Abstract
AIM: To assess whether antibiotic resistance varies between the antrum and corpus of the stomach of patients that are either Helicobacter pylori (H. pylori) therapy-naive or pre-treated.
METHODS: H. pylori strains were isolated from antrum and corpus biopsies from 66 patients that received a diagnostic gastroduodenoscopy for variant clinical indications. Antimicrobial susceptibility to amoxicillin, clarithromycin, tetracycline, metronidazole, levofloxacin and rifabutin was tested with the E-test method on Iso-Sensitest agar with 10 vol% defibrinated horse blood. In patients with a different antibiotic susceptibility pattern between the isolates from the antrum and corpus, DNA fingerprinting via random amplified polymorphic DNA analysis was performed to detect differences among DNA patterns of H. pylori isolates.
RESULTS: Primary, secondary and tertiary resistance to clarithromycin was 6.9%, 53.8% and 83.3%, retrospectively. Metronidazole and levofloxacin resistance also increased according to the number of previous treatments (17.2%, 69.2%, 83.3%; 13.8%, 23.1%, 33.3%). Tertiary resistance to rifabutin was detected in 12.5% of patients. In none of the 66 patients a resistance against amoxicillin or tetracycline was detectable. Discordant antibiotic susceptibility between antrum and corpus isolates for different antibiotics was seen in 15.2% (10/66) of the patients. Two out of those ten patients were naive to any H. pylori antibiotic treatment. The remaining eight patients previously received at least one eradication therapy. DNA fingerprinting analysis revealed no substantial differences among DNA patterns between antrum and corpus isolates in the majority of patients suggesting an infection with a single H. pylori strain.
CONCLUSION: Different antibiotic susceptibility between antrum and corpus biopsies is a common phenomenon and a possible explanation for treatment failure. Resistant H. pylori strains may be missed if just one biopsy from one anatomic site of the stomach is taken for H. pylori susceptibility testing.
Keywords: Helicobacter pylori treatment, Antibiotic susceptibility, Heteroresistance, Antibiotic susceptibility
Core tip: Different antibiotic susceptibility between antrum and corpus biopsies of the stomach represents a phenomenon and a possible explanation for treatment failure. Resistant Helicobacter pylori (H. pylori) strains, as a possible reason for treatment failure may be missed in a reasonable number if just one biopsy from one anatomic site of the stomach is taken for H. pylori susceptibility testing.
INTRODUCTION
Helicobacter pylori (H. pylori) is a gram-negative bacterium that chronically infects the human gastric mucosa. Infection with H. pylori is the main cause of chronic gastritis, peptic ulcer disease, low grade gastric mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric adenocarcinoma[1]. The recognition of H. pylori as the key pathogen in various gastroduodenal diseases with various clinical manifestations has posed an increasing demand for therapeutic strategies. The development of effective treatment options for H. pylori infection was leading to an enormous change in the clinical management of gastroduodenal diseases with curative antibiotic therapeutic strategies for H. pylori related diseases such as low-grade gastric MALT lymphoma and H. pylori related peptic ulcer disease[2]. The worldwide and frequent use of antibiotic therapies in general and for H. pylori infection in particular is the reason for an increasing number of H. pylori eradication failures over the last decade[3]. The explanation for this phenomenon is the increasing antimicrobial resistance seen in many countries, in particular against clarithromycin[4]. Several national and international guidelines provide clear recommendations for first and second line treatment options for the treatment of H. pylori infection[1,5-7]. According to the European Maastricht consensus, following second line treatment failure, rescue treatment should be guided according to antimicrobial susceptibility testing, if available. However, there are no recommendations whether one or more gastric biopsies need to be taken for the assessment of H. pylori resistance to antibiotics. Because of the patchy distribution of H. pylori in the gastric mucosa, biopsies from several sites of the stomach might increase the diagnostic yield for the detection of H. pylori[8,9]. Additionally, H. pylori strains from one individual patient have usually either an antibiotic resistant or susceptible phenotype. However, both antibiotic susceptible and -resistant H. pylori strains may be present in the same patient[10]. H. pylori infection with differing antimicrobial susceptibilities in different parts of the stomach is likely to negatively affect the success of eradication therapies. This led to the hypothesis whether biopsies from the antrum and corpus can increase the diagnostic yield in order to optimise H. pylori antibiotic susceptibility guided therapy.
Therefore, we examined H. pylori antibiotic resistance in patients from biopsies simultaneously taken from the antrum and corpus to detect possible different antibiotic resistance pattern in individual patients.
MATERIALS AND METHODS
This study presents a retrospective analysis to evaluate the presence of H. pylori antibiotic resistance by analysing H. pylori isolates from antrum and corpus.
Data collection and patient characteristics
The electronic patient documentation system of the Department of Gastroenterology, Hepatology and Infectious Diseases at the University Hospital of Magdeburg, Germany, was used to retrieve all patients that underwent upper gastrointestinal endoscopy for H. pylori antibiotic susceptibility testing in the period between January 2010 and October 2012. Furthermore, patients were just considered for inclusion in the study that received biopsies for antibiotic susceptibility testing from the antrum and corpus of the stomach. In total, 66 patients, 50 females and 16 males met these criteria. The mean age of the patients was 52.54 ± 14.50 years. Out of these patients, 43 had a chronic gastritis, 18 had an atrophic gastritis, 2 had gastric ulcer and 3 suffered from adenocarcinoma of the stomach. Most of the patients included in the study (n = 37; 56.1%) were previously treated for H. pylori infection. Twenty-nine patients (43.9%) were naive to any H. pylori eradication therapy. Thirteen (19.7%) and 24 patients (36.4%) underwent one and 2 or more eradication therapies, respectively, before antibiotic susceptibility testing.
Ethics
The study was designed with respect to the corresponding sections of the “World Medical Association Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects”. Due to the retrospective character of our study that analyzed already realized medical treatments, approval of an IRB was not obtained. Written informed consent prior to endoscopic treatment and prior to data analyzing was given by all patients.
Diagnosis of H. pylori and histopathological assessment
Histopathological alterations of the gastric mucosa were assessed according to the updated Sydney system. Biopsies were processed by routine methods. One section was stained with hematoxilin and eosin, modified Giemsa for diagnosis of H. pylori, and PAS stain. H. pylori infection status was further assessed by rapid urease test (Campylobacter-like organism-test, Kimberly-Clark, Rosewell, United States).
Gastric biopsy specimen (one biopsy each from the antrum and corpus) were carried in Portagerm pylori® tubes (bioMerieux, France) and culture was performed on Columbia-agar-based medium that contained 10 vol% washed human erythrocytes and 10 vol% heat inactivated horse serum (purchased from the NRZ Nationale Referenzzentrum Helicobacter Freiburg, Germany) without and with antibiotic supplement (vancomycin 10 mg/mL; nystatin 1 mg/mL and trimethoprim 5 mg/mL) to suppress the overgrowth of the oral flora[11,12]. Incubation of the plates was performed under microaerophilic conditions at 37° Celsius by using CampyGen™ gasbags (Oxoid, Germany) and examination was done every 2-3 d for up to 10 d. Identification of H. pylori was performed by typical morphology on Gram stain and positive urease, oxidase, and catalase tests.
Susceptibility testing to amoxicillin, clarithromycin, tetracycline, metronidazole, levofloxacin and rifabutin was performed with the Etest method (bioMerieux, France) on Iso-Sensitest agar with 10 vol% defibrinated horse blood (Oxoid, Germany)[13,14]. This test is useful for analysing slow-growing bacteria with fastidious growth requirements. It is minimally affected by the size of the inoculum or the duration of incubation, and therefore this method is reliable and simple enough to be used routinely. The Etest is able to detect antibiotic-resistant subpopulations. In this study, agar plates were treated with suspensions of H. pylori after adjustment to a turbidity approximately equaI to that of a McFarland standard No. 3. Determination of the Minimum inhibitory concentrations (MICs) of the antibiotics was carried out after 3 d of incubation or until the inhibition zone became visible. A resistant isolate was definedif the MIC was above the following breakpoints: amoxicillin, clarithromycin, tetracycline and levofloxacine > 1 mg/L; rifabutin > 4 mg/L and metronidazole > 8 mg/L[12].
DNA fingerprinting via random amplified polymorphic DNA analysis
In patients with a different antibiotic susceptibility pattern between the isolates from the antrum and corpus, H. pylori DNA was extracted from single colony isolates using QIAamp DNA mini Kit (Qiagen, Hilden, Germany). The specific H. pylori decanucleotide primers 1254, 1281, and 1290 were applied for DNA fingerprinting via random amplified polymorphic DNA analysis (RAPD-PCR). RAPD-PCR was carried out in a 27 μL volume, using 20 ng of H. pylori genomic DNA, 20 pmol of each primer, 1.5 U of AmpliTaq DNA polymerase, 250 mmol/L of deoxynucleotide triphosphate, and 3 mmol/L MgCl2 in standard PCR-buffer. Amplifications consisted of 40 cycles of 94 °C (1 min), 36 °C (1 min), 72 °C (2 min), and 72 °C (10 min). All RAPD-PCR amplifications were performed three times to confirm reproducibility of the results. Analysis of the PCR fragments was performed by electrophoresis in a 2% agarose gel under 95 voltages for 3 h. The following criteria were used for the classification of the fingerprint patterns: (1) identical when fingerprint patterns had exactly the same pattern; (2) similar or degenerate when fingerprints had the same patterns with additional or different size of bands; or (3) different when the fingerprints had totally different patterns.
Statistical analysis
The Fisher’s exact test with a 95% confidence interval was used to define differences in the proportion of patients with susceptible and resistant H. pylori strains. P values were considered significant when P < 0.05.
RESULTS
Overall antibiotic susceptibility
All 132 successfully cultured H. pylori isolates from the two gastric sites (antrum and corpus) of the 66 patients were assayed against amoxicillin, clarithromycin, metronidazole, levofloxacin, tetracyclin, and rifabutin (Table 1). Antibiotic resistance to clarithromycin was seen in 29 patients (43.9%) and in 34 patients (51.5%) a resistance to metronidazole was detected. Antibiotic resistance to both clarithromycin and metronidazole was found in 18 of the 29 strains. Levofloxacin resistance was found in 15 (22.7%) patients, while rifabutin resistance was seen in 3 (4.5%) patients. In none of the patients a resistance against amoxicillin or tetracycline was detectable.
Table 1.
Overall antibiotic resistance of 132 isolated Helicobacter pylori strains isolated from 66 patients n (%)
| Antibiotic | Patients with resistant Helicobacter pylori strains | 95%CI |
| Amoxicillin | 0 (0) | |
| Clarithromycin | 29 (43.9) | 0.32-0.56 |
| Metronidazole | 34 (51.5) | 0.39-0.64 |
| Tetracycline | 0 (0) | |
| Levofloxacin | 15 (22.7) | 0.09-0.28 |
| Rifabutin | 3 (4.5) | 0.01-0.04 |
In the 29 patients (43.9%) that have not received any eradication therapy before H. pylori susceptibility testing, primary resistance against clarithromycin was 6.9% (n = 2). Primary resistance against metronidazole and levofloxacin was a bit higher with 17.2% (n = 5) and 13.8% (n = 4) in those patients. Thirteen patients underwent one eradication attempt before resistance testing. In those patients resistance against clarithromycin and metronidazole was high with 53.8% (n = 7) and 69.2% (n = 9). Levofloxacin resistance was detectable in three out of those 13 patients (23.1%).
Twenty-four patients (36.4%) received at least more than two eradication therapies. In those patients, clarithromycin resistance was expectably high with 83.3% (n = 20). A comparably high rate with 83.3% (n = 20) and 33.3% (n = 8) was also seen for metronidazole and levofloxacin. Rifabutin resistance was detected in three patients (12.5%) that all received multiple eradication therapies. The overall antibiotic resistance increased significantly in accordance to the number of eradication therapies (P < 0.001). Table 2 gives an overview about the resistance data regarding the number of previous eradication attempts.
Table 2.
Antibiotic resistance of Helicobacter pylori strains depending on the number of eradication therapies n (%)
| Patients with resistant Helicobacter pylori strains | 95%CI | |
| No previous eradication therapy (n = 29) | ||
| Clarithromycin | 2 (6.9) | 0.03-0.18 |
| Metronidazole | 5 (17.2) | 0.03-0.34 |
| Levofloxacin | 4 (13.8) | 0.02-0.24 |
| One previous eradication therapy (n = 13) | ||
| Clarithromycin | 7 (53.8) | 0.30-0.98 |
| Metronidazole | 9 (69.2) | 0.30-0.98 |
| Levofloxacin | 3 (23.1) | 0.04-0.59 |
| Two or more previous eradication therapies (n = 24) | ||
| Clarithromycin | 20 (83.3) | 0.63-0.97 |
| Metronidazole | 20 (83.3) | 0.69-0.99 |
| Levofloxacin | 8 (33.3) | 0.09-0.47 |
| Rifabutin | 3 (12.5) | 0.04-0.12 |
Antibiotic susceptibility at different gastric sites (antrum and corpus)
The overall prevalence of patients infected simultaneously with susceptible and resistant H. pylori strains was 15.2% (10/66). Two out of those ten patients were naive to any H. pylori antibiotic treatment. The remaining eight patients previously received at least one eradication therapy. Six patients had in the past at least two eradication therapies, while the remaining two patients had one eradication attempt.
A different antibiotic susceptibility pattern between antrum and corpus biopsies against two different antibiotics was detected in three patients, while seven patients showed a discordant susceptibility against one antibiotic agent. In two patients resistance against clarithromycin was detectable in the antrum, while the corpus biopsy was susceptible against clarithromycin. A comparable result was found for metronidazole. Four patients showed a resistance in the antrum, while metronidazole sensitivity was seen in the corpus. In five patients a discordant antibiotic resistance against levofloxacin was detectable. Two levofloxacin-resistant strains were detected in the antrum and three resistant strains in the corpus, while the biopsies from the different stomach site were susceptible for levofloxacin. Different antibiotic resistance against rifabutin was determined in two patients (2.8%) that showed susceptibility in the antrum and resistance in the corpus. All ten patients simultaneously infected with susceptible and resistant H. pylori strains were successfully treated with different antibiotic regimens which were adapted to their individual resistance pattern. The results of the patients with different antibiotic susceptibility between antrum and corpus biopsies are outlined in Table 3.
Table 3.
Antibiotic susceptibility in patients with discordant resistance pattern between antrum and corpus biopsies
| Patients | Clarithromycin | Metronidazole | Levofloxacin | Rifabutin |
| Patient 1 | ||||
| Antrum | Resistant | Resistant | Susceptible | Susceptible |
| Corpus | Susceptible | Resistant | Resistant | Susceptible |
| Patient 2 | ||||
| Antrum | Susceptible | Resistant | Susceptible | Susceptible |
| Corpus | Susceptible | Resistant | Resistant | Susceptible |
| Patient 3 | ||||
| Antrum | Resistant | Resistant | Susceptible | Susceptible |
| Corpus | Susceptible | Susceptible | Susceptible | Susceptible |
| Patient 4 | ||||
| Antrum | Resistant | Resistant | Susceptible | Susceptible |
| Corpus | Resistant | Resistant | Resistant | Resistant |
| Patient 5 | ||||
| Antrum | Susceptible | Resistant | Susceptible | Susceptible |
| Corpus | Susceptible | Susceptible | Susceptible | Susceptible |
| Patient 6 | ||||
| Antrum | Resistant | Resistant | Susceptible | Susceptible |
| Corpus | Resistant | Susceptible | Susceptible | Susceptible |
| Patient 7 | ||||
| Antrum | Susceptible | Susceptible | Resistant | Susceptible |
| Corpus | Susceptible | Susceptible | Susceptible | Susceptible |
| Patient 8 | ||||
| Antrum | Resistant | Resistant | Resistant | Susceptible |
| Corpus | Resistant | Resistant | Resistant | Resistant |
| Patient 9 | ||||
| Antrum | Resistant | Susceptible | Resistant | Susceptible |
| Corpus | Resistant | Susceptible | Susceptible | Susceptible |
| Patient 10 | ||||
| Antrum | Resistant | Resistant | Susceptible | Susceptible |
| Corpus | Resistant | Susceptible | Susceptible | Susceptible |
DNA fingerprinting
Due to the retrospective character of our study, DNA isolation of H. pylori isolates from patients with resistance and sensitivity to the same antibiotic in the same host was available for nine out of the ten patients. In eight patients who where all previously treated for H. pylori infection besides one patient no substantial differences from the DNA pattern of the antrum and corpus isolates were detectable suggesting an infection with a single H. pylori strain in the stomach. In only one patient a difference in the DNA pattern between antrum and corpus was proven suggesting a mixed infection in this patient. This patient received one eradication attempt previously (Figure 1). From a clinical point of view, eight patients suffered from chronic gastritis, while one patient was diagnosed with gastric cancer.
Figure 1.
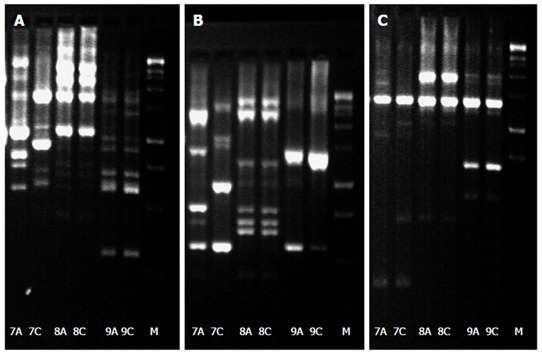
DNA fingerprinting via random amplified polymorphic DNA analysis profiles of Helicobacter pylori isolates from the antrum (A) and corpus (C) of patient 7-9 with 1254, 1281, and 1290 random primers. A clear difference between antrum and corpus is only observed in patient 7.
Epidemiological data
No statistical significance was observed for differences in age, gender and gastric diseases of the patients included in the study. Furthermore, the analysis of the distribution and histological grade of activity of the gastritis (antrum vs corpus) did not reveal any difference for the patients with discordant antibiotic susceptibility between antrum and corpus isolates (data not shown).
DISCUSSION
In our study, we demonstrate that different antibiotic susceptibility between antrum and corpus biopsies is a common phenomenon and occurs in eradication naive as well in previously treated patients.
In agreement with previous studies, we found a comparably high discordant antibiotic susceptibility to clarithromycin and metronidazole between antrum and corpus biopsies[10,15,16]. Interestingly, clarithromycin resistance in the heteroresistance cases was exclusively detected in antrum biopsies, which is in disagreement with previous studies that showed a higher rate of resistant isolates in the corpus[15,17]. In particular, the resistance against clarithromycin affects the treatment success of standard triple therapy and the knowledge of clarithromycin resistance is crucial for the recommendation of first-line therapy[18]. In regions with a clarithromycin resistance higher than 20%, clarithromycin based triple therapy should not be used. In this case, quadruple therapy should be applied[19]. Our data clearly confirm previous studies that demanded multiple gastric biopsies from different sites of the stomach to detect clarithromycin resistant strains[15,16]. According to our results there is a reasonable chance to underestimate or miss clarithromycin resistance if just one biopsy is taken for antimicrobial susceptibility testing. This might certainly negatively affect the treatment success of H. pylori therapy. This is of particular clinical interest both for patients receiving first-line and patients receiving antibiotic susceptibility guided rescue therapy. In our study, we took biopsies and tested them for antibiotic susceptibility separately from the antrum and corpus of the stomach. From a practical point of view and considering economical aspects, the biopsies from the antrum and corpus can be cultured together.
Another interesting aspect of our study is the detection of discordant antibiotic susceptibility to levofloxacin and rifabutin. To our knowledge, this is one the first reports that describes this discordance between antrum and corpus biopsies for these two antibiotic agents. After failure of first-line options, levofloxacin based triple therapy is currently recommended as a possible second-line therapy. Thus, it is a widely used antibiotic agent for H. pylori infection and recent data show increasing resistance rates to levofloxacin[4]. Rifabutin is widely accepted as an important part of H. pylori rescue therapy[20]. The detected discordant antibiotic susceptibility to the two antibiotics is in our opinion of great clinical importance for patients receiving second-line or rescue therapies.
One of the most intriguing aspects of the study is the result of the DNA-fingerprinting analysis of the H. pylori isolates of the patients with a discordant antibiotic susceptibility result between antrum and corpus isolates. Eight of the nine patients showed identical fingerprinting patterns suggesting an infection with a single H. pylori strain. Only in one patient substantial difference in the DNA pattern was detectable suggesting a mixed infection. This finding is in agreement with a previous study showing that DNA fingerprinting patterns of each pairs of isolates from the antrum and corpus are identical or at least similar suggesting that antibiotic resistant H. pylori strains typically develop from a pre-existing susceptible strain rather than co-infection with a different strain[10]. This finding might be also an explanation for our results, because the majority of patients (seven out of eight) with a single infection, but discordant antibiotic susceptibility were pre-treated and under these stress conditions (previous antibiotic treatment) alterations in the genomic DNA occur, which may affect the phenotype of H. pylori. However our results are in disagreement with another study demonstrating substantial differences among DNA patterns between isolates of the proximal and distal stomach suggesting a mixed infection[21]. However, in this study the majority of patients were never treated before testing which might give an explanation for the disagreement.
A weakness of our study represents the fact that the information about the exact composition of previous H. pylori eradication therapies that a single patient received is unavailable. It is well known, that the resistance rates are increasing significantly in dependency of the number of previously prescribed eradication therapies[22,23]. The primary resistance rates in our study was comparable to other studies conducted in Europe[4,22,23], but it has to be noted that the primary resistance rate against clarithromycin was more than three times higher than in a study that was conducted in our department from the year 1995 till 2000[24]. The knowledge about previous H. pylori eradication therapies could at least partially explain the high resistance rates against clarithromycin, metronidazole and levofloxacin in our patient population. One can just speculate that the majority of patients included in our study received as first-line therapy clarithromycin based standard triple and as second line levofloxacin based triple therapy, as recommended in the German S3-guidelines[7]. It is well known, that especially in case of clarithromycin, the resistance rate is dramatically increasing after its first use and thus this might give the explanation for the high clarithromycin resistance in our study population. Another important point is that we could not gather an accurate history on the intake of other medications due to the retrospective character of our study. We carefully analysed our electronic patient data system, however it is possible that we missed important information about previous antibiotic intake for other clinical indications. Clarithromycin and levofloxacin are important antibiotic agents often used for other infectious diseases, such as respiratory tract infections[25], and this may be an explanation for the high resistance rates seen in our study.
In conclusion, different antibiotic susceptibility between antrum and corpus biopsies can occur in naive and pre-treated patients and in our population it is caused by an infection with a single H. pylori strain. This finding indicates that H. pylori antibiotic resistant strains may be missed if biopsies for culture are taken only from one location and offers a possible explanation for treatment failure. Thus antibiotic susceptibility guided therapy by taking just one biopsy from one single site of the stomach might not be considered representative. Therefore, we suggest taking one separate biopsy from the antrum and corpus in patients undergoing endoscopy for H. pylori antibiotic susceptibility guided therapy.
ACKNOWLEDGMENTS
The authors wish to thank Professor Luigina Cellini and Leonardo Marzio (University “G. d’Annunzio”, Chieti-Pescara, Italy) for providing their expertise and primer sequences for the RAPD-PCR.
COMMENTS
Background
Helicobacter pylori (H. pylori) eradication rates with standard triple therapy which represents the recommended first-line therapeutic option decrease constantly over the last years. Responsible for this is the increasing resistance rate against various antibiotic agents. Therefore, antibiotic susceptibility guided therapy is recommended in patients receiving rescue therapy.
Research frontiers
There are no reports whether only one biopsy from one anatomic site of the stomach is sufficient to detect antibiotic resistant H. pylori strains.
Innovations and breakthroughs
Discordant antibiotic susceptibility between antrum and corpus isolates for different antibiotics was seen in a reasonable number of patients. This might represent an explanation for H. pylori treatment failure.
Applications
By taking biopsies from the antrum and corpus of the stomach for H. pylori susceptibility testing, heteroresistance between antrum and corpus can be easily detected and treatment failure might be prevented.
Terminology
H. pylori eradication therapy consists of a mixture of different antibiotics that are given for at least one week. Resistance to antibiotics is the main reason for treatment failure, which became a problem in the last years. Therefore the knowledge of antibiotic resistance is important, especially in patients receiving rescue therapies.
Peer review
In this retrospective study authors aimed at assessing whether antibiotic resistance varies between antrum and corpus of the stomach in 66 patients that were either H. pylori therapy-naive or pre-treated. Discordant antibiotic susceptibility between antrum and corpus isolates for different antibiotics has been found in 10 (15%) patients while DNA fingerprinting analysis revealed no substantial differences between antrum and corpus isolates from samples of 8 of 9 available patients. Authors conclude that different antibiotic susceptibility between antrum and corpus biopsies is a common phenomenon and a possible explanation for treatment failure.
Footnotes
Supported by In part supported by a grant from the BMBF, BMBF-0315905D in the frame of ERA-NET PathoGenoMics
P- Reviewer: D‘Elios MM, Galgoczy L, Luzza F S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
References
- 1.Malfertheiner P, Megraud F, O’Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, et al. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P, Chan FK, McColl KE. Peptic ulcer disease. Lancet. 2009;374:1449–1461. doi: 10.1016/S0140-6736(09)60938-7. [DOI] [PubMed] [Google Scholar]
- 3.Selgrad M, Malfertheiner P. Treatment of Helicobacter pylori. Curr Opin Gastroenterol. 2011;27:565–570. doi: 10.1097/MOG.0b013e32834bb818. [DOI] [PubMed] [Google Scholar]
- 4.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 5.Asaka M, Kato M, Takahashi S, Fukuda Y, Sugiyama T, Ota H, Uemura N, Murakami K, Satoh K, Sugano K. Guidelines for the management of Helicobacter pylori infection in Japan: 2009 revised edition. Helicobacter. 2010;15:1–20. doi: 10.1111/j.1523-5378.2009.00738.x. [DOI] [PubMed] [Google Scholar]
- 6.Fock KM, Katelaris P, Sugano K, Ang TL, Hunt R, Talley NJ, Lam SK, Xiao SD, Tan HJ, Wu CY, et al. Second Asia-Pacific Consensus Guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24:1587–1600. doi: 10.1111/j.1440-1746.2009.05982.x. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach W, Malfertheiner P, Hoffmann JC, Bolten W, Bornschein J, Götze O, Höhne W, Kist M, Koletzko S, Labenz J, et al. S3-guideline “helicobacter pylori and gastroduodenal ulcer disease” of the German society for digestive and metabolic diseases (DGVS) in cooperation with the German society for hygiene and microbiology, society for pediatric gastroenterology and nutrition e. V., German society for rheumatology, AWMF-registration-no. 021 / 001. Z Gastroenterol. 2009;47:1230–1263. doi: 10.1055/s-0028-1109855. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin CS, Worsley BW. Microbiology of Helicobacter pylori. Gastroenterol Clin North Am. 1993;22:5–19. [PubMed] [Google Scholar]
- 9.Bayerdörffer E, Oertel H, Lehn N, Kasper G, Mannes GA, Sauerbruch T, Stolte M. Topographic association between active gastritis and Campylobacter pylori colonisation. J Clin Pathol. 1989;42:834–839. doi: 10.1136/jcp.42.8.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JJ, Kim JG, Kwon DH. Mixed-infection of antibiotic susceptible and resistant Helicobacter pylori isolates in a single patient and underestimation of antimicrobial susceptibility testing. Helicobacter. 2003;8:202–206. doi: 10.1046/j.1523-5378.2003.00145.x. [DOI] [PubMed] [Google Scholar]
- 11.Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wueppenhorst N, Stueger HP, Kist M, Glocker E. Identification and molecular characterization of triple- and quadruple-resistant Helicobacter pylori clinical isolates in Germany. J Antimicrob Chemother. 2009;63:648–653. doi: 10.1093/jac/dkp003. [DOI] [PubMed] [Google Scholar]
- 13.Cederbrant G, Kahlmeter G, Ljungh A. The E test for antimicrobial susceptibility testing of Helicobacter pylori. J Antimicrob Chemother. 1993;31:65–71. doi: 10.1093/jac/31.1.65. [DOI] [PubMed] [Google Scholar]
- 14.Glupczynski Y, Labbé M, Hansen W, Crokaert F, Yourassowsky E. Evaluation of the E test for quantitative antimicrobial susceptibility testing of Helicobacter pylori. J Clin Microbiol. 1991;29:2072–2075. doi: 10.1128/jcm.29.9.2072-2075.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayala G, Galván-Portillo M, Chihu L, Fierros G, Sánchez A, Carrillo B, Román A, López-Carrillo L, Silva-Sánchez J. Resistance to antibiotics and characterization of Helicobacter pylori strains isolated from antrum and body from adults in Mexico. Microb Drug Resist. 2011;17:149–155. doi: 10.1089/mdr.2010.0154. [DOI] [PubMed] [Google Scholar]
- 16.Rimbara E, Noguchi N, Tanabe M, Kawai T, Matsumoto Y, Sasatsu M. Susceptibilities to clarithromycin, amoxycillin and metronidazole of Helicobacter pylori isolates from the antrum and corpus in Tokyo, Japan, 1995-2001. Clin Microbiol Infect. 2005;11:307–311. doi: 10.1111/j.1469-0691.2005.01099.x. [DOI] [PubMed] [Google Scholar]
- 17.Yousfi MM, Reddy R, Osato MS, Graham DY. Is antrum or corpus the best site for culture of Helicobacter pylori? Helicobacter. 1996;1:88–91. doi: 10.1111/j.1523-5378.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 18.Selgrad M, Bornschein J, Malfertheiner P. Guidelines for treatment of Helicobacter pylori in the East and West. Expert Rev Anti Infect Ther. 2011;9:581–588. doi: 10.1586/eri.11.80. [DOI] [PubMed] [Google Scholar]
- 19.Malfertheiner P, Bazzoli F, Delchier JC, Celiñski K, Giguère M, Rivière M, Mégraud F. Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazole, and tetracycline given with omeprazole versus clarithromycin-based triple therapy: a randomised, open-label, non-inferiority, phase 3 trial. Lancet. 2011;377:905–913. doi: 10.1016/S0140-6736(11)60020-2. [DOI] [PubMed] [Google Scholar]
- 20.Gisbert JP, Castro-Fernandez M, Perez-Aisa A, Cosme A, Molina-Infante J, Rodrigo L, Modolell I, Cabriada JL, Gisbert JL, Lamas E, et al. Fourth-line rescue therapy with rifabutin in patients with three Helicobacter pylori eradication failures. Aliment Pharmacol Ther. 2012;35:941–947. doi: 10.1111/j.1365-2036.2012.05053.x. [DOI] [PubMed] [Google Scholar]
- 21.Marzio L, Cellini L, Amitrano M, Grande R, Serio M, Cappello G, Grossi L. Helicobacter pylori isolates from proximal and distal stomach of patients never treated and already treated show genetic variability and discordant antibiotic resistance. Eur J Gastroenterol Hepatol. 2011;23:467–472. doi: 10.1097/MEG.0b013e328345d40f. [DOI] [PubMed] [Google Scholar]
- 22.Karczewska E, Wojtas-Bonior I, Sito E, Zwolińska-Wcisło M, Budak A. Primary and secondary clarithromycin, metronidazole, amoxicillin and levofloxacin resistance to Helicobacter pylori in southern Poland. Pharmacol Rep. 2011;63:799–807. doi: 10.1016/s1734-1140(11)70592-8. [DOI] [PubMed] [Google Scholar]
- 23.Raymond J, Lamarque D, Kalach N, Chaussade S, Burucoa C. High level of antimicrobial resistance in French Helicobacter pylori isolates. Helicobacter. 2010;15:21–27. doi: 10.1111/j.1523-5378.2009.00737.x. [DOI] [PubMed] [Google Scholar]
- 24.Wolle K, Leodolter A, Malfertheiner P, König W. Antibiotic susceptibility of Helicobacter pylori in Germany: stable primary resistance from 1995 to 2000. J Med Microbiol. 2002;51:705–709. doi: 10.1099/0022-1317-51-8-705. [DOI] [PubMed] [Google Scholar]
- 25.Torres A, Liapikou A. Levofloxacin for the treatment of respiratory tract infections. Expert Opin Pharmacother. 2012;13:1203–1212. doi: 10.1517/14656566.2012.688952. [DOI] [PubMed] [Google Scholar]


