Background: The orphan receptor isoforms RORβ1 and RORβ2 encoded by Rorb are differentially expressed during retinal differentiation.
Results: The RORβ2 isoform is photoreceptor-specific and induces the rod differentiation factor NRL, which, in turn, induces RORβ2 through positive feedback.
Conclusion: NRL induces the RORβ2-specific promoter of Rorb, its own inducer gene.
Significance: Positive feedback between Nrl and Rorb genes reinforces the commitment to a rod differentiation fate.
Keywords: Cell Differentiation, Neurodevelopment, Photoreceptor, Retina, Transcription Factor
Abstract
Vision requires the generation of cone and rod photoreceptors that function in daylight and dim light, respectively. The neural retina leucine zipper factor (NRL) transcription factor critically controls photoreceptor fates as it stimulates rod differentiation and suppresses cone differentiation. However, the controls over NRL induction that balance rod and cone fates remain unclear. We have reported previously that the retinoid-related orphan receptor β gene (Rorb) is required for Nrl expression and other retinal functions. We show that Rorb differentially expresses two isoforms: RORβ2 in photoreceptors and RORβ1 in photoreceptors, progenitor cells, and other cell types. Deletion of RORβ2 or RORβ1 increased the cone:rod ratio ∼2-fold, whereas deletion of both isoforms in Rorb−/− mice produced almost exclusively cone-like cells at the expense of rods, suggesting that both isoforms induce Nrl. Electroporation of either RORβ isoform into retinal explants from Rorb−/− neonates reactivated Nrl and rod genes but, in Nrl−/− explants, failed to reactivate rod genes, indicating that NRL is the effector for both RORβ isoforms in rod differentiation. Unexpectedly, RORβ2 expression was lost in Nrl−/− mice. Moreover, NRL activated the RORβ2-specific promoter of Rorb, indicating that NRL activates Rorb, its own inducer gene. We suggest that feedback activation between Nrl and Rorb genes reinforces the commitment to rod differentiation.
Introduction
Vision in mammals is mediated by cone and rod photoreceptors that function in bright light and dim light, respectively. Cones also confer color vision, which, in most mammals, is mediated by cone subpopulations with peak sensitivity to short (S)3 or medium/longer (M) wavelengths of light (1). Retinal progenitor cells generate cones, rods, and other retinal cell types with distinct kinetics and in characteristic proportions (2–5). In mice, 97% of photoreceptors are rods, and ∼3% are cones (6).
The developmental controls that generate photoreceptor diversity are incompletely understood but involve a key role for transcription factors (7, 8). NRL (neural retina leucine zipper factor) drives photoreceptor precursors to a rod fate and suppresses a cone fate. Nrl−/− mice lack rods but, instead, display excess S cones (9). An immediate action of NRL is the induction of orphan nuclear receptor NR2E3 (10), which, together with NRL, induces rod genes and suppresses cone genes (11–15). Posttranslational modification has also been reported to modulate the NRL-NR2E3 pathway (16, 17). Cones lack NRL and diversify as M or S cones in response to thyroid hormone receptor β2 (TRβ2) and other factors (18–20). We have proposed a model for mammalian photoreceptor diversity in which an initial binary decision between a default cone fate or acquired rod fate hinges on NRL (8, 21, 22). The induction of Nrl is, therefore, critical for the rod versus cone decision but remains poorly delineated.
The retinoid-related orphan nuclear receptor β gene (Rorb) mediates circadian, motor, and visual functions in mice (23, 24), and human RORB mutations have recently been associated with intellectual disability and epilepsy (25, 26). Rorb−/− mice lack Nrl expression in the retina and resemble Nrl−/− mice because they lack rods but possess excess cone-like cells (27). Interestingly, the Nrl gene contains binding sites for RORβ and the homeodomain proteins OTX2 and CRX (28, 29). Rorb differentially expresses two N-terminal isoforms, RORβ1 and RORβ2 (30), from distinct promoters, suggesting a complex mode of action in the retina. We reported previously that RORβ1 is expressed in retinal progenitor cells and promotes the differentiation of amacrine and horizontal cells (31), but the function of RORβ2 is unknown. Here we show that RORβ2 is expressed in rod photoreceptors and that its targeted deletion increases the cone:rod ratio in the mouse retina. Further loss and gain of function analyses indicate that both RORβ2 and RORβ1 induce Nrl. Surprisingly, RORβ2 itself was identified as a target of NRL, indicating that NRL promotes positive feedback over Rorb, its own inducer gene. We propose that a mutual activation loop between the Rorb and Nrl genes reinforces the commitment to a rod fate.
EXPERIMENTAL PROCEDURES
Targeted Mutagenesis
The RORβ2-specific exon was identified by alignment of mouse genomic and cDNA sequences (accession no. NM_146095). The RORβ2 exon was replaced with lacZ, and homology arms isolated from W9.5 ES cell DNA using Phusion polymerase (New England Biolabs) were inserted into pACN carrying a neomycin resistance gene that self-excises upon germ line transmission by male chimeric mice (32) (5′ arm coordinates, −1965 to −1 relative to the RORβ2 ATG start codon and 3′ arm, +103 to +4153). Targeted W9.5 ES cell clones were used to generate chimeric mice at the Genetics Core Facility, Mount Sinai School of Medicine. Chimeras were crossed with C57BL/6J mice, and then Rorb+/2z mice were intercrossed to generate +/+, +/2z, and 2z/2z progeny on a mixed C57BL/6J × 129/Sv background for analysis. The targeted allele structure was confirmed by Southern blot using probes external to the targeting construct. Genotypes were determined by Southern blot or PCR using the following primers: common forward, CCA CTG CTG AAT GAA GTC TGC G; wild-type reverse, GCC TCT TCT ACC CAA AGT CAC; and 2z mutant reverse, GTA AAA CGA CGG GAT CCC CG; yielding wild-type and mutant bands of 710 and 463 bp, respectively. Genotyping has been described previously for the Rorb− total deletion (27), Rorb1gfp allele (31), and Nrl deletion (9). For the Nr2e3 rd7 allele, genotype primers were as follows: wild-type forward, CAG CCA CTG AAC TCC AAA CCT C; mutant forward, TGT GCT GAT TTT TAT ACA ATG; and common reverse, AAC ATA CTT CCA CTG TCA CTG CCC. Rorb2z and Rorb1g alleles were on C57BL/6J × 129/svJ backgrounds, and the Nrl null allele was on a C57BL/6J background. The studies were performed in accordance with protocols approved by the Animal Care and Use Committee at the NIDDK, National Institutes of Health.
Analysis of RNA by PCR
Reverse transcription PCR analysis of RORβ1 and RORβ2 mRNAs from pooled mouse retinas (≥ 3 mice/pool) was performed using the following primers: β1 forward, AAC AAA ACC CAG GCA CCA G; β2 forward, CAG ACC TCA AGT GAA ACG G; and common reverse AAC AGT TTC TCT GCC TTG G; yielding RORβ1 and RORβ2 bands of 442 and 249 bp, respectively. Gapdh was used as a control (primers: forward, ACC TGC CGC CTG GAG AAA CC; reverse, GAC CAT GAG GTC CAC CAC CCT G), yielding a 252-bp product. Real-time quantitative PCR (qPCR) analysis of mRNA levels of Opn1mw, Opn1sw, Rho, Nrl, Crx, Nr2e3, Gnat1, Pde6b, Mef2c, Chx10, and the two Rorb isoforms were normalized to Gapdh RNA levels. The primer pairs for qPCR were as follows: Opn1mw, TCG AAA CTG CAT CTT ACA TCT C and GGA GGT AAA ACA TGG CCA AA; Opn1sw, TGC TGG GGA TCT GAG ATG AT and AAT GAG GTG AGG CCA TTC TG; Rho, CCC TTC TCC AAC GTC ACA GG and TGA GGA AGT TGA TGG GGA AGC; Nrl, TCA CCC ACC TTC AGT GAG C and CCC GAG AAC CTC ATC CGA C; Nr2e3, ACT TGT GCT AAA CTG GAG CCA GAG and AAG CTC ATT CCA TGC CTC TTC C; Gnat1, GAT GCC CGC ACT GTG AAA C and CCA GCG AAT ACC CGT CCT G; Pde6b, GCA GCA CTT TTT GAA CTG GTG and CAT TGC GCT GGC GGT ACA TA; Mef2c, TGC CAG TTA CCA TCC CAG TGT C and ATC AGA CCG CCT GTG TTA CCT G; and Chx10, GCT CCG ATT CCG AAG ATG TTT C and AAA GAT TGT CCT GTG TCG CCG C. The transcription initiation site was mapped using an Ambion 5′ rapid amplification of cDNA ends kit (Ambion, catalog no. AM1700), followed by cloning and sequencing of product bands.
In Situ Hybridization, Immunohistochemistry, and Histology
Tissues were fixed in 4% paraformaldehyde, and 10-μm-thick retinal cryosections were hybridized with digoxigenin-labeled antisense and sense riboprobes as described previously (21). β-Galactosidase encoded by lacZ was detected on cryosections after incubation at 37 °C overnight with 1 mg/ml X-gal. For histology, tissues were fixed in 3% glutaraldehyde/2% paraformaldehyde. 3-μm-thick methacrylate plastic sections were stained with hematoxylin and eosin as described previously (21). Cells were counted on 160-μm lengths of ONL in the central retina on three sections each from six eyes for each group at an age of ∼3 months. Data represent mean ± S.D. Statistical analysis used Student's two tailed t test to compare +/+ with the test genotype.
Western Blot Analysis
Western blot analysis was performed on 20-μg samples of retinal protein using antibodies for RORβ (rabbit polyclonal, 1/1000, Diagenode, catalog no. pAbRORbHS100), NRL (rabbit polyclonal, 1/1000) (21), and actin (mouse monoclonal, 1/5000, Millipore, catalog no. MAb1501) using chemiluminescence detection.
Immunofluorescence analyses
Eyes or explanted retinal cultures were fixed in 1% paraformaldehyde for 4 h at 4 °C, cryoprotected in 30% sucrose overnight at 4 °C then embedded in Optimal Cutting Temperature. Ten μm thick cryosections were incubated in blocking buffer in PBS containing 1% BSA, 2.5% normal goat serum, and 0.1% Triton X-100 for 1 h at room temperature. Antibody dilutions and sources: M opsin rabbit polyclonal (1:1000, Millipore AB5405), S opsin rabbit polyclonal (1:1000, Millipore AB5407), rhodopsin mouse monoclonal (1:1000, Chemicon MAb5316), NRL (1:500) and NR2E3 (1:1000) rabbit polyclonals (21), TRβ2 rabbit polyclonal (1:1000) (21), GFP rabbit polyclonal (1:1000, Millipore AB3080). Imaging was performed on a Leica TCS SPE scanning confocal microscope. Images represented scans of 0.5 μm thickness. Brightness, contrast and merged images were adjusted using ImageJ (http://rsb.info.nih.gov/ij/).
Electroretinogram Analysis
The electroretinogram was recorded using an Espion electrophysiology system (Diagnosys LLC) on 8- to 10-week-old mice anesthetized with 25 mg/g of body weight ketamine, 10 mg/g of body weight xylazine, and 1000 mg/g of body weight urethane, as described previously (21, 33). Briefly, photopic responses were measured with rod responses suppressed using constant green light (520 nm) at 10 candelas/m2. S opsin responses were evoked with a light stimulus peak at 367 nm at intensities of 0.0001, 0.0002, 0.00032, 0.00056, 0.001, 0.0018, 0.0032, 0.0056, 0.01, 0.0178, 0.0316, and 0.056 candelas/m2. M opsin responses were evoked with a stimulus peak at 520 nm at intensities of 0.5, 1, 1.58, 2.51, 3.16, 5.01, 6.31, 10, 15.84, 25.12, and 38 candelas/m2. Scotopic rod responses were evoked after overnight dark adaptation using a stimulus peak at 520 nm at intensities of 1 × 10−6, 1 × 10−5, 1 × 10−4, 1 × 10−3, and 1 × 10−2 candelas/m2. Groups consisted of five to eight mice with males and females in each group. Data represent mean ± S.E. Statistical analysis used Student's t test to compare +/+ with the test genotype.
Electroporation and Retinal Explant Culture
Retinal explants from neonatal mice were immersed in DNA solution (1 μg/μl in PBS), and DNA plasmids were introduced by electroporation using five square pulses (30 V) of 50-ms duration with 950-ms intervals using a pulse generator (BTX Harvard apparatus) (34). Retinas were cultured at 37 °C on Nucleopore polycarbonate filters (Whatman, 0.2-μm pore size) with 1:1 mixed Ham's F-12 medium (Invitrogen) and DMEM (Invitrogen) containing 10% FCS (HyClone). After 8–10 days, retinas were analyzed by immunohistochemistry or qPCR analysis. RORβ1, RORβ2, NRL, and tdTomato were expressed from the pUb vector (35). The Rorb2p-GFP reporter contained the RORβ2-specific promoter (−4959 to −1, ATG start codon as +1) and a 0.6-kb conserved intron region (+1519 to +2109) that was inserted after the poly-A signal of the GFP cassette. Cell count data represent mean ± S.D. Statistical analysis used Student's two tailed t test to compare +/+ with the test genotype.
Isolation of Rod Cells and Strand-specific RNA Sequencing
Retinas were dissected from mice carrying the Nrlp-GFP transgene (36) at P2, P4, P6, P10, P14, and P28 in Hanks' balanced salt solution (Invitrogen), and cells were dissociated at 37 °C for 10 min using Accutase (Invitrogen). After incubation, cells were washed with 4 ml of PBS. Dissociated cells were collected by centrifugation at 1600 rpm for 5 min and resuspended in 1 ml of PBS. GFP+ cells were collected by flow sorter using FACS AriaII (BD Biosciences) in “precision mode” to minimize contamination from different retinal cell types. After flow sorting, samples were reanalyzed with the same settings to check purity. Samples over 97% purity were analyzed. Flow-sorted GFP+ cells were lysed in TRIzol LS (Invitrogen) to isolate total RNA. RNA quality was evaluated by Bioanalyzer RNA 6000 Pico assay (Agilent Technologies). Using 20 ng of total RNA, mRNA sequencing libraries were generated with poly-A selection using a TruSeq RNA sample prep kit v2 (catalog no. 15025062, Illumina, San Diego, CA). Strand-specific sequencing libraries were generated from at least two biological replicates as described previously (37) and sequenced on a Genome Analyzer IIx platform (Illumina). Transcript quantitation was performed with eXpress v1.3.1 by streaming pass filter reads to Bowtie2 v2.1.0 for alignment to GRCm38.p2/Ensembl v73 annotation.
RESULTS
Expression of RORβ2 and RORβ1 in Differentiating Rod Cell Populations
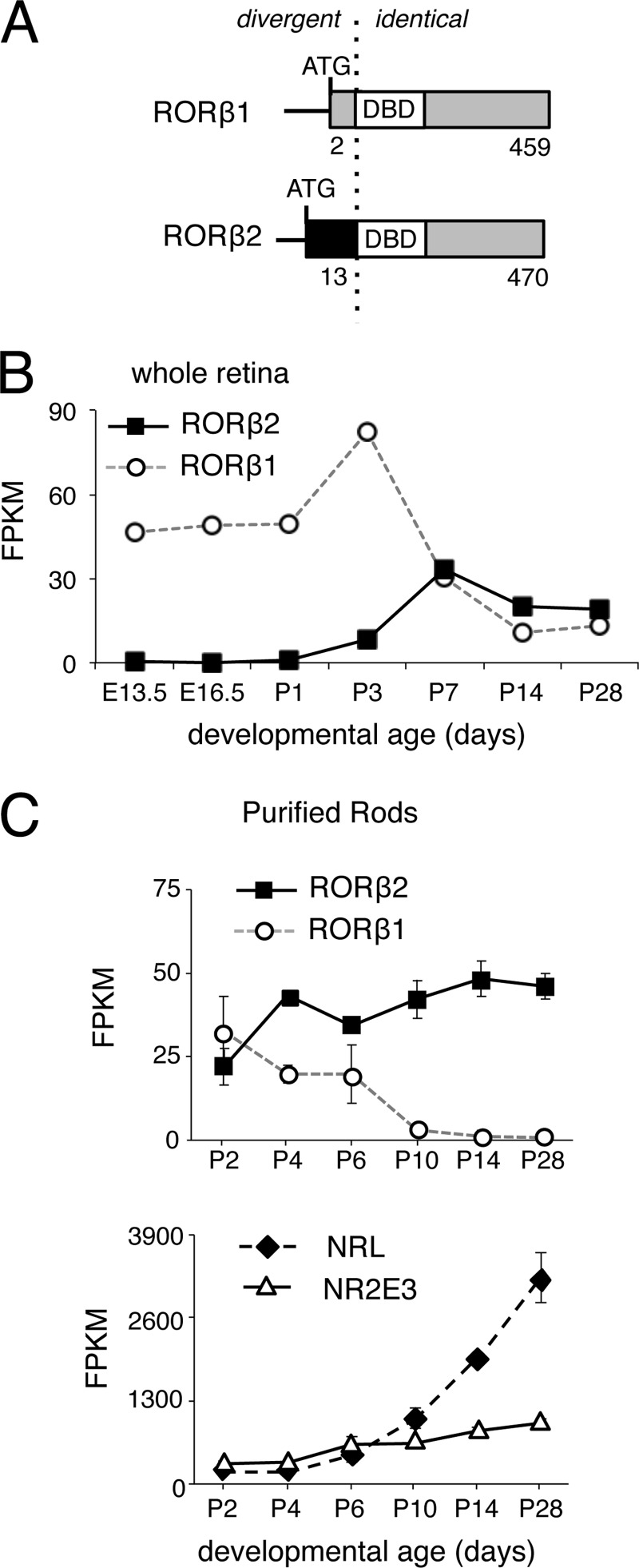
RORβ2 and RORβ1 isoforms possess identical DNA binding and C-terminal domains but divergent N termini encoded by unique 5′ exons, each with its own promoter (30, 31) (Fig. 1A). RNA sequencing analysis during retinal development revealed that RORβ2 mRNA expression rose at late embryonic stages and attained a plateau in the second postnatal week (Fig. 1B), a temporal pattern that correlated with peak generation of rod photoreceptors in mice (2, 3). In contrast, RORβ1 displayed an earlier rise and broader peak of expression, partly reflecting expression in undifferentiated progenitor cells and in horizontal and amacrine precursor cells (31). These results were in accord with a previous quantitative PCR analysis of RORβ2 and RORβ1 mRNA levels during retinal development (31).
FIGURE 1.
Differential temporal expression of RORβ isoforms. A, diagram of RORβ1 and RORβ2 isoforms showing the divergent N termini but identical DNA binding domain (DBD) and C terminus. The numbers refer to amino acid coordinates. B, expression profiles of RORβ1 and RORβ2 mRNA during retinal development, determined by RNA sequencing analysis of whole retina. RNA samples represented pooled retinas (from more than three mice) at each time point. FPKM, fragments per kilobase of transcript per million mapped reads; E13.5, embryonic day 13.5. C, expression profiles of RORβ1 and RORβ2 mRNA in rod photoreceptors, determined by RNA sequencing analysis of purified rod photoreceptors from Nrlp-GFP transgenic mice at the ages shown. The expression of NRL and NR2E3 rod factors rose over this period.
To investigate the role of RORβ isoforms specifically in rod differentiation, cells were purified by fluorescence-activated flow sorting from mice carrying a transgene that expresses GFP from the Nrl promoter during the final mitosis in rod genesis (36). Transcriptome analysis by RNA sequencing of these GFP+ rod populations revealed RORβ1 expression at postnatal day 2 (P2), the earliest stage examined, with a decline to undetectable levels by P14 (Fig. 1C). In contrast, RORβ2 continued to rise after P2 to a plateau by P14. Stages earlier than P2 could not be examined by this method because of the small, limiting numbers of rod precursors at embryonic stages. NRL and NR2E3 mRNA levels rose as expected in these differentiating rod populations (Fig. 1C). Rod sample purity was ascertained by detection of little or no expression of amacrine and horizontal (Calb1), bipolar (Prkca), and ganglion cell (Brn3b/Pou4f2) markers (data not shown). Coexpression of RORβ1 and RORβ2 at neonatal stages, when Nrl expression is amplified and large numbers of rods differentiate, implicated both isoforms with a role in rod differentiation fate.
Cellular Expression of RORβ2 Traced with a Targeted lacZ Marker
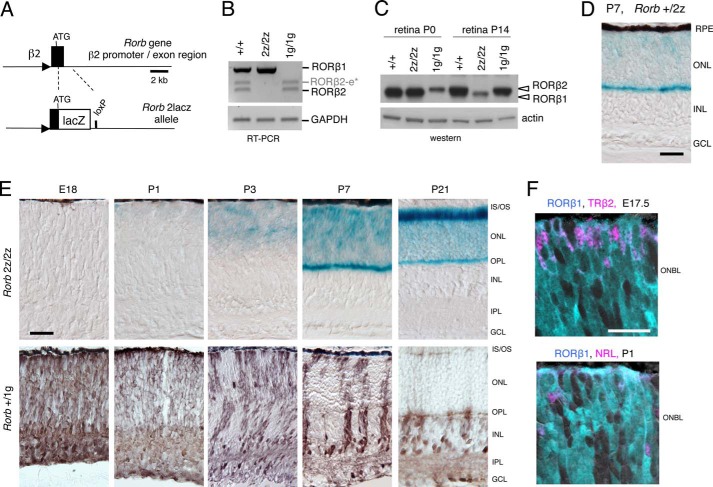
To determine RORβ2 function in mice, targeted mutagenesis (Fig. 2A) was used to replace the RORβ2-specific exon with lacZ, which also facilitated detection of expression. It has not been possible previously to distinguish cellular expression of RORβ2 and RORβ1 because these isoforms differ by only 13 and two unique N-terminal amino acids, respectively (Fig. 1A). RT-PCR analysis showed that Rorb2lacZ/2lacZ (2z/2z) mice lacked RORβ2 mRNA, whereas previously generated RORβ1-deficient mice (Rorb1gfp/1gfp, 1g/1g) (31) lacked RORβ1 mRNA (Fig. 2B). PCR bands for RORβ1, RORβ2, and an alternatively spliced, non-functional product, RORβ2-e* (24), were sequenced to confirm that the predicted products were deleted in 2z/2z mice. Western blot analysis of the retina at P14, when RORβ2 expression is normally prominent and exceeds that of RORβ1, confirmed loss of the 53-kDa RORβ2 protein and retention of the 52-kDa RORβ1 in 2z/2z mice (Fig. 2C). Conversely, analysis at P0, when RORβ1 expression is normally more prominent than that of RORβ2, confirmed loss of RORβ1 protein but retention of RORβ2 in 1g/1g mice.
FIGURE 2.
Targeted deletion of RORβ2 and cellular expression of RORβ isoforms. A, deletion of the RORβ2-specific exon and replacement with lacZ fused at the RORβ2 ATG codon, creating the Rorb2lacz (2z) allele. Triangle, RORβ2 promoter; loxP, residual loxP site after deletion of the ACN selection cassette. B, RT-PCR showing loss of RORβ2-specific mRNA (249-bp band) in 2z/2z mice and loss of RORβ1-specific mRNA (442-bp band) in RORβ1-deficient (1g/1g) mice. The alternatively spliced RORβ2-e* mRNA (302-bp band) was also deleted in 2z/2z mice. RORβ2-e* encodes a non-functional, 16-amino acid peptide without a DNA binding domain (24). C, Western blot analysis showing loss of RORβ2 and retention of RORβ1 protein in 2z/2z mice at the ages indicated. In 1g/1g mice, RORβ1 is lost, but RORβ2 is retained. A control blot for actin is shown below. Arrowheads, RORβ1 ∼52-kDa and RORβ2 ∼53-kDa bands. D, expression of RORβ2 in the immature photoreceptor layer (ONL) detected by histochemistry for β-galactosidase (turquoise) in +/2z mice at P7. Control +/+ mice gave no detectable β-galactosidase signal. RPE, retinal pigmented epithelium; INL, inner nuclear layer; GCL, ganglion cell layer. E, developmental expression of RORβ2 and RORβ1 in retinas monitored by, respectively, histochemistry for β-galactosidase (turquoise) in 2z/2z mice and immunohistochemistry for GFP (brown) in +/1g mice. IS/OS, inner/outer segments; OPL, outer plexiform layer; IPL, inner plexiform layer. F, RORβ1 in photoreceptor precursors detected by double fluorescence in Rorb+/1gfp mice for GFP (turquoise) with TRβ2 (purple) for cones or NRL (purple) for rods in the outer neuroblast layer (ONBL) at the indicated ages. Scale bars = 25 μm.
RORβ2 expression traced by histochemistry for β-galactosidase activity was detected specifically in the photoreceptor layer (ONL) of the retina at postnatal stages in heterozygous +/2z mice (Fig. 2D). No expression was detected in other retinal cell layers. Expression throughout the ONL was consistent with expression in the rod population, which spans the entire width of the ONL. Expression was weak in +/2z mice but was more readily detected in 2z/2z mice, with a similar spatial pattern in both genotypes. Developmental analysis in 2z/2z mice revealed an initially weak expression in the neonatal retina, with a marked increase in levels in the ONL by P7 and later stages (Fig. 2E). In contrast, RORβ1 expression traced by immunostaining for GFP in Rorb+/1g mice was prominent in embryonic and neonatal neuroblast layers and in horizontal and amacrine precursors. After P3, GFP expression declined but also spread at low levels to Müller glia and bipolar cells, as reported previously (31). GFP expression was transient in the photoreceptor layer but could be colocalized with the TRβ2 cone marker at embryonic day 17.5 and the NRL rod marker at P1 (Fig. 2F). The results indicated that RORβ2 was photoreceptor-specific, whereas RORβ1 was more widely expressed in photoreceptors and other retinal cell types.
Retinal Morphology and Excess Cones in RORβ2- and RORβ1-deficient Mice
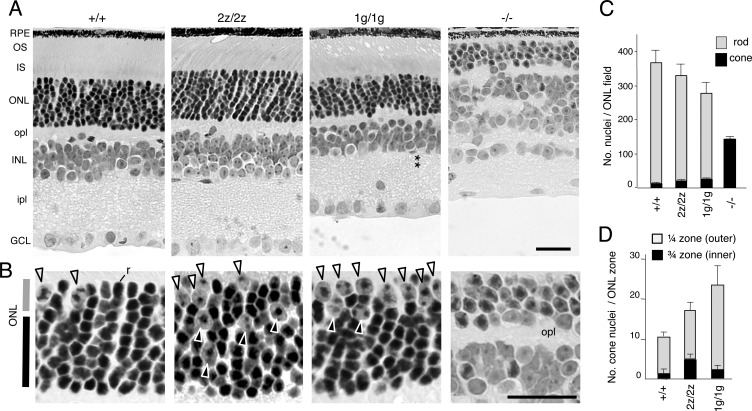
The expression of RORβ2 in rods at early postnatal stages suggested a role in determining the rod versus cone differentiation outcome. This hypothesis was supported by detection of ∼2-fold increased cone numbers in 2z/2z mice compared with +/+ mice at 3 months of age (p < 0.001) (Fig. 3, A and B). Cones are distinguished histologically by their large nuclei and dispersed chromatin, whereas rods possess smaller nuclei with condensed chromatin (6). Moreover, in 2z/2z mice, ∼30% of cone nuclei were misplaced in the inner ¾ zone of the ONL (Fig. 3, B and D) compared with the typical location in the outer ¼ zone of the ONL in +/+ mice (6).
FIGURE 3.
Retinal morphology and gain of cones in mice lacking RORβ isoforms. A, histological sections revealed a normal retinal structure in RORβ2-deficient (2z/2z) mice but with an ∼2-fold excess of cone nuclei. RORβ1-deficient (1g/1g) mice also displayed excess cone nuclei and, as reported, loss of horizontal and amacrine cells and disorganized inner and outer plexiform layers. The asterisks indicate the missing amacrine cell layer in the inner nuclear layer. In comparison, Rorb−/− mice have almost exclusively cone-like cells instead of rods, lack photoreceptor segments, lack horizontal and amacrine cells, and display disorganized plexiform layers. RPE, retinal pigmented epithelium; OS, outer segment; IS, inner segment; opl, outer plexiform layer; INL, inner nuclear layer; ipl, inner plexiform layer; GCL, ganglion cell layer. B, higher magnification indicating cone nuclei (large with dispersed chromatin, arrowheads) and a representative rod nucleus (r, small with dense chromatin in +/+ mice (left panel)). Excess cone nuclei are present in 2z/2z and 1g/1g mice. Many cones in 2z/2z mice are misplaced in the inner zone of the ONL, but, in 1g/1g mice, most are in the outer zone. C, counts (mean ± S.D.) of cone and rod nuclei in 160-μm-long ONL fields showing excess cones in 2z/2z and 1g/1g genotypes (each p < 0.001 versus +/+) and an almost exclusive presence of cones in Rorb−/− mice. D, counts (mean ± S.D.) of cone nuclei in outer ¼ and inner ¾ zones of the ONL showing substantial dispersal of cones in the inner zone of 2z/2z mice (p < 0.001 versus +/+). Rorb−/− mice were not included in this comparison because they possess a thin ONL that cannot be compared with the ONL in the other mouse strains and because almost all cells in the ONL in Rorb−/− mice are cone-like. Scale bars = 25 μm.
Although RORβ2-deficient mice displayed a 2-fold excess of cones, mice lacking both RORβ isoforms (Rorb−/−) possessed almost exclusively cone-like cells and no rods (Fig. 3A). We therefore tested whether RORβ1 also contributes to the rod versus cone outcome. RORβ1-deficient mice also displayed ∼2-fold increased cone numbers (p < 0.001). Most of these cones localized to the outer ¼ zone of the ONL, a differing distribution from that in 2z/2z mice, suggesting that subtle distinctions exist in the excess cone populations in 2z/2z and 1g/1g mice. Cones represented 2.8 ± 0.5% of total photoreceptor numbers (cones and rods) in +/+ mice, 5.2 ± 1.7% in 2z/2z mice, 8.4 ± 1.1% in 1g/1g mice, and 99.7 ± 6.5% in Rorb−/− mice (mean ± S.D.) (Fig. 3C). The cone excess in 2z/2z and 1g/1g mice was accompanied by decreased rod numbers (p < 0.001 each versus +/+), although the decreases were moderate as a proportion of the very large rod population.
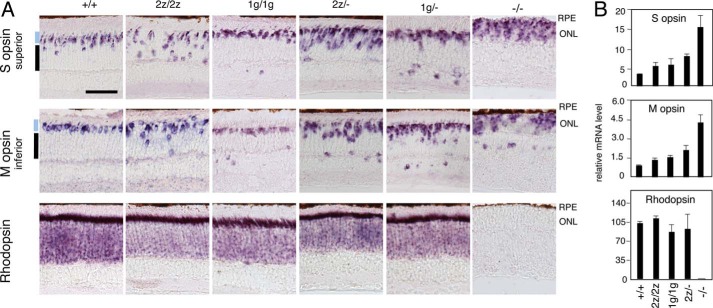
In accord with the histological evidence, in situ hybridization also revealed increases in cells expressing cone S and M opsins (Fig. 4A) and cone arrestin 3 (data not shown) in both 2z/2z and 1g/1g mice. Many S opsin+ and M opsin+ cells in 2z/2z mice were dispersed in the inner ¾ zone of the ONL. In 1g/1g mice, most S opsin+ and M opsin+ cells resided in the outer ¼ zone, although a few cells were misplaced in the outer plexiform layer and inner nuclear layer. Overt changes in rhodopsin expression were not detected in 2z/2z or 1g/1g mice, in contrast to the nearly complete lack of rhodopsin in Rorb−/− mice.
FIGURE 4.
Excess cone opsin-expressing cells in mice lacking RORβ isoforms. A, in situ hybridization revealed ∼2-fold increases in S opsin+ and M opsin+ cells, with many cones misplaced in the inner zones of the ONL in 2z/2z mice at 3 months of age. 1g/1g mice also showed increased cone opsin+ cells, most of which were in a normal location in the outer zone of the ONL. Stepwise deletion of additional RORβ isoforms in 2z/− or 1g/− compound heterozygotes further increased cone opsin+ cell numbers. In Rorb−/− mice lacking both RORβ isoforms, cone opsins were overexpressed, and rhodopsin was almost completely lacking. RPE, retina pigment epithelium. Scale bar = 50 μm. B, analysis by qPCR of S opsin, M opsin, and rhodopsin mRNA in the retina of 3-month-old mice of the indicated genotypes.
Retinal morphology, apart from the excess cones, was largely normal in 2z/2z mice. In contrast, 1g/1g mice lacked horizontal and amacrine cells and displayed disorganized plexiform layers, as described previously (31) (Fig. 3A). In Rorb−/− mice, deleted for both RORβ isoforms, photoreceptors lacked inner and outer segments, but both 2z/2z and 1g/1g mice retained segments, suggesting that RORβ2 and RORβ1 serve overlapping functions in promoting segment biosynthesis.
Photoreceptor Phenotypes following Combined Deletion of RORβ Isoforms
The moderate, ∼2-fold increase in the cone:rod ratio resulting from loss of either RORβ isoform compared with the extreme predominance of cone-like cells caused by the absence of all RORβ isoforms suggested that RORβ1 and RORβ2 substitute for each other in rod versus cone differentiation. Therefore, we investigated whether a more pronounced phenotype would be unmasked in compound heterozygotes in which the Rorb2z (RORβ2-deficient) allele was combined with the Rorb− null allele (lacking both isoforms). In comparison with Rorb2z/2z mice, the stepwise removal of additional RORβ isoforms of both types in Rorb2z/− mice further increased the proportion of S opsin+ and M opsin+ cells 2.0- and 1.5-fold, respectively (each p < 0.005 versus 2z/2z) (Fig. 4A). Similarly, the combination of the Rorb1g (RORβ1-deficient) allele with the null allele in Rorb1g/− compound heterozygotes also resulted in increased proportions of S opsin+ and M opsin+ cells (Fig. 4A).
These results, together with the evidence of an almost completely cone-dominated retina in Rorb−/− mice lacking all RORβ isoforms, indicate that RORβ2 and RORβ1 cooperate during retinal development and are capable of substantial substitution for each other in rod versus cone differentiation. Analysis by qPCR confirmed the increased cone opsin mRNA levels in 2z/2z and 1g/1g mice and a much exacerbated increase in Rorb−/− mice lacking all RORβ isoforms (Fig. 4B).
Electroretinogram Analysis of Cone and Rod Function
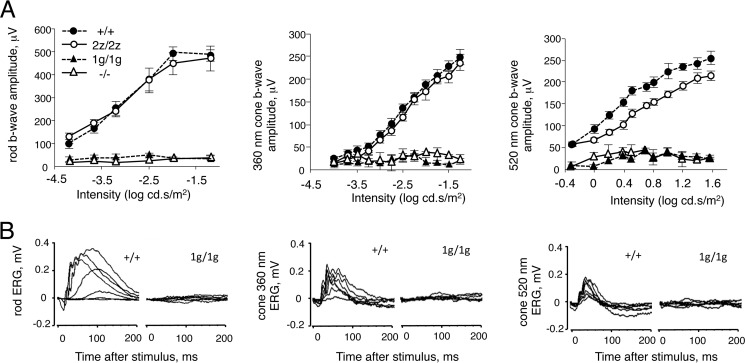
We examined whether 2z/2z and 1g/1g mice exhibit altered cone or rod function in accord with the increased cone:rod ratio because Nrl−/− mice display enhanced S cone function and loss of rod function (9). The scotopic electroretinogram revealed rod responses in the normal range in 2z/2z mice (Fig. 5A). The photopic electroretinogram evoked with light wavelengths optimal for stimulation of S cones (360 nm) and M cones (520 nm) in mice (33) revealed no difference for S cone responses and a slight decrease in M cone responses. Therefore, despite the ∼2-fold increased cone numbers, no enhanced cone response was evident.
FIGURE 5.
Electroretinogram analysis of mice lacking RORβ isoforms. A, intensity-response curves of electroretinogram responses for groups of five to eight mice (means ± S.E.) at ∼8 weeks of age. cd.s, candela second. Rod (left panel), S cone (center panel), and M cone (right panel) responses were absent in 1g/1g and −/− mice. B, representative electroretinogram traces for +/+ and 1g/1g mice. Left panel, scotopic rod responses. Center and right panels, photopic cone responses to 360- and 520-nm wavelengths that optimally activate S and M opsins, respectively. Families of traces are shown for stimuli of varying intensities.
In 1g/1g mice, no scotopic or phototopic responses could be evoked, even with high intensity stimuli. No a wave or b wave was detected (Fig. 5B). The lack of an a wave in the electroretinograms suggested the contribution of a photoreceptor-intrinsic defect because the a wave reflects the response of photoreceptors rather than the indirect response of bipolar neurons. Therefore, RORβ1 is essential for the acquisition of phototransduction function in all rod and cone photoreceptors. Rorb−/− mice, deficient for both RORβ1 and RORβ2, also lacked all rod and cone responses.
RORβ Isoforms Induce Nrl and Rod Differentiation Genes
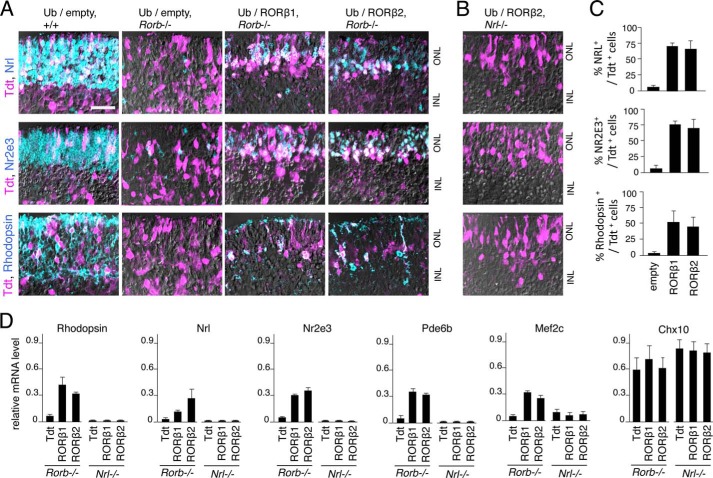
To demonstrate, by gain of function, that both RORβ isoforms promote a rod versus cone outcome and do so by induction of Nrl, vectors expressing RORβ1 and RORβ2 were electroporated into retinal explants from neonatal Rorb−/− mice that contain almost exclusively cone-like cells instead of rods (27). RORβ protein has been shown to bind to the Nrl gene promoter (28, 29), but a functional induction of Nrl has not been demonstrated. Electroporated cells were identified by coelectroporation of a vector for the Tdt marker. Control retinal explants from +/+ pups electroporated with empty expression vector (Ub/empty) expressed NRL, NR2E3, and rhodopsin in many cells in the ONL, whereas explants from Rorb−/− mice lacked almost all expression of these rod markers (Fig. 6A). Electroporation of the Ub/RORβ1 and Ub/RORβ2 expression vectors into explants from Rorb−/− mice resulted in the detection of NRL, NR2E3, and rhodopsin in many cells in the ONL (Fig. 6, A and C). Analysis by qPCR of explants electroporated in parallel demonstrated the induction of mRNA for many rod genes, including Rho, Nrl, Nr2e3, Pde6b, Mef2c (Fig. 6D), and Gnat1 (data not shown). As a negative control, the Chx10 bipolar cell marker showed no change in response to RORβ1, RORβ2, or empty control vector.
FIGURE 6.
Induction of Nrl and rod gene expression by RORβ isoforms. A, retinal explants from +/+ or Rorb−/− mice at P0 electroporated with RORβ1 or RORβ2 expression vectors. After 8 days in vitro, electroporated cells were identified with Tdt marker (purple), and NRL, NR2E3, and rhodopsin were detected by immunostaining (turquoise; or double-positive, whitish). Rorb−/− explants severely lacked NRL+, NR2E3+, and rhodopsin+ cells, but substantial expression of these markers was recovered by electroporation of Ub/RORβ1 or Ub/RORβ2. INL, inner nuclear layer. Scale bar = 25 μm. B, similar electroporations with RORβ2 or RORβ1 (data not shown) expression vectors in Nrl−/− explants failed to recover NR2E3 or rhodopsin expression. C, percentage of electroporated cells that express rod markers in explants from Rorb−/− mice counted per 160-μm length of retina. p < 0.001 for Ub/RORβ1 or Ub/RORβ2, each versus Ub/empty. D, analysis by qPCR showing reinduction of rod genes (rhodopsin, Nrl, Nr2e3, Pde6b, and Mef2c) after electroporation of Ub/RORβ1 or Ub/RORβ2 in Rorb−/− explants but not in Nrl−/− explants. A control bipolar cell gene, Chx10, did not vary in response to RORβ isoforms in any genotype. In a control electroporation in Nrl−/− retina, Ub/NRL expression vector reinduced mRNA for rod markers (data not shown).
To demonstrate that both RORβ1 and RORβ2 promote rod differentiation through induction of NRL, electroporations were performed in retinal explants from Nrl−/− pups (Fig. 6B). Neither RORβ isoform induced expression of any rod gene examined by immunofluorescence or qPCR analysis (Fig. 6D). These results indicate that the activation of rod genes in cone-like precursors by RORβ1 and RORβ2 depends upon the induction of NRL.
NRL-dependent Expression of RORβ2
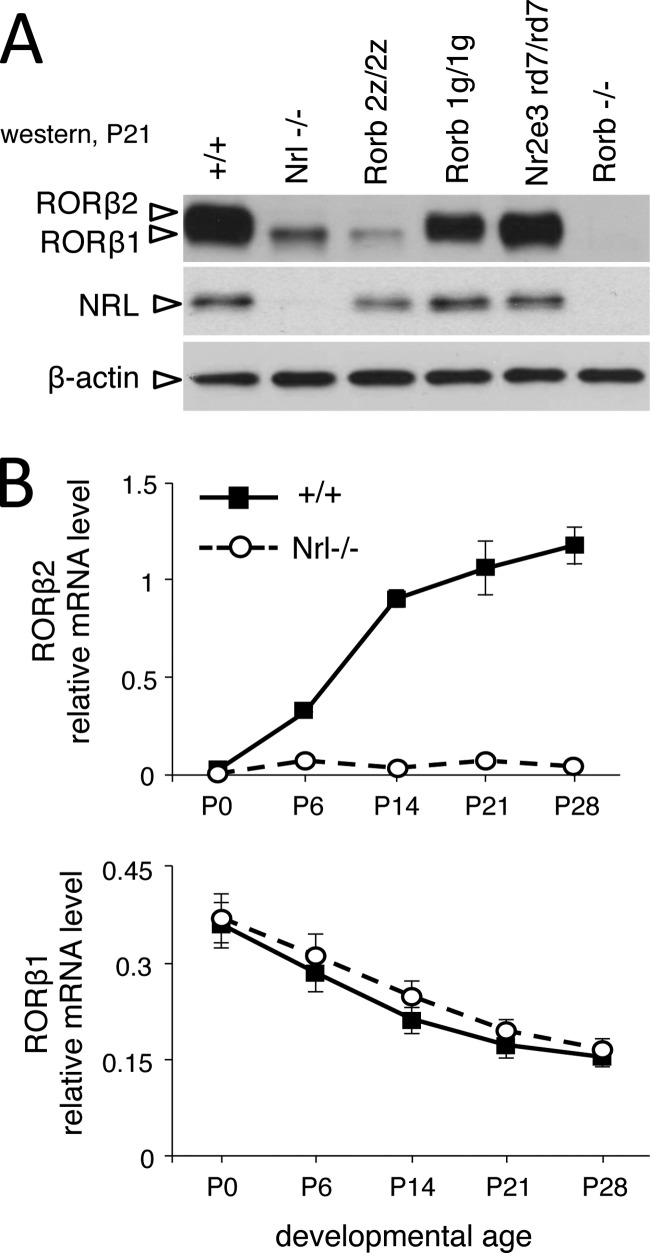
Rod differentiation requires the sequential induction of NRL and NR2E3, but the place of RORβ2 and RORβ1 in this hierarchy is undefined. The Rorb gene is required for Nrl expression, but Western blot analysis demonstrated, unexpectedly, that RORβ2 was undetectable in Nrl−/− mice (Fig. 7A), suggesting that RORβ2 was downstream of NRL. The residual RORβ protein band on the Western blot represented RORβ1, as indicated by comparison with 2z/2z mice that express only RORβ1. Analysis by qPCR confirmed the severe loss of RORβ2 mRNA in Nrl−/− mice in postnatal development (Fig. 7B). In contrast, RORβ1 protein and mRNA levels were largely unchanged in Nrl−/− mice at any postnatal stage (Fig. 7, A and B), consistent with RORβ1 being upstream of NRL in photoreceptor precursor cells. Neither RORβ1 nor RORβ2 expression was altered in Nr2e3rd7/rd7 mutant mice, indicating that expression of both RORβ isoforms was independent of NR2E3. These results suggested that RORβ isoforms were sequentially expressed relative to NRL in the order RORβ1-NRL-RORβ2 and, surprisingly, that RORβ2 itself was a target of NRL.
FIGURE 7.
NRL-dependent expression of RORβ2 in the retina. A, Western blot analysis showing lack of RORβ2 in the retina in Nrl−/− mice at P21. The slightly smaller RORβ1 isoform was retained, identified by comparison with 2z/2z mice that express only RORβ1. Control lanes for 1g/1g and Rorb−/− show loss of RORβ1 and loss of all RORβ bands, respectively. NR2E3-deficient mice showed no obvious loss of RORβ2 or RORβ1. B, quantitative PCR analysis showing severe loss of RORβ2 but not RORβ1 mRNA in retinas of Nrl−/− mice during postnatal development.
NRL-dependent Promoter in the Rorb Gene
To determine whether RORβ2 was induced at the transcriptional level by NRL, we first demonstrated the existence of a transcriptional start site in the putative RORβ2-specific promoter of the Rorb gene. A major 5′ end point of RORβ2 mRNA was localized 74 nucleotides upstream of the ATG initiator codon of RORβ2 by rapid amplification of 5′ complementary DNA ends made from mouse retinal RNA (Fig. 8A). Further analysis of ChIP sequencing (ChIPseq) data of NRL-bound chromatin from mouse retinas at P21 (38) revealed two peaks located in a 5-kb region upstream and a 2-kb region downstream of the RORβ2-specific exon of the Rorb gene. No other major NRL-bound peaks were detected over the ∼200-kb span of the Rorb gene.
FIGURE 8.
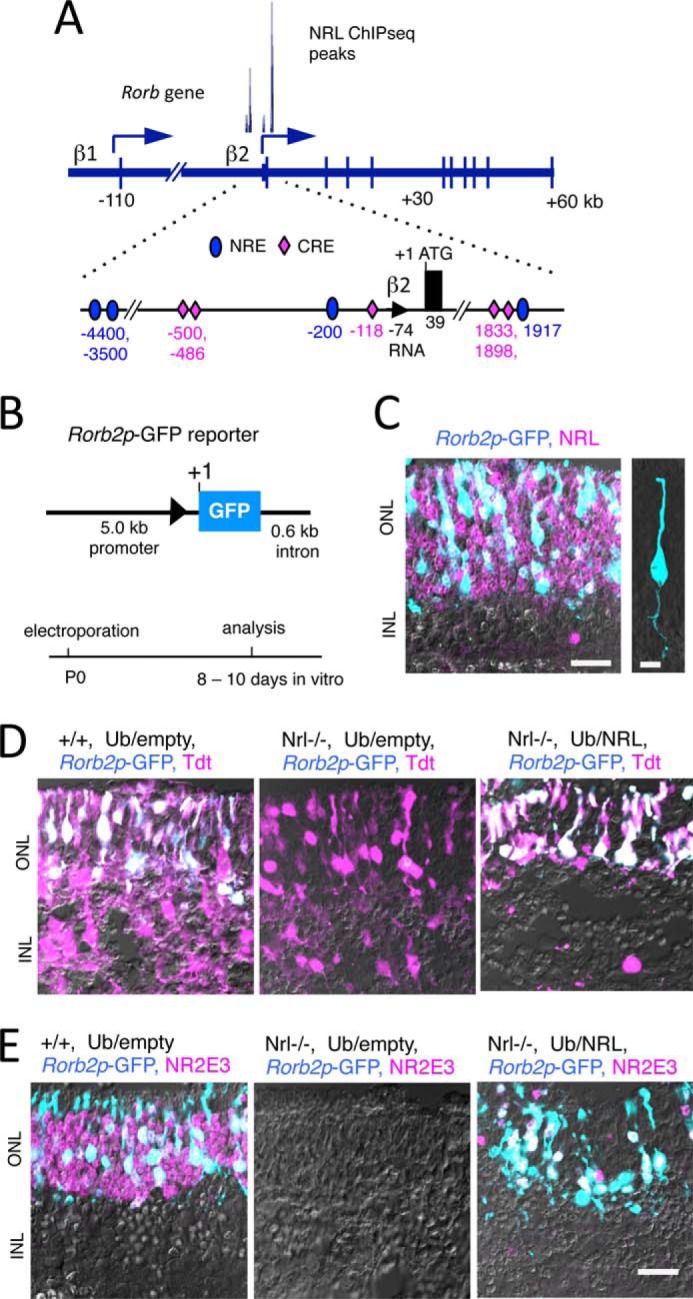
NRL-dependent promoter in the Rorb gene. A, diagram of the Rorb gene showing RORβ1- and β2-specific promoters and NRL-bound peaks detected by ChIPseq. Location of potential NRL response element (NRE, turquoise ovals) and CRX response element (CRE, purple diamonds) in the promoter and intron around the RORβ2-specific exon with nucleotide coordinates noted relative to the ATG (+1). Location of NREs is on the basis of NRL ChIPseq peaks and CREs on a consensus TAATCC motif and location of CRX ChIPseq peaks (39). Triangle, RORβ2 mRNA 5′ end mapped by 5′ rapid amplification of cDNA ends. B, RORβ2 promoter-GFP reporter (Rorb2p-GFP) carrying promoter and intron regions with the NREs and CREs shown in A. A schematic for electroporation and culture of retinal explants is shown below. C, Rorb2p-GFP expression (turquoise) in rod precursors in electroporated retina of +/+ mice. Almost all GFP+ cells were NRL+ (purple, or whitish for double-positive). Scale bar = 20 μm. INL, inner nuclear layer. Right panel, GFP+ cell showing rod-like morphology. Scale bar = 5 μm. D, Rorb2p-GFP lacked activity in the retinas from Nrl−/− pups but was reinduced by coelectroporation with the Ub/NRL expression vector. Electroporated cells were identified by the Tdt marker (purple, or whitish for double-positive). E, expression of electroporated Rorb2p-GFP (turquoise) and endogenous NR2E3 (purple) was not detected in Nrl−/− mice, but both were recovered in the same cells (whitish) by coelectroporation of Ub/NRL. Scale bar = 20 μm.
To investigate the functional consequence of NRL interaction with the RORβ2 promoter, a reporter (Rorb2p-GFP) carrying 5.0-kb promoter and 0.6-kb intron fragments that included the two NRL-bound regions mapped by ChIPseq was electroporated into retinal explants of neonatal +/+ mice (Fig. 8B). After 8 days in culture, GFP expression was detected in the ONL and colocalized with rhodopsin (data not shown) and NRL, indicating expression in rods (Fig. 8C). GFP+ cells displayed a rod-like morphology with projections extending to the outer edge of the ONL and to the outer plexiform layer. Rorb2p-GFP expression was not detected in explants from Nrl−/− mice but was recovered by coelectroporation with an NRL expression plasmid (Ub/NRL) (Fig. 8D). Most of these GFP+ cells also displayed a recovered expression of endogenous NR2E3, indicating that NRL induced the expression of both the RORβ2 promoter of the Rorb gene and the endogenous Nr2e3 gene in the same rod precursor cells (Fig. 8E).
DISCUSSION
Mutual Activation of Rorb and Nrl Genes in Rod Differentiation
We report that the Rorb gene displays a novel form of sequential expression of two orphan receptor isoforms, RORβ1 and RORβ2, during the process of induction of Nrl, a key gene that dictates rod differentiation (Fig. 9). We propose that, after the initiation of NRL expression by the RORβ1 isoform together with the homeodomain factors OTX2 and CRX (39, 40), NRL exerts dual actions to stabilize rod fate. First, NRL induces RORβ2, which, in turn, reinforces the induction of NRL through a mutual activation loop. This positive feedback might elevate NRL expression to a threshold, or sustain expression at a threshold, that biases the precursor cell to a rod fate. The second action of NRL is the induction of NR2E3 (9, 10), which promotes the activation of downstream rod genes and suppression of cone genes (11–15). On the basis of the developmental expression patterns of RORβ1 and RORβ2, it is possible that, in early differentiating rods, NRL is induced by RORβ1 and, in the later expanding rod population, by both RORβ isoforms because RORβ2 is induced by NRL in the neonatal phase.
FIGURE 9.
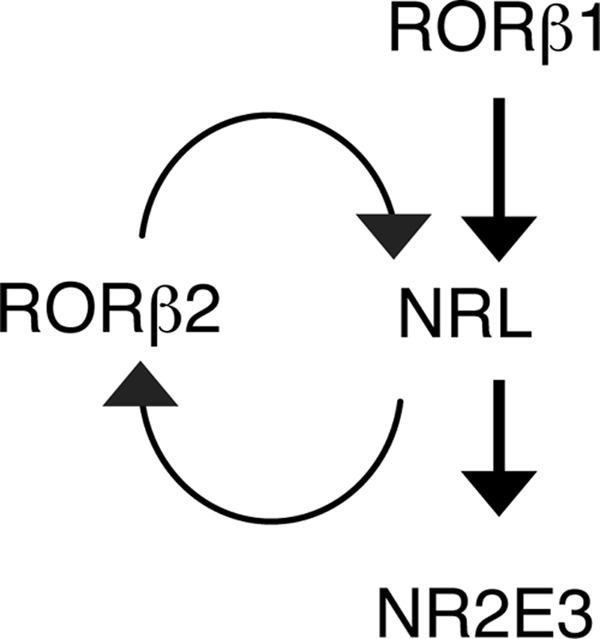
Model for the induction of the NRL rod differentiation factor by differentially expressed RORβ isoforms. In this proposed model, RORβ1 initiates the induction of NRL in immature precursors. NRL then exerts dual actions to stabilize the rod outcome. First, it induces RORβ2, which, in turn, reinforces NRL expression by positive feedback. Secondly, NRL induces NR2E3, which, together with NRL, activates rod genes and suppresses cone genes. Other factors not shown here, including OTX2 and CRX, probably enhance the expression of NRL and RORβ2.
The proposed model resembles other systems in which feedback loops bias a poised cell to commit to one of two alternative fates (41). Feedback loops have also been reported to stabilize photoreceptor cell fates in the Drosophila compound eye (42). An interesting speculation is that the self-reinforcing activation of the Rorb and Nrl genes may be part of the counting mechanism that produces a greater number of rods than cones in the mouse retina. In contrast, a simple linear induction process without such feedback may be less decisive in directing a rod outcome, a proposal that is supported by the elevated cone:rod ratio resulting from loss of either one of the two RORβ isoforms.
Our model predicts that deletion of either RORβ isoform might give a greater increase in the cone:rod ratio than the ∼2-fold shift observed in 1g/1g and 2z/2z mice. Why is this not the case, and why is there not a major loss of Nrl expression? A likely explanation is that transcriptional compensation masks a full-blown phenotype. For example, in RORβ1-deficient mice, OTX2 and CRX may still be able to induce a small amount of NRL that is adequate to initiate the expression of RORβ2, which, in turn, would more fully induce NRL. We showed that RORβ1 and RORβ2 individually are potent inducers of Nrl in retinal explants and can largely substitute for each other in directing the differentiation of the majority of rods in vivo. A technical limitation is that we are currently unable to quantify the fluctuations in levels of component transcription factors in an individual cell and cannot monitor the individual cellular transitions that occur in changing populations of precursors over many days in a complex tissue. Our current data concern average gene expression levels.
NRL also maintains the rod phenotype at mature stages (38, 43), but, because RORβ1 is no longer present in mature rods, how is NRL expression sustained? Potentially, CRX, which stimulates many rod and cone genes (44, 45), contributes to the expression of both NRL and RORβ2. Crx−/− mice have partly reduced RORβ2 expression (data not shown), and the Rorb gene contains CRX binding sites near to the NRL binding sites in the RORβ2 promoter (39) (Fig. 8A). Crx−/− mice also have reduced expression of NRL (40). It is also possible that the mutual activation of NRL and RORβ2 eventually becomes self-sustaining so that the initial stimulus, RORβ1, is no longer necessary for NRL expression at later stages.
Differential Functions for RORβ Isoforms in Retinal Development
Our evidence indicates that the functions of the Rorb gene are extended by its ability to express distinct isoforms in highly cell-specific and temporally distinct patterns. Rorb shares this property with other developmentally critical nuclear receptor genes, including thyroid hormone receptor β (19) and retinoid-related orphan receptor γ (46) genes. A unique feature of the Rorb gene is its sequential expression of both RORβ1 and RORβ2 isoforms in rod photoreceptors, which is coordinated by positive feedback by the downstream target, NRL rod differentiation factor. Another form of autoinduction of a nuclear receptor isoform during eye morphogenesis has been observed for the RARβ2-specific promoter of the retinoic acid receptor β gene in response to retinoic acid and the orphan nuclear receptor TLX (homolog of the Drosophila tailless protein) (47). Our results highlight the role of nuclear receptors, along with basic helix-loop-helix, homeodomain, and other classes of transcription factors (48–50), in retinal cell differentiation.
Although RORβ2 is restricted to photoreceptors, RORβ1 also promotes the differentiation of horizontal and amacrine interneurons (31). An intriguing parallel between the differentiation pathways for rod photoreceptors and interneurons is that, in both cases, RORβ isoforms act by induction of a key effector protein: NRL in rods and Ptf1a, a basic-helix-loop-helix factor, in horizontal and amacrine interneurons (51). The ability of RORβ isoforms to initiate two such different pathways is facilitated by cooperation with distinct cell-specific factors. Therefore, RORβ1 induces the Ptf1a gene synergistically with forkhead factor FOXN4 (52), whereas it initially induces Nrl in conjunction with the homeodomain factor OTX2 (28, 29, 40).
RORβ1 is also required for the development of phototransduction properties in all photoreceptors because rod or cone electroretinogram responses, including both a and b waves, were absent in RORβ1-deficient mice. In contrast, RORβ2-deficient mice retain rod and cone responses. However, cone responses are not enhanced in 2z/2z mice, suggesting that the excess cones lack function for reasons that remain unclear. Nr2e3 mutant mice also possess excess S cones but lack enhanced S cone function (53). In contrast, human NR2E3 mutations produce enhanced S cone responses (54), suggesting that the precise roles of factors immediately downstream of NRL may vary somewhat between species.
Another distinction between 2z/2z and 1g/1g mice is that the excess cones in 1g/1g mice align in the outer zone of the ONL but, in 2z/2z mice, are dispersed over the inner zone of the ONL, perhaps reflecting derivation from differing subpopulations of precursor cells, in accord with the temporally distinct expression patterns of RORβ1 and RORβ2. It has been reported that the final zonal residence of a mature photoreceptor in the ONL correlates with its time of generation from undifferentiated progenitor cells (2, 3). Further study of RORβ isoforms may yield insights into the events that control the timing of differentiation and the migration of photoreceptor precursor cells during maturation of the mammalian neuroretina.
Acknowledgments
We thank Mario Capecchi, Colin Stewart, Michael Becker-Andre, and Margarita Dubocovich for materials and Kevin Kelley at the Genetics Core Facility of Mount Sinai School of Medicine for microinjections.
This work was supported, in whole or in part, by the intramural research programs of the NIDDK (to D.F.) and NEI (to A.S.) at the National Institutes of Health. This work was also supported by the KaroBio Foundation (to D.F.).
- S
- short
- M
- medium/longer
- qPCR
- quantitative PCR
- ONL
- outer nuclear layer
- P2
- postnatal day 2
- ChIPseq
- ChIP sequencing
- CRX
- Cone-Rod homeobox protein
- OTX2
- Orthodenticle homeobox 2 protein.
REFERENCES
- 1. Nathans J. (1999) The evolution and physiology of human color vision: insights from molecular genetic studies of visual pigments. Neuron 24, 299–312 [DOI] [PubMed] [Google Scholar]
- 2. Carter-Dawson L. D., LaVail M. M. (1979) Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 188, 263–272 [DOI] [PubMed] [Google Scholar]
- 3. Young R. W. (1985) Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205 [DOI] [PubMed] [Google Scholar]
- 4. Brzezinski J., Reh T. A. (2010) Encyclopedia of the Eye, pp. 73–80, Elsevier, Amsterdam [Google Scholar]
- 5. Livesey F. J., Cepko C. L. (2001) Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2, 109–118 [DOI] [PubMed] [Google Scholar]
- 6. Carter-Dawson L. D., LaVail M. M. (1979) Rods and cones in the mouse retina: I: structural analysis using light and electron microscopy. J. Comp. Neurol. 188, 245–262 [DOI] [PubMed] [Google Scholar]
- 7. Adler R., Raymond P. A. (2008) Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain Res. 1192, 134–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swaroop A., Kim D., Forrest D. (2010) Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001) Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447–452 [DOI] [PubMed] [Google Scholar]
- 10. Oh E. C., Cheng H., Hao H., Jia L., Khan N. W., Swaroop A. (2008) Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 1236, 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng H., Khanna H., Oh E. C., Hicks D., Mitton K. P., Swaroop A. (2004) Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 13, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 12. Chen J., Rattner A., Nathans J. (2005) The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 25, 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng G. H., Ahmad O., Ahmad F., Liu J., Chen S. (2005) The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14, 747–764 [DOI] [PubMed] [Google Scholar]
- 14. Haider N. B., Mollema N., Gaule M., Yuan Y., Sachs A. J., Nystuen A. M., Naggert J. K., Nishina P. M. (2009) Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp. Eye Res. 89, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng H., Khan N. W., Roger J. E., Swaroop A. (2011) Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum. Mol. Genet. 20, 4102–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Onishi A., Peng G. H., Hsu C., Alexis U., Chen S., Blackshaw S. (2009) Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61, 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roger J. E., Nellissery J., Kim D. S., Swaroop A. (2010) Sumoylation of bZIP transcription factor NRL modulates target gene expression during photoreceptor differentiation. J. Biol. Chem. 285, 25637–25644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujieda H., Bremner R., Mears A. J., Sasaki H. (2009) Retinoic acid receptor-related orphan receptor α regulates a subset of cone genes during mouse retinal development. J. Neurochem. 108, 91–101 [DOI] [PubMed] [Google Scholar]
- 19. Ng L., Hurley J. B., Dierks B., Srinivas M., Saltó C., Vennström B., Reh T. A., Forrest D. (2001) A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98 [DOI] [PubMed] [Google Scholar]
- 20. Roberts M. R., Hendrickson A., McGuire C. R., Reh T. A. (2005) Retinoid X receptor γ is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 46, 2897–2904 [DOI] [PubMed] [Google Scholar]
- 21. Ng L., Lu A., Swaroop A., Sharlin D. S., Swaroop A., Forrest D. (2011) Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J. Neurosci. 31, 11118–11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oh E. C., Khan N., Novelli E., Khanna H., Strettoi E., Swaroop A. (2007) Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc. Natl. Acad. Sci. U.S.A. 104, 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. André E., Conquet F., Steinmayr M., Stratton S. C., Porciatti V., Becker-André M. (1998) Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Srinivas M., Ng L., Liu H., Jia L., Forrest D. (2006) Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor β. Mol. Endocrinol. 20, 1728–1741 [DOI] [PubMed] [Google Scholar]
- 25. Baglietto M. G., Caridi G., Gimelli G., Mancardi M., Prato G., Ronchetto P., Cuoco C., Tassano E. (2014) RORB gene and 9q21.13 microdeletion: report on a patient with epilepsy and mild intellectual disability. Eur. J. Med. Genet. 57, 44–46 [DOI] [PubMed] [Google Scholar]
- 26. Boudry-Labis E., Demeer B., Le Caignec C., Isidor B., Mathieu-Dramard M., Plessis G., George A. M., Taylor J., Aftimos S., Wiemer-Kruel A., Kohlhase J., Annerén G., Firth H., Simonic I., Vermeesch J., Thuresson A. C., Copin H., Love D. R., Andrieux J. (2013) A novel microdeletion syndrome at 9q21.13 characterised by mental retardation, speech delay, epilepsy and characteristic facial features. Eur. J. Med. Genet. 56, 163–170 [DOI] [PubMed] [Google Scholar]
- 27. Jia L., Oh E. C., Ng L., Srinivas M., Brooks M., Swaroop A., Forrest D. (2009) Retinoid-related orphan nuclear receptor RORβ is an early-acting factor in rod photoreceptor development. Proc. Natl. Acad. Sci. U.S.A. 106, 17534–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kautzmann M. A., Kim D. S., Felder-Schmittbuhl M. P., Swaroop A. (2011) Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. J. Biol. Chem. 286, 28247–28255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montana C. L., Lawrence K. A., Williams N. L., Tran N. M., Peng G. H., Chen S., Corbo J. C. (2011) Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J. Biol. Chem. 286, 36921–36931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. André E., Gawlas K., Becker-André M. (1998) A novel isoform of the orphan nuclear receptor RORβ is specifically expressed in pineal gland and retina. Gene 216, 277–283 [DOI] [PubMed] [Google Scholar]
- 31. Liu H., Kim S. Y., Fu Y., Wu X., Ng L., Swaroop A., Forrest D. (2013) An isoform of retinoid-related orphan receptor β directs differentiation of retinal amacrine and horizontal interneurons. Nat. Commun. 4, 1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bunting M., Bernstein K. E., Greer J. M., Capecchi M. R., Thomas K. R. (1999) Targeting genes for self-excision in the germ line. Genes Dev. 13, 1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyubarsky A. L., Falsini B., Pennesi M. E., Valentini P., Pugh E. N., Jr. (1999) UV- and midwave-sensitive cone-driven retinal responses of the mouse: a possible phenotype for coexpression of cone photopigments. J. Neurosci. 19, 442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsuda T., Cepko C. L. (2008) Analysis of gene function in the retina. Methods Mol. Biol. 423, 259–278 [DOI] [PubMed] [Google Scholar]
- 35. Kim D. S., Matsuda T., Cepko C. L. (2008) A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J. Neurosci. 28, 7748–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akimoto M., Cheng H., Zhu D., Brzezinski J. A., Khanna R., Filippova E., Oh E. C., Jing Y., Linares J. L., Brooks M., Zareparsi S., Mears A. J., Hero A., Glaser T., Swaroop A. (2006) Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 103, 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brooks M. J., Rajasimha H. K., Swaroop A. (2012) Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol. Biol. 884, 319–334 [DOI] [PubMed] [Google Scholar]
- 38. Hao H., Kim D. S., Klocke B., Johnson K. R., Cui K., Gotoh N., Zang C., Gregorski J., Gieser L., Peng W., Fann Y., Seifert M., Zhao K., Swaroop A. (2012) Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 8, e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corbo J. C., Lawrence K. A., Karlstetter M., Myers C. A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L. G., Weber B. H., Langmann T. (2010) CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roger J. E., Hiriyanna A., Gotoh N., Hao H., Cheng D. F., Ratnapriya R., Kautzmann M. A., Chang B., Swaroop A. (2014) OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest. 124, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Losick R., Desplan C. (2008) Stochasticity and cell fate. Science 320, 65–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jukam D., Desplan C. (2010) Binary fate decisions in differentiating neurons. Curr. Opin. Neurobiol. 20, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Montana C. L., Kolesnikov A. V., Shen S. Q., Myers C. A., Kefalov V. J., Corbo J. C. (2013) Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 110, 1732–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen S., Wang Q. L., Nie Z., Sun H., Lennon G., Copeland N. G., Gilbert D. J., Jenkins N. A., Zack D. J. (1997) Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 45. Furukawa T., Morrow E. M., Li T., Davis F. C., Cepko C. L. (1999) Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 23, 466–470 [DOI] [PubMed] [Google Scholar]
- 46. Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. (2006) The orphan nuclear receptor RORγ t directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 47. Kobayashi M., Yu R. T., Yasuda K., Umesono K. (2000) Cell-type-specific regulation of the retinoic acid receptor mediated by the orphan nuclear receptor TLX. Mol. Cell. Biol. 20, 8731–8739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Forrest D., Swaroop A. (2012) Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol. Endocrinol. 26, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vetter M. L., Brown N. L. (2001) The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin. Cell Dev. Biol. 12, 491–498 [DOI] [PubMed] [Google Scholar]
- 50. Xiang M. (2013) Intrinsic control of mammalian retinogenesis. Cell Mol. Life Sci. 70, 2519–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R. J., Furukawa T., Fujikado T., Magnuson M. A., Xiang M., Wright C. V. (2006) Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133, 4439–4450 [DOI] [PubMed] [Google Scholar]
- 52. Li S., Mo Z., Yang X., Price S. M., Shen M. M., Xiang M. (2004) Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43, 795–807 [DOI] [PubMed] [Google Scholar]
- 53. Ueno S., Kondo M., Miyata K., Hirai T., Miyata T., Usukura J., Nishizawa Y., Miyake Y. (2005) Physiological function of S-cone system is not enhanced in rd7 mice. Exp. Eye. Res. 81, 751–758 [DOI] [PubMed] [Google Scholar]
- 54. Haider N. B., Jacobson S. G., Cideciyan A. V., Swiderski R., Streb L. M., Searby C., Beck G., Hockey R., Hanna D. B., Gorman S., Duhl D., Carmi R., Bennett J., Weleber R. G., Fishman G. A., Wright A. F., Stone E. M., Sheffield V. C. (2000) Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 24, 127–131 [DOI] [PubMed] [Google Scholar]