYP1 and AtXYP2, BLAST searches (E-value < 10−7) were adopted to identify the OsXYLPs at the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/). The results of three searches were integrated and then the redundant sequences were removed. The remaining protein sequences were submitted to InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) to make sure the presence of non-specific lipid transfer protein-like (nsLTP) domains. The presence of N-terminal signal peptide, GPI-anchored signal, and N-glycosylation sites were predicted on SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/), Big-PI Plant Predictor (http://mendel.imp.ac.at/gpi/plant_server.html), and NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). Putative AG glycomodules were predicted mainly followed the criterion described in the papers [9,14,16]. Then, the protein sequences of identified OsXYLPs were used for BLASTP to ensure that all XYLP genes in available databases are identified. The characteristics of OsXYLP sequences were listed in Additional file 2: Table S1.
Sequence and phylogenetic analysis
The sequences of non-specific lipid transfer protein-like (nsLTP) domains and full-length of OsXYLPs and AtXYLPs were aligned using DNAMAN and Clustal X (version 1.83) program, respectively. An un-rooted phylogenetic tree was generated in Clustal X using neighbor-joining method, the bootstrap value was 1000.
Chromosomal localization and gene duplications
The approximate locations of OsXYLP genes were marking on the skeleton maps of rice chromosomes using the Mapchart software. Tandem duplicates genes were considered to be separated by no more than five genes. Genes belong to segmental duplicates were obtained from the “Segmental genome duplication of rice” at RGAP database (http://chibba.agtec.uga.edu/duplication/).
Digital expression analysis
The EST expression data of OsXYLP genes were acquired from the UniGene database at NCBI (http://www.ncbi.nlm.nih.gov/unigene/). Genes whose EST number of any tissue occupied more than a half of the total values were deemed to specifically express.
The microarray data of OsXYLPs were obtained from the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE). Several tissues were chose for temporal and spatial analysis (GSE6893): YR, young roots; ML, mature leaf; YL, young leaf; SAM, shoot apical meristem; P1, 0–3 cm panicle, floral transition and floral organ development; P2 and P3, 3–5 cm and 5–10 cm panicle, meiotic stage; P4, 10–15 cm panicle, young microspore stage; P5, 15–22 cm panicle, vacuolated pollen stage; P6, 22–30 cm panicle, mature pollen stage; S1, 0–2 DAP (days after pollination) seed, early globular embryo; S2, 3–4 DAP seed, middle and late globular embryo; S3, 5–10 DAP seed, embryo morphogenesis; S4, 11–20 DAP seed, embryo maturation; S5, 21–29 DAP seed, dormancy and desiccation tolerance [48,55]. For abiotic stress analysis, rice seedlings were transferred to 200 mM NaCl solution for salt stress, dried on filter paper for drought stress, and stayed at 4°C for cold stress, for 3 h treatment respectively. The expression data of AtXYLPs were obtained from “Bulk Gene Download” at Nottingham Arabidopsis Stock Centre (http://www.ncbi.nlm.nih.gov/geo/info/download.html). Several tissues compared to those used in rice were selected: developmental stages (GSE5629-5633) and abiotic stresses treatments (GSE5620-5621 and 5623–5624). The absolute value of a gene in one tissue is divided by the average of all absolute values of all genes, and then the logarithms of the ratios from above procedure were used as input for cluster display take advantage of the Cluster and Treeview software.
The MPSS (massively parallel signature sequencing) data of OsXYLP and AtXYLP genes were obtained from the MPSS project (http://mpss.udel.edu). MPSS expression data representing different organs and tissues (9 in rice and 5 in Arabidosis) were used further analysis. The description of rice organs and tissues is: NYR, 14 day young roots; NRA, 60 day mature roots; NST, 60 day stems; NYL, 14 day young leaves; NME, 60 day meristem tissue; NPO, mature pollens; NOS, ovaries and mature stigmas; NIP, 90 day immature panicles; NCA, 35 day callus. The organs and tissues of Arabidopsis are: Ca, actively growing callus; In, inflorescence, mixed stages; L, 21 day leave; R, 21 day roots; Si, 24–48 hour post-fertilization siliques.
Real-time PCR analysis
To confirm the expression of OsXYLP genes in rice tissues at different developmental stages and stress treatments identified by digital data analysis, quantitative real-time PCR (qRT-PCR) was performed by using SYBR-green fluorescence under a Rotor-Gene Q machine (Qiagen). The primer sequences are listed in Additional file 8: Table S5. The expression of genes in different samples was normalized to the expression of UBQ5 housekeeping gene [56]. The relative expression levels were calculated using the standard curve method, a stand curve for each gene was built by using three times of 1, 3, 9, and 27 (from low to high) diluted series of a mixed cDNA pools [57]. At least two independent biological samples and three technical replicates of each biological sample were used for real-time PCR analysis.
Availability of supporting data
Here we are with the supporting data (including alignments and protein sequences) as additional files. The phylogenetic data (alignments, phylogenetic trees, and protein structures) were deposited in Dryad (http://datadryad.org/). DOI: doi:10.5061/dryad.44tj3.
Acknowledgments
This research was supported by the National Basic Research Program of China (2012CB944801, 2013CB126903), National Natural Science Foundation of China (31170171), and the Special Doctorial Program Funds of the Ministry of Education of China (20130141130008).
Abbreviations
- AGPs
Arabinogalactan proteins
- AG
Arabinogalactan
- FLAs
Fasciclin-like AGPs
- TE
Tracheary elements
- RAP-DB
Rice annotation project database
- MPSS
Massively parallel signature sequencing
- EST
Expressed sequence tag
- NAA
1-naphthylacetic acid
- 6-BA
6-Benzylaminopurine
- GA
Gibberellin A3
- β-GlcY
β-glucosyl Yariv reagent
- DAP
Day after pollination
Additional files
Multiple sequence alignments of the nsLTP domain of OsXYLPs and AtXYLPs. Identical (100%), conservative (75-99%) and block (50-74%) of similar amino acid residues are shaded in black, red and light blue, respectively.
Protein backbones of XYLPs in rice and Arabidopsis.
ESTs expression profiles of OsXYLP genes.
MPSS analysis of OsXYLP and AtXYLP genes.
Microarray analysis of OsXYLP and AtXYLP genes.
Analyses of T-DNA insertion in the OsXYLP7 and OsXYLP16 genes. (A) Genomic drawing of OsXYLP7 locus and the position of T-DNA insertion. The T-DNA insertion of xylp7 allele represented in triangle inserted in the second exon (E2). Bar = 500 bp. (B) Genomic drawing of OsXYLP16 locus and the position of T-DNA insertion. The T-DNA insertion of xylp16 allele represented in triangle inserted in the first exon (E1). Bar = 500 bp. (C) and (E) PCR analysis of T-DNA insert in xylp7 and xylp16 mutant, respectively; wild type (WT) is as the control test. (D) and (F) RT-PCR analysis of expression level in xylp7 and xylp16 mutant, respectively; wild type (WT) is as the control test.
The expression levels of OsXYLP7 in different stages of stems.
Primers used in real-time PCR and mutants identification.
Footnotes
Tengfei Ma and Haoli Ma contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TFM performed experimental work, contributed bioinformatic analyses, and wrote the manuscript. HLM contributed to the bioinformatic analyses and manuscript editing. HMZ performed the RNA extraction and contributed advice. HDQ performed the qRT-PCR validation. JZ guided all aspects of the project and helped to draft and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Tengfei Ma, Email: matengfei@whu.edu.cn.
Haoli Ma, Email: mahaoli@whu.edu.cn.
Heming Zhao, Email: zhaoheming@whu.edu.cn.
Huandong Qi, Email: qi_huandong@163.com.
Jie Zhao, Email: jzhao@whu.edu.cn.
References
- 1.Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 2.Majewska-Sawka A, Nothnagel EA. The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 2000;122:3–10. doi: 10.1104/pp.122.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- 4.Ellis M, Egelund J, Schultz CJ, Bacic A. Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 2010;153:403–419. doi: 10.1104/pp.110.156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer-Knudsen J, Clarke AE, Bacic A. Proline- and hydroxyproline-rich gene products in the sexual tissues of flowers. Sex Plant Reprod. 1997;10:253–260. doi: 10.1007/s004970050095. [DOI] [Google Scholar]
- 6.Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of arabidopsis. Plant Cell. 2000;12:1751–1768. doi: 10.1105/tpc.12.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol. 2001;47:161–176. doi: 10.1023/A:1010683432529. [DOI] [PubMed] [Google Scholar]
- 8.Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM. The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant Cell Physiol. 2005;46:975–984. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- 9.Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y, Motose H, Iwamoto K, Fukuda H. Expression and genome-wide analysis of the xylogen-type gene family. Plant Cell Physiol. 2011;52:1095–1106. doi: 10.1093/pcp/pcr060. [DOI] [PubMed] [Google Scholar]
- 13.Mashiguchi K, Asami T, Suzuki Y. Genome-wide identification, structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Biosci Biotechnol Biochem. 2009;73:2452–2459. doi: 10.1271/bbb.90407. [DOI] [PubMed] [Google Scholar]
- 14.Ma HL, Zhao HM, Liu Z, Zhao J. The phytocyanin gene family in rice (Oryza sativa L.): genome-wide identification, classification and transcriptional analysis. PLoS One. 2011;6:e25184. doi: 10.1371/journal.pone.0025184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faik A, Abouzouhair J, Sarhan F. Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analysis. Mol Genet Genomics. 2006;276:478–494. doi: 10.1007/s00438-006-0159-z. [DOI] [PubMed] [Google Scholar]
- 16.Ma HL, Zhao J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.) J Exp Bot. 2010;1(6):2647–2668. doi: 10.1093/jxb/erq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yariv J, Lis H, Katchalski E. Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem J. 1967;105:1c–2c. doi: 10.1042/bj1050001c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettolino F, Liao ML, Ying Z, Mau SL, Bacic A. Structure, function and cloning of arabinogalactan-proteins (AGPs): an overview. Food Food Ingredients J Japan. 2006;211:12–25. [Google Scholar]
- 19.Knox JP. The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int J Cytol. 1997;171:79–120. doi: 10.1016/S0074-7696(08)62586-3. [DOI] [PubMed] [Google Scholar]
- 20.Willats WG, Knox JP. A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 1996;9:919–925. doi: 10.1046/j.1365-313X.1996.9060919.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee KJD, Sakata Y, Mau SL, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell. 2005;17:3051–3065. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Showalter AM. Expression and localization of AtAGP18, a lysine-rich arabinogalactan-protein in Arabidopsis. Planta. 2007;226:169–179. doi: 10.1007/s00425-007-0478-2. [DOI] [PubMed] [Google Scholar]
- 23.Serpe MD, Nothnagel EA. Effects of Yariv phenylglycosides on rose cell-suspensions—evidence for the involvement of arabinogalactan-proteins in cell proliferation. Planta. 1994;193:542–550. doi: 10.1007/BF02411560. [DOI] [Google Scholar]
- 24.Langan KJ, Nothnagel EA. Cell surface arabinogalactan-proteins and their relation to cell proliferation and viability. Protoplasma. 1997;196:87–98. doi: 10.1007/BF01281062. [DOI] [Google Scholar]
- 25.Yang J, Sardar HS, McGovern KR, Zhang Y, Showalter AM. A lysine-rich arabinogalactan protein in Arabidopsisis essential for plant growth and development, including cell division and expansion. Plant J. 2007;49:629–640. doi: 10.1111/j.1365-313X.2006.02985.x. [DOI] [PubMed] [Google Scholar]
- 26.Gao M, Showalter AM. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan-protein involvement. Plant J. 1999;19:321–331. doi: 10.1046/j.1365-313X.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 27.Chaves I, Regalado AP, Chen M, Ricardo CP, Showalter AM. Programmed cell death induced by (β-D-galactosyl)3 Yariv reagent in Nicotiana tabacum BY-2 suspension-cultured cells. Physiol Plant. 2002;116:548–553. doi: 10.1034/j.1399-3054.2002.1160414.x. [DOI] [Google Scholar]
- 28.Lamport DT, Kieliszewski MJ, Showalter AM. Salt stress up regulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 2006;169:479–492. doi: 10.1111/j.1469-8137.2005.01591.x. [DOI] [PubMed] [Google Scholar]
- 29.Park MH, Suzuki Y, Chono M, Knox JP, Yamaguchi I. CsAGP1, a gibberellin-responsive gene from Cucumber Hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol. 2003;131:1450–1459. doi: 10.1104/pp.015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H, Kim YS, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Hengel AJ, Roberts K. AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J. 2003;36:256–270. doi: 10.1046/j.1365-313X.2003.01874.x. [DOI] [PubMed] [Google Scholar]
- 32.Acosta-Garcia G, Vielle-Calzada JP. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell. 2004;16:2614–2628. doi: 10.1105/tpc.104.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Y, Zhao J. Localization of arabinogalactan-proteins in different stages of embryos and their role in cotyledon formation of Nicotiana tabacum L. Sex Plant Reprod. 2007;20:213–224. doi: 10.1007/s00497-007-0058-4. [DOI] [Google Scholar]
- 34.Li J, Yu M, Geng LL, Zhao J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010;64:482–497. doi: 10.1111/j.1365-313X.2010.04344.x. [DOI] [PubMed] [Google Scholar]
- 35.Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J. 2008;56:351–363. doi: 10.1111/j.1365-313X.2008.03607.x. [DOI] [PubMed] [Google Scholar]
- 36.Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. Pollen grain development is compromised in Arabidopsisagp6 agp11 null mutants. J Exp Bot. 2009;60:3133–3142. doi: 10.1093/jxb/erp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu HM, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;82:395–403. doi: 10.1016/0092-8674(95)90428-X. [DOI] [PubMed] [Google Scholar]
- 38.Pereira LG, Coimbra S, Monteiro HOL, Sottomayor M. Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta. 2006;223:374–380. doi: 10.1007/s00425-005-0137-4. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Qin Y, Zhao J. Localization of an arabinogalactan protein epitope and the effects of Yariv phenyl glycoside during zygotic embryo development of Arabidopsis thaliana. Protoplasma. 2006;229:21–31. doi: 10.1007/s00709-006-0185-z. [DOI] [PubMed] [Google Scholar]
- 40.Qin Y, Zhao J. Localization of arabinogalactan proteins in egg cells, zygotes, and two-celled proembryos and effects of β-D-glucosyl Yariv reagent on egg cell fertilization and zygote division in Nicotiana tabacum L. J Exp Bot. 2006;57:2061–2074. doi: 10.1093/jxb/erj159. [DOI] [PubMed] [Google Scholar]
- 41.Yu M, Zhao J. The cytological changes of tobacco zygote and proembryo cells induced by beta-glucosyl Yariv reagent suggest the involvement of arabinogalactan proteins in cell division and cell plate formation. BMC Plant Biol. 2012;12:126. doi: 10.1186/1471-2229-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong J, Ren YJ, Yu M, Ma TF, Zhang XL, Zhao J. Roles of arabinogalactan proteins in cotyledon formation and cell wall deposition during embryo development of Arabidopsis. Protoplasma. 2011;248:551–563. doi: 10.1007/s00709-010-0204-y. [DOI] [PubMed] [Google Scholar]
- 43.Kader JC, Julienne M, Vergnolle C. Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur J Biochem. 1984;139:411–416. doi: 10.1111/j.1432-1033.1984.tb08020.x. [DOI] [PubMed] [Google Scholar]
- 44.Yeats TH, Rose JK. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs) Protein Sci. 2008;17:191–198. doi: 10.1110/ps.073300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motose H, Fukuda H, Sugiyama M. Involvement of local intercellular communication in the differentiation of zinnia mesophyll cells into tracheary elements. Planta. 2001;213:121–131. doi: 10.1007/s004250000482. [DOI] [PubMed] [Google Scholar]
- 46.Mashiguchi K, Yamaguchi I, Suzuki Y. Isolation and Identification of glycosylphosphatidylinositol-anchored arabinogalactan proteins and novel β-glucosyl yariv-reactive proteins from seeds of rice (Oryza sativa) Plant Cell Physiol. 2004;45:1817–1829. doi: 10.1093/pcp/pch208. [DOI] [PubMed] [Google Scholar]
- 47.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 48.Itoh JI, Nonomura KI, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- 49.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ridoutt BG, Pharis RP, Sands R. Fibre length and gibberellins A1 and A20 are decreased in Eucalyptus globules by acylcyclohexanedione injected into the stem. Physiol Plant. 1996;96:559–566. doi: 10.1111/j.1399-3054.1996.tb00227.x. [DOI] [Google Scholar]
- 51.Eriksson ME, Israelsson M, Olsson O, Moritz T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol. 2000;18:784–788. doi: 10.1038/77355. [DOI] [PubMed] [Google Scholar]
- 52.Dettmer J, Elo A, Helariutta Y. Hormone interactions during vascular development. Plant Mol Biol. 2009;69:347–360. doi: 10.1007/s11103-008-9374-9. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki Y, Kitagawa M, Knox JP, Yamaguchi I. A role for arabinogalactan proteins in gibberellin-induced α-amylase production in barley aleuron cells. Plant J. 2002;29:733–741. doi: 10.1046/j.1365-313X.2002.01259.x. [DOI] [PubMed] [Google Scholar]
- 54.Ashikari M, Wu JZ, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 57.Yuan J, Chen D, Ren Y, Zhang X, Zhao J. Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 2008;146:1637–1650. doi: 10.1104/pp.107.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]


