Background: Pathogenic bacteria avoid killing by phagocytes through inhibition of C5a chemotactic activity.
Results: Periodontopathogen Porphyromonas gingivalis expresses unique peptidylarginine deiminase, which inactivates C5a by converting C-terminal arginine to citrulline.
Conclusion: Citrullination of C5a constitutes a novel virulence strategy that may contribute to immune evasion by P. gingivalis.
Significance: P. gingivalis peptidylarginine deiminase is a potential target for drug development.
Keywords: Bacteria, Chemotaxis, Complement System, Inflammation, Periodontal Disease
Abstract
Evasion of killing by the complement system, a crucial part of innate immunity, is a key evolutionary strategy of many human pathogens. A major etiological agent of chronic periodontitis, the Gram-negative bacterium Porphyromonas gingivalis, produces a vast arsenal of virulence factors that compromise human defense mechanisms. One of these is peptidylarginine deiminase (PPAD), an enzyme unique to P. gingivalis among bacteria, which converts Arg residues in polypeptide chains into citrulline. Here, we report that PPAD citrullination of a critical C-terminal arginine of the anaphylatoxin C5a disabled the protein function. Treatment of C5a with PPAD in vitro resulted in decreased chemotaxis of human neutrophils and diminished calcium signaling in monocytic cell line U937 transfected with the C5a receptor (C5aR) and loaded with a fluorescent intracellular calcium probe: Fura-2 AM. Moreover, a low degree of citrullination of internal arginine residues by PPAD was also detected using mass spectrometry. Further, after treatment of C5 with outer membrane vesicles naturally shed by P. gingivalis, we observed generation of C5a totally citrullinated at the C-terminal Arg-74 residue (Arg74Cit). In stark contrast, only native C5a was detected after treatment with PPAD-null outer membrane vesicles. Our study suggests reduced antibacterial and proinflammatory capacity of citrullinated C5a, achieved via lower level of chemotactic potential of the modified molecule, and weaker cell activation. In the context of previous studies, which showed crosstalk between C5aR and Toll-like receptors, as well as enhanced arthritis development in mice infected with PPAD-expressing P. gingivalis, our findings support a crucial role of PPAD in the virulence of P. gingivalis.
Introduction
Porphyromonas gingivalis is a major causative agent of periodontitis, a chronic inflammatory disease of tooth-supporting structures that affects up to 30% of the world's population (1). This Gram-negative, anaerobic bacterium uses a large arsenal of virulence factors such as hemagglutinins/adhesins, fimbriae, and proteolytic enzymes to facilitate colonization of the gingival sulcus, to generate nutrients and growth factors, and to provide protection from the host immune system (2). The latter is achieved by sophisticated manipulation of the host inflammatory response through activation of coagulation factors and contact activation systems, corrupting complement functions, shedding receptors, and modifying cytokines and intracellular signaling (3). In this way, P. gingivalis maintains the local chronic inflammatory reaction and thrives in this environment to access host components essential for bacterial growth.
Recently, P. gingivalis peptidylarginine deiminase (PPAD),4 an enzyme absent in other prokaryotes, which converts Arg residues in polypeptide chains into citrulline (Cit), has been hypothesized to be a potential virulence factor (4). Post-translational modification mediated by the deiminase activity of PPAD may change protein function in a similar manner to that described for citrullination of chemokines (5) and antibacterial peptide LL-37 (6) by endogenous PADs. However, in stark contrast to mammalian enzymes, PPAD has a strong preference for C-terminal Arg residues, probably to neutralize the positive charge at the C terminus of (poly)peptide fragments generated by degradation of proteins by P. gingivalis Arg-specific gingipains (7). Furthermore, PPAD can also abrogate essential biological activities of host proteins and peptides, which are dependent on the C-terminal Arg residues. This hypothesis is supported by the finding that C-terminal citrullination of EGF by PPAD impaired biological activity of EGF (8).
Anaphylatoxin C5a is a polypeptide of 74 residues released from C5 by C5 convertase during complement activation. Widespread expression of two C5a receptors (C5aR and C5L2) throughout the body ensures a variety of biological responses, including chemotaxis of inflammatory cells, phagocytosis, respiratory burst, vascular permeability, and releases of pro-inflammatory cytokines and chemokines. The C-terminal Arg residue is crucial for C5a function, and in vivo, the molecule is rapidly converted by carboxypeptidases to the far less potent C5a-desArg of significantly lower affinity for C5aR (9). Because C5a is an essential component of the inflammatory response to bacterial infection, it was of interest to determine whether PPAD can abolish its biological activity. Here, we showed that PPAD efficiently deiminated C-terminal Arg in C5a in vitro and that citrullinated Arg-74 of C5a can be found in C5 samples treated with outer membrane vesicles isolated from P. gingivalis culture. Moreover, this modification decreased the ability of C5a to induce calcium influx in a monocytic cell line expressing C5aR and strongly reduced its chemotactic potential for neutrophils.
EXPERIMENTAL PROCEDURES
Ethics Statement
The regional ethical board in Lund has approved collection of blood from healthy volunteers after informed consent.
Proteins
As a member of the C-terminal domain protein family of P. gingivalis, native PPAD was engineered to be secreted from a mutant of P. gingivalis W83 as a soluble form with a hexahistidine affinity tag using the same molecular strategy as was reported previously for the RgpB protease in the mutant 662i6H (10). Subsequently, PPAD was purified from the culture medium via ion-exchange and gel filtration chromatography. Briefly, the bacteria were cultured in enriched tryptic soy broth medium for 72 h, cells were removed by centrifugation, and proteins in cell-free culture medium were precipitated with acetone, resuspended in phosphate buffer (pH 6.5), dialyzed, and passed through a DE-52 column (Whatman) to remove excess hemin. The flow-through was dialyzed against 50 mm Tris-HCl, 0.02% NaN3, pH 8.0, and loaded on a Mono Q column (GE Healthcare). Adsorbed proteins were eluted with NaCl gradient, and fractions containing PPAD activity were pooled. The final PPAD purification was achieved by gel filtration chromatography using Superdex 75 column (GE Healthcare). The purity of PPAD was evaluated by SDS-PAGE followed by silver staining. Activity of PPAD was tested using a colorimetric assay as reported previously (11). Before any assay, PPAD was preactivated by incubation for 20 min 37 °C in 10 mm Tris, pH 8.0, buffer in presence of 1 mm l-cysteine. Because a very minor contamination (less than 0.5%) with arginine-specific gingipain (Rgp) was observed in some batches of purified PPAD, assay buffers were supplemented with 1 μm KYT-1, a specific Rgp inhibitor (Peptide International). At the concentrations used, KYT-1 and l-cysteine did not influence the assays (data not shown). C5a and C5a-desArg were purchased from Complement Technology.
Isolation of Outer Membrane Vesicles (OMV) from P. gingivalis Strains
P. gingivalis strains W83 and mutant Δppad (in W83 background) (12) were cultivated in enriched Schaedler broth (supplemented with 5 mg/ml hemin, 0.5 mg/ml menadione, and 5 mm l-cysteine, and for mutant strain, supplemented with 5 μg/ml erythromycin) overnight in anaerobic conditions to reach optical density = 1. An aliquot (100 ml) of each culture was gently sonicated in a sonicator bath to release OMV to the cell supernatant. Bacteria were removed by centrifugation (20 min, 10,000 × g), and the remaining supernatant from each strain was subjected to ultracentrifugation (1 h, 150,000 × g, 4 °C). Pellets containing OMV fraction were resuspended in 20 mm Bis-Tris, pH 6.8, 150 mm NaCl, 5 mm CaCl2 and analyzed for PPAD- and Arg-specific gingipain (Rgp) activity. Rgp activity was tested using a spectrophotometric assay with Nα-benzoyl-l-arginine 4-nitroanilide hydrochloride as a substrate (13), and Rgp concentration was calculated based on the activity of the purified enzyme.
HPLC Analysis of Full-length Form of C5a
Samples (100 μl) containing 10 μg of C5a or C5a were treated with PPAD (in buffer containing 100 mm Tris, 5 mm l-cysteine, 5 μm KYT-1, pH 7.6) for 3 h at 37 °C. Samples were subsequently acidified with TFA (Sigma) and separated through a 4.6 × 150-mm Phenomenex Aeris C18 Widepore (Phenomenex) on ÄKTAmicro (GE Healthcare). Peptides were then eluted in H2O/0.1% TFA (A) and 80% acetonitrile/0.08% TFA (B) gradient and monitored at 215 nm.
Deglycosylation and Proteolytic Treatment
Samples of C5a and C5a-Cit prepared in the same way as for HPLC analysis were lyophilized and dissolved in 20 mm Tris, 100 mm NaCl, pH 8.0, containing 5 mm DTT and denatured at 95 °C for 1 h. To remove N-linked glycosylation, samples were treated with 0.25 units of N-glycosidase F (Sigma) per μg of C5a and incubated at 37 °C for 3 h. To generate two different variants of the C-terminal peptide, the deglycosylated sample was treated with either trypsin (1:25 ratio) or clostripain (4 units/μg of C5a) (both from Sigma) in the presence of 1 mm calcium acetate and incubated at 37 °C for 16 h. To uniformly modify all cysteine residues, samples were reduced by 5 mm DTT for 30 min at 25 °C followed by 30 min with 15 mm iodoacetamide. The samples were micropurified on StageTips (Thermo Scientific) according to the manufacturer's instructions.
Modification of C5 by Enzymes Present in OMVs from Different Strains of P. gingivalis
Purified C5 (Complement Technology) was mixed at 25:1 molar ratio to Rgp with OMVs isolated from P. gingivalis strains W83 and Δppad (in PBS buffer, pH 7.4, supplemented with 5 mm l-cysteine). Samples were incubated for 30 min 37 °C, and the reaction was stopped by the addition of TFA. Citrullination of the C-terminal Arg residue in C5a was assessed by mass spectrometry.
Mass Spectrometry
nLC-MS/MS analyses were performed on an EASY-nLC II system (Thermo Scientific) connected to a TripleTOF 5600+ mass spectrometer (AB SCIEX) equipped with a NanoSpray III source (AB SCIEX) operated under Analyst TF 1.5.1 control. The lyophilized samples were suspended in 0.1% formic acid, injected, trapped, and desalted isocratically on a Biosphere C18 column (5 μm, 2 cm × 100-μm inner diameter; Nano Separations). Peptides were eluted using 250 nl/min and a 20-min (for C5 samples treated with OMVs) or 50-min (for C5a samples treated with purified PPAD) gradient from 5 to 35% phase B (0.1% formic acid and 90% acetonitrile). Eluted peptide samples were separated on a 15-cm analytical column (75-μm inner diameter) with RP ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH). The collected MS files were converted to Mascot generic format (MGF) using the AB SCIEX MS Data Converter beta 1.1 (AB SCIEX) and the “protein pilot MGF” parameters. The generated peak lists were searched against an in-house database containing the C5a sequence using the Mascot search engine (Matrix Science). Search parameters were either trypsin or Arg-C as protease allowing three missed cleavage sites. Carbamidomethyl was set as fixed modification, and citrullination of Arg, deamidation of Asn or Gln, and oxidation of Met residues were set as variable modifications. Peptide tolerance and MS/MS tolerance were set to 10 ppm and 0.2 Da, respectively.
Cell Culture Conditions
U937 cells expressing C5a receptor (U937-C5aR) were maintained in RPMI medium containing 10% FCS and G418 (400 μg/ml) at 37 °C in a humidified 5% CO2 atmosphere (14).
Calcium Mobilization Assay
U937-C5aR cells were harvested by centrifugation, washed twice with PBS, resuspended in HBSS (0.137 m NaCl, 5.4 mm KCl, 0.25 mm Na2HPO4, 0.44 mm KH2PO4, 1.0 mm MgSO4, 4.2 mm NaHCO3, 1 g/liter glucose) with 1.3 mm Ca2+ at 2 × 106 cells/ml, and incubated in the presence of a cell-permeable fluorescent intracellular calcium probe (5 μm Fura-2 AM, Invitrogen) for 30 min 37 °C with gentle agitation. After double washing with HBSS/Ca2+, cells were resuspended in the same buffer at 1.25 × 106 cells/ml and kept on ice until use. Cell response to a test ligand was monitored with continuous fluorescence measurements at λexcitation = 340 and 380 nm and λemission = 510 nm, using the Infinite 200 PRO microplate reader (Tecan) and i-Control software. After a few seconds of the basal signal measurement, C5a (50 μl, final concentration 10 nm) or C5a pretreated for 1 h at 37 °C with PPAD was mixed with 200 μl of cell suspension, and measurement was continued for 5 min. Values of fluorescence intensity for each time point were recalculated as the ratio of fluorescence intensity: λemission = 510 nm at λexcitation = 340 nm/λemission = 510 nm at λexcitation = 380 nm. As controls, C5a-desArg and C5a-desArg preincubated with PPAD were used.
Neutrophil Chemotaxis
Human neutrophils were isolated from healthy donors with Histopaque-1119/Percoll (15), washed twice, resuspended in PBS supplemented with 0.5% human serum albumin (Sigma-Aldrich) at 106 cells/ml, and labeled with 1 μm carboxyfluorescein succinimidyl ester (Fluka) for 15 min at room temperature with gentle agitation. After staining, cells were washed twice and resuspended in PBS with 0.5% human serum albumin and 4% heat-inactivated (30 min, 56 °C) hirudin-treated human plasma at a concentration of 5 × 106 cells/ml prior to use. The purity of neutrophil population (defined as CD14 low/CD15+/CD16+) was determined with a CyFlow Space (Partec) flow cytometry system using α-CD14-PE (IgG2a) (BD Biosciences) and α-CD15-FITC (IgM) and α-CD16-APC (IgG1) (ImmunoTools) antibodies and confirmed as 90–95%.
Migration of neutrophils was assessed using a disposable 96-well cell migration system with 3-μm polycarbonate membranes (ChemoTx; Neuro Probe). C5a was preincubated with serial dilutions of PPAD for 1 h at 37 °C. The samples were then supplemented with heat-inactivated human plasma at the same concentration as for neutrophil medium and applied to the wells of ChemoTx plate. C5a at a final concentration of 12.5 nm was used as a positive control, whereas 125 nm C5a-desArg, treated and untreated with PPAD, served as a negative control. Neutrophil suspension (50 μl) was then applied to each well of the upper filter, and the plate was incubated for 60 min 37 °C in humidified air with 5% CO2. The membrane filter was then removed, and the cell suspensions from the bottom wells were transferred to a flat-bottom, 98-well black plate (Nunc). The bottom wells of the ChemoTx plate were washed twice with 30 μl of PBS, and the washes were pooled with the corresponding cell suspensions from the bottom wells. Fluorescence intensity signals from cells were measured for 0.1 s using a Wallac Victor 2 1420 multilabel counter (PerkinElmer) using λexcitation = 485 nm and λemission = 535 nm.
Statistical Analysis
One-way analysis of variance with Dunnett's post test was used to estimate whether the observed differences between groups were statistically significant. Data were analyzed using GraphPad Prism 5.0.
RESULTS
PPAD Citrullinates C5a
The purity of PPAD was determined with SDS-PAGE followed by silver staining (Fig. 1A). To confirm citrullination of C5a by PPAD, native and PPAD-treated C5a were analyzed using HPLC. C5a in the native form eluted in two peaks from the C-18 column, whereas PPAD-treated C5a eluted in four peaks, at retention times different from those of the native peptide. This indicated citrullination of arginine residue(s) in C5a by PPAD (Fig. 1B). To determine which Arg residue(s) were deiminated, the native and PPAD-treated C5a were digested with trypsin or clostripain and subsequently analyzed by mass spectrometry on-line with RP-HPLC. Among tryptic peptides derived from PPAD-incubated C5a, one abundant peptide showed a clear shift in retention time as compared with the control C5a-derived peptides. Similarly, a single peptide with shifted retention time was also found in clostripain-digested, PPAD-treated C5a versus native C5a (data not shown). Subsequently, the peptides were identified by MS/MS as derived from the C terminus of C5a (DMQLGR and ANISHKDMQLGR). The citrullination of terminal Arg residue in these peptides was indicated by a 1-Da mass shift, peptide score (sequence information from MS), as well as the retention time shifts (Fig. 1C). In PPAD-treated C5a, only the citrullinated version of the C terminus was detected, indicating complete modification at the C terminus. Native C5a did not exhibit significant amounts of this modification. Interestingly, we confirmed endoarginine deiminase activity of PPAD by detecting modifications of positions 40 (AAR*ISLGPR) and 46 (ISLGPR*CIK) in the tryptic digest of PPAD-treated C5a. These, however, were much less abundant than the C-terminal modification.
FIGURE 1.
Citrullination of C5a by P. gingivalis PPAD. A, Laemmli SDS-PAGE followed by silver staining of PPAD purified from genetically modified P. gingivalis strain W83 culture medium. LMW, low molecular weight marker. B, C5a (10 μg), incubated in the reaction buffer alone or with 3.22 μm PPAD for 3 h at 37 °C, was analyzed with HPLC on a C18 column. C, samples (the same as for B digested with trypsin/clostripain) were analyzed with nLC-MS/MS. Results indicated significant citrullination of the C-terminal Arg in C5a treated with PPAD. D, C5 (10 μg), incubated with OMVs from P. gingivalis W83 wild-type strain and Δppad mutant strain at 25:1 molar ratio to Rgp for 30 min at 37 °C, was analyzed with nLC-MS/MS after trypsin digestion. Results demonstrate significant citrullination of the Arg-74 residue in C5a formed from C5 by Rgp and modified by PPAD present in OMVs from W83 strain. C5a was not citrullinated in the sample treated with OMVs from the Δppad mutant strain. mAU, milliabsorbance units.
OMVs Released from P. gingivalis Generate Citrullinated form of C5a from C5
Based on previous studies showing the release of active C5a from C5 by Arg-specific gingipains (16) and the presence of both Rgp and PPAD in OMVs from P. gingivalis (17, 18) we incubated intact C5 with OMVs from the wild-type P. gingivalis and the isogenic mutant strain lacking PPAD. Mass spectrometry analysis revealed full citrullination of the residue Arg-74 of C5a in C5 samples treated with wild-type strain OMVs. No such modification was present in the C5 sample treated with OMVs derived from Δppad. In this sample, only the native C5a C terminus was detected (Fig. 1D). Significantly, the gingipain activity in wild-type and PPAD-null vesicles was identical, and both degraded C5 to the same extant, releasing C5a. Nevertheless citrullination of generated C5a was observed only after treatment with OMVs from wild-type P. gingivalis. This clearly indicates that PPAD very efficiently modifies C5a released by gingipains.
Citrullinated C5a Has Decreased Chemotactic Activity
C-terminal arginine of C5a anaphylatoxin is crucial for biological activity of this peptide. We hypothesized that deimination of this residue by PPAD, resulting in generation of neutral citrulline, should suppress proinflammatory activity of C5a, in a manner similar to physiological removal of C-terminal arginine by carboxypeptidases. Indeed, preincubation of C5a with PPAD strongly reduced its chemotactic activity for neutrophils, in a concentration-dependent manner, whereas it had no effect on the activity of C5a-desArg (Fig. 2A). This result suggests that C5a-Cit, similar to C5a-desArg, has significantly lower affinity for C5aR on neutrophils. In keeping with this finding, treatment of C5a with PPAD also impaired its ability to induce calcium influx in a myeloid-derived cell line transfected with C5aR (Fig. 2, B and C). Interestingly, at higher concentrations and/or prolonged incubation with PPAD, the enzyme totally abrogated the capacity of C5a to activate neutrophils and U937 C5aR cells. In contrast, C5a-desArg treated with PPAD did not show altered potential to stimulate neutrophil chemotaxis and calcium release in U937-C5aR cells. As expected, native C5a-desArg had much lower activity than C5a in these assays.
FIGURE 2.
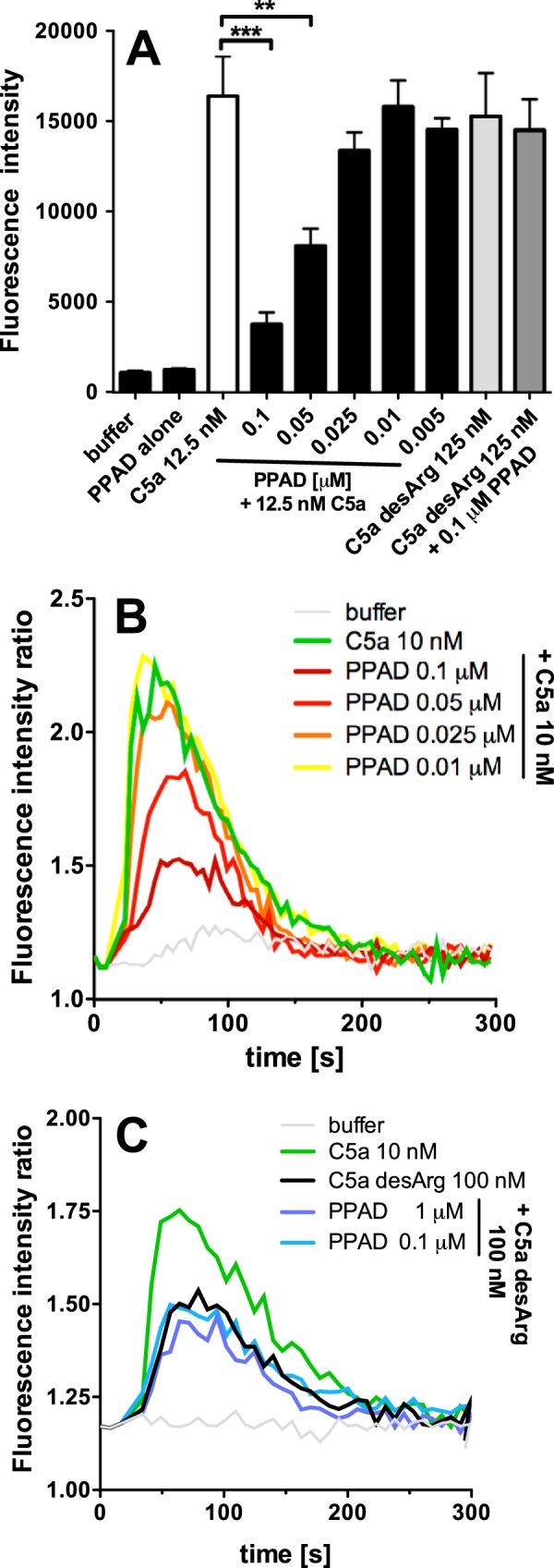
PPAD citrullinates C5a, decreasing its chemotactic ability against neutrophils and calcium influx into U937 C5aR cells. A, C5a (12.5 nm) was incubated with serial dilutions of PPAD for 1 h at 37 °C. Neutrophil migration was measured after 1 h of incubation as fluorescence intensity of carboxyfluorescein succinimidyl ester-labeled cells in the lower chamber of the transmigration assay plate. Buffer with KYT-1 and l-cysteine, PPAD alone, and C5a-desArg (125 nm) incubated with 0.1 μm PPAD or without were used as negative controls, and human C5a (12.5 nm) was used as the positive control. Results are the average of three independent experiments with S.D. Statistical significance was calculated using a one-way analysis of variance with Dunnett's post test. **, p < 0.01; ***, p < 0.001 B, C5a (10 nm) was incubated with increasing PPAD concentrations for 1 h at 37 °C. Changes in [Ca2+] were monitored in U937 C5aR cells loaded with Fura-2 AM in the presence of 1.3 mm calcium. FI ratio, fluorescence intensity ratio. C, C5a-desArg (100 nm) was incubated with PPAD or alone in buffer supplemented with KYT-1 (used as negative control). Depicted are the results of one representative experiment.
DISCUSSION
The complement system constitutes an essential part of innate immunity. Its activation, in a cascade-like manner, unleashes a spectrum of molecules aimed at destroying invading microbes. The membrane attack complex perforates cell membranes, anaphylatoxins C3a and C5a attract and activate neutrophils, opsonization with fragments of C3b facilitates phagocytosis and intracellular killing, and finally, activated components of complement enhance adaptive immune response. Therefore, it is not surprising that successful human pathogens developed a variety of mechanisms to inhibit complement activation and/or to neutralize activated components. Several of the latter strategies target C5a or C5aR. Proteolytic inactivation of C5a by Serratia marcescens 56K protease, protease ScpA of Streptococcus pyogenes, and ScpA-like enzymes of group B streptococci results in inhibition of C5a-mediated pro-inflammatory and chemotactic signaling that slows the influx of inflammatory cells and hinders removal of bacteria from the initial site of invasion (19). The importance of ScpA as a specialized virulence factor is underscored by the narrow specificity of this protease. The enzyme has no activity against intact C5 or other proteins but cleaves exclusively C5a at the C terminus, removing part of the region that interacts with C5aR on neutrophils (20).
In contrast to other bacterial pathogens for which suppression of the inflammatory response is the immune evasion strategy, the key periodontal pathogen P. gingivalis seems to thrive in the inflammatory milieu. Through proteolytic degradation of C3 and C5 (21) and hijacking complement regulator C4BP (22), the bacterium efficiently blocks assembly of membrane attack complex and opsonization. However, the cleavage of C5 by gingipains is associated with the release of fully functional C5a (16). The release of this potent chemoattractant and activator of neutrophils has limited effects on P. gingivalis, which is fairly resistant to killing by granulocytes and macrophages. It has been reported that in murine macrophages, C5a induces a crosstalk between C5aR-dependent signaling and activated Toll-like receptor-2 (TLR-2) resulting in P. gingivalis survival in vivo (23). The importance of C5a signaling for P. gingivalis survival was verified by the finding that specific blockade of C5aR enabled eradication of infection (24). This mechanism described for murine macrophages and the murine model of periodontitis is at odds with two countermeasures P. gingivalis takes to temper C5a biological activity. First, the bacterium proteolytically inactivates C5aR on human phagocytes (25). Second, as we report here, P. gingivalis can use PPAD to attenuate proinflammatory functions of C5a by deimination of the C-terminal Arg residue. This reaction is likely facilitated by close proximity of Rgps and PPAD on both the P. gingivalis cell surface as well as in OMVs released by this bacterium, in the same manner as citrullination of C-terminal Arg residues of fibrinogen-derived peptides generated during incubation of P. gingivalis with fibrinogen (7).
In comparison with native C5a, the citrullinated anaphylatoxin shows significantly reduced chemotactic activity for neutrophils and ability to activate Ca2+ influx in U937 cells expressing C5aR. This appears to be mediated mostly by complete C-terminal citrullination and not by modifications of intrapeptide arginines as C5a-desArg showed no further attenuation by PPAD treatment. It is likely that C5a-Cit and C5a-desArg, the physiological attenuated form of C5a generated by endogenous carboxypeptidase(s) (26), elicit similar biological effects by signaling through C5aR on immune cells and activating receptor-associated G-proteins. Taking into account the abundance of carboxypeptidases on immune cell surface (27) and the expression of PPAD by P. gingivalis (4), it is tempting to speculate that paralysis of bactericidal activity of murine macrophages is induced not by the native anaphylatoxin crosstalk but by generated in situ C-terminally modified C5a. Such a hypothetical scenario, which needs to be experimentally verified, reconciles PPAD as an important virulence factor contributing to P. gingivalis survival within the inflammatory site in vivo.
Modification of C5a by PPAD may generate a potential antigenic epitope due to the presence of C-terminal citrulline. Together with other C-terminally citrullinated peptides derived from bacterial and host proteins, including those derived from Rgps-degraded fibrinogen and enolase, C5a-Cit adds to the burden of post-translationally modified antigens. Occurring within a chronically inflamed periodontal tissue, these citrullinated epitopes may initiate breakdown of immune tolerance against host citrullinated proteins and the generation of autoantibodies in susceptible individuals, eventually leading to symptoms associated with rheumatoid arthritis (28). This contention is supported by the finding that collagen-induced arthritis was exacerbated in mice infected with wild-type P. gingivalis as manifested by earlier onset, accelerated progression, and enhanced severity of the disease, including significantly increased bone and cartilage destruction (12). The ability of P. gingivalis to augment arthritis was dependent on the expression of PPAD and associated with increased levels of citrullinated epitopes.
Taken together, in this study, we described a novel pathogenic strategy of bacterial pathogen to inactivate the antibacterial, proinflammatory activity of C5a by deimination of its C-terminal arginine. Such an approach is, thus far, unique for P. gingivalis, which is the sole bacterium able to express peptidyl arginine deiminase, with strong preference for C-terminal arginine. In this context, PPAD emerges as an important virulence factor, which warrants further investigation.
Acknowledgment
The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of structural funds from the European Union (POIG.02.01.00-12-064/08).
This work was supported by the Swedish Research Council (Grant K2012-66X-14928-09-5), the Swedish Government Funds for Clinical Research (ALF), the Torsten Söderberg Foundation, and the Österlund Foundation, Greta and Johan Kock Foundation, King Gustav V's 80-year Anniversary Fund, Knut and Alice Wallenberg Foundation, and Inga-Britt and Arne Lundberg Foundation.
- PPAD
- peptidylarginine deiminase
- C5aR
- C5a receptor
- Cit
- citrulline
- Rgp
- arginine-specific gingipain
- OMV
- outer membrane vesicle
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- nLC-MS/MS
- nano-LC-MS/MS.
REFERENCES
- 1. Brown L. J., Löe H. (1993) Prevalence, extent, severity and progression of periodontal disease. Periodontology 2000 2, 57–71 [DOI] [PubMed] [Google Scholar]
- 2. Lamont R. J., Jenkinson H. F. (1998) Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62, 1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Curtis M. A., Slaney J. M., Aduse-Opoku J. (2005) Critical pathways in microbial virulence. J. Clin. Periodontol. 32, Suppl. 6, 28–38 [DOI] [PubMed] [Google Scholar]
- 4. McGraw W. T., Potempa J., Farley D., Travis J. (1999) Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67, 3248–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loos T., Mortier A., Gouwy M., Ronsse I., Put W., Lenaerts J. P., Van Damme J., Proost P. (2008) Citrullination of CXCL10 and CXCL11 by peptidylarginine deiminase: a naturally occurring posttranslational modification of chemokines and new dimension of immunoregulation. Blood 112, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 6. Kilsgård O., Andersson P., Malmsten M., Nordin S. L., Linge H. M., Eliasson M., Sörenson E., Erjefält J. S., Bylund J., Olin A. I., Sørensen O. E., Egesten A. (2012) Peptidylarginine deiminases present in the airways during tobacco smoking and inflammation can citrullinate the host defense peptide LL-37, resulting in altered activities. Am. J. Respir. Cell Mol. Biol. 46, 240–248 [DOI] [PubMed] [Google Scholar]
- 7. Wegner N., Wait R., Sroka A., Eick S., Nguyen K. A., Lundberg K., Kinloch A., Culshaw S., Potempa J., Venables P. J. (2010) Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62, 2662–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pyrc K., Milewska A., Kantyka T., Sroka A., Maresz K., Koziel J., Nguyen K. A., Enghild J. J., Knudsen A. D., Potempa J. (2013) Inactivation of epidermal growth factor by Porphyromonas gingivalis as a potential mechanism for periodontal tissue damage. Infect. Immun. 81, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monk P. N., Scola A. M., Madala P., Fairlie D. P. (2007) Function, structure and therapeutic potential of complement C5a receptors. Br. J. Pharmacol. 152, 429–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou X. Y., Gao J. L., Hunter N., Potempa J., Nguyen K. A. (2013) Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol. Microbiol. 89, 903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao Y. F., Hsieh H. C., Liu G. Y., Hung H. C. (2005) A continuous spectrophotometric assay method for peptidylarginine deiminase type 4 activity. Anal. Biochem. 347, 176–181 [DOI] [PubMed] [Google Scholar]
- 12. Maresz K. J., Hellvard A., Sroka A., Adamowicz K., Bielecka E., Koziel J., Gawron K., Mizgalska D., Marcinska K. A., Benedyk M., Pyrc K., Quirke A. M., Jonsson R., Alzabin S., Venables P. J., Nguyen K. A., Mydel P., Potempa J. (2013) Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD). PLoS Pathog. 9, e1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potempa J., Pike R., Travis J. (1995) The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63, 1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kew R. R., Peng T., DiMartino S. J., Madhavan D., Weinman S. J., Cheng D., Prossnitz E. R. (1997) Undifferentiated U937 cells transfected with chemoattractant receptors: a model system to investigate chemotactic mechanisms and receptor structure/function relationships. J. Leukocyte Biol. 61, 329–337 [DOI] [PubMed] [Google Scholar]
- 15. Jusko M., Potempa J., Karim A. Y., Ksiazek M., Riesbeck K., Garred P., Eick S., Blom A. M. (2012) A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J. Immunol. 188, 2338–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wingrove J. A., DiScipio R. G., Chen Z., Potempa J., Travis J., Hugli T. E. (1992) Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 267, 18902–18907 [PubMed] [Google Scholar]
- 17. Nakao R., Takashiba S., Kosono S., Yoshida M., Watanabe H., Ohnishi M., Senpuku H. (2014) Effect of Porphyromonas gingivalis outer membrane vesicles on gingipain-mediated detachment of cultured oral epithelial cells and immune responses. Microbes Infect. 16, 6–16 [DOI] [PubMed] [Google Scholar]
- 18. Veith P. D., Chen Y. Y., Gorasia D. G., Chen D., Glew M. D., O'Brien-Simpson N. M., Cecil J. D., Holden J. A., Reynolds E. C. (2014) Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J. Proteome Res. 13, 2420–2432 [DOI] [PubMed] [Google Scholar]
- 19. Potempa M., Potempa J. (2012) Protease-dependent mechanisms of complement evasion by bacterial pathogens. Biol. Chem. 393, 873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cleary P. P., Prahbu U., Dale J. B., Wexler D. E., Handley J. (1992) Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 60, 5219–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Popadiak K., Potempa J., Riesbeck K., Blom A. M. (2007) Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J. Immunol. 178, 7242–7250 [DOI] [PubMed] [Google Scholar]
- 22. Potempa M., Potempa J., Okroj M., Popadiak K., Eick S., Nguyen K. A., Riesbeck K., Blom A. M. (2008) Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J. Immunol. 181, 5537–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang M., Krauss J. L., Domon H., Hosur K. B., Liang S., Magotti P., Triantafilou M., Triantafilou K., Lambris J. D., Hajishengallis G. (2010) Microbial hijacking of complement-Toll-like receptor crosstalk. Sci. Signal. 3, ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abe T., Hosur K. B., Hajishengallis E., Reis E. S., Ricklin D., Lambris J. D., Hajishengallis G. (2012) Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J. Immunol. 189, 5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jagels M. A., Travis J., Potempa J., Pike R., Hugli T. E. (1996) Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect. Immun. 64, 1984–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reis E. S., Chen H., Sfyroera G., Monk P. N., Köhl J., Ricklin D., Lambris J. D. (2012) C5a receptor-dependent cell activation by physiological concentrations of desarginated C5a: insights from a novel label-free cellular assay. J. Immunol. 189, 4797–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krause S. W., Rehli M., Andreesen R. (1998) Carboxypeptidase M as a marker of macrophage maturation. Immunol. Rev. 161, 119–127 [DOI] [PubMed] [Google Scholar]
- 28. Willemze A., Trouw L. A., Toes R. E., Huizinga T. W. (2012) The influence of ACPA status and characteristics on the course of RA. Nat. Rev. Rheumatol. 8, 144–152 [DOI] [PubMed] [Google Scholar]



