Background: The therapeutic potential of induced neural stem cells (iNSCs) that are directly converted from fibroblasts still remains elusive.
Results: Engrafted iNSCs enhance functional recovery after spinal cord injury in the rat model.
Conclusion: The directly converted iNSCs hold therapeutic potential.
Significance: This is the first report describing the therapeutic potential of iNSCs.
Keywords: Differentiation, Neural Stem Cell (NSC), Reprogramming, Stem Cells, Transplantation
Abstract
The spinal cord does not spontaneously regenerate, and treatment that ensures functional recovery after spinal cord injury (SCI) is still not available. Recently, fibroblasts have been directly converted into induced neural stem cells (iNSCs) by the forced expression defined transcription factors. Although directly converted iNSCs have been considered to be a cell source for clinical applications, their therapeutic potential has not yet been investigated. Here we show that iNSCs directly converted from mouse fibroblasts enhance the functional recovery of SCI animals. Engrafted iNSCs could differentiate into all neuronal lineages, including different subtypes of mature neurons. Furthermore, iNSC-derived neurons could form synapses with host neurons, thus enhancing the locomotor function recovery. A time course analysis of iNSC-treated SCI animals revealed that engrafted iNSCs effectively reduced the inflammatory response and apoptosis in the injured area. iNSC transplantation also promoted the active regeneration of the endogenous recipient environment in the absence of tumor formation. Therefore, our data suggest that directly converted iNSCs hold therapeutic potential for treatment of SCI and may thus represent a promising cell source for transplantation therapy in patients with SCI.
Introduction
The spinal cord composes millions of nerve cells including neurons and oligodendrocytes. They form multiple bundles to convey electronic signals from the brain to the rest of the body (1). Once the spinal cord is injured, these nerve cells located at the injured site are severely damaged and eventually dead. The dead cells are cleared away by the immune system, and in turn a cavity remains (1). Glia scar tissue forms around at the cavity area that prevents axonal regeneration, and as a consequence of this, no more electronic signals can be transmitted, resulting in the loss of motor, sensory, and autonomic functions (2).
More than 20 million individuals over the world suffer from paralysis caused by spinal cord injury (SCI)4 (3). The number of SCI patients is continuously increasing, and each year, an estimated 180,000 individuals over the world get a new injury (3). Unfortunately, no effective treatments that ensure functional recovery after SCI are available yet. Although there have been a few medications in the market, they are no longer widely used because of the limited efficacy and severe side effects. Furthermore, surgical treatments and rehabilitation may lighten minor symptoms but cannot substantially induce neurological and functional recovery. Thus, the cell-based therapy has been proposed as a promising treatment strategy for SCI. To develop the cell-based therapy, various donor cell types have been engrafted, such as Schwann cells, oligodendrocytes precursor cells (OPCs), and neural stem cells (NSCs), into injured sites of animal models (4). Although the transplantation of these cell types substantially promotes the functional recovery of SCI, there are several limitations that may impede its clinical translation, such as limited accessibility and postmitotic feature of primary cell types precluding the production of sufficient amounts of cells required for the transplantation.
Because of the limitations of primary cell types, induced pluripotent stem cells (iPSCs), which are equivalent to embryonic stem cells, have been considered as an alternative source for the transplantation study (5, 6). Although patient-specific iPSCs might be an unlimited source for various cell types in regenerative medicine, however, the iPSC-based clinical application continuously precipitates a concern regarding the tumor formation because of the residual undifferentiated iPSCs even after inducing differentiation (7). As an alternative, the ectopic expression of cell type-specific transcription factors has been reported to directly switch cell fates between somatic cells, circumventing the pluripotent state and eliminating the risk of tumorigenesis.
Previously, we and other groups generated self-renewable and functional induced NSCs (iNSCs) from easily accessible somatic fibroblasts using the direct conversion technology (7–9). Directly converted iNSCs closely resemble their in vivo counterpart in terms of the gene expression pattern, epigenetic status, and both in vitro and in vivo differentiation potential (8). Although iNSCs have been considered as a feasible, effective, and autologous source for clinical applications, its therapeutic ability has not yet been fully addressed. In the current study, we investigated the therapeutic potential of iNSCs in a rat SCI model. Engrafted iNSCs could differentiate into all neuronal lineages including different subtypes of neuron and restore axonal regeneration of SCI models, resulting in recovery of motor, sensory, and autonomic functions. Time course analysis of both engrafted donor cells and host environment further revealed that the functional recovery is mediated by the combined effects of neuroprotection, immunomodulation, cell replacement, and stimulation of the endogenous host environment. Taken together, our data suggest that directly converted iNSCs hold therapeutic potential that is comparable with control NSCs for the treatment of SCI.
EXPERIMENTAL PROCEDURES
Derivation of Fibroblasts and Control NSCs
Mouse embryonic fibroblasts (MEFs) were derived from C3H mouse strain embryos at embryonic day 13.5 after removing the head and all internal organs including the gonads and the spinal cord. MEFs were maintained in DMEM (Biowest) high glucose, 10% FBS (Biowest), 1× MEM nonessential amino acids, 1× l-glutamine with Pen/Strep, and 0.1 mm β-mercaptoethanol (all Invitrogen). For the derivation of control NSCs, brain tissue was collected from embryonic day 16.5 OG2/ROSA26 heterozygous female mice according to a previous protocol (10). Isolated NSCs were grown in standard NSC medium: DMEM/F-12 supplemented with N2 or B27 supplements (Invitrogen), 10 ng/ml EGF, 10 ng/ml bFGF (both from Invitrogen), and 1× l-glutamine with Pen/Strep (Invitrogen).
Generation of iNSCs
To generate iNSCs, 5 × 104 fibroblasts were transduced with replication-defective retroviral particles coding for Sox2, Klf4, c-Myc, and Brn4. After 48 h, the transduced fibroblasts were cultured in standard NSC medium: DMEM/F-12 supplemented with N2 or B27 supplements (Invitrogen), 10 ng/ml EGF, 10 ng/ml bFGF (both from Invitrogen), and 1× l-glutamine with Pen/Strep (Invitrogen). iNSC clusters were observed 4–5 weeks after transduction and expanded as previously described (8). iNSC clusters were observed 4–5 weeks after viral transduction. Finally, iNSC cell lines were established and cultured on gelatin-coated dishes.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (Sigma) for 20 min at room temperature, washed three times with PBS (Biowest), and then incubated in blocking solution (0.3% Triton X-100 (Sigma) plus 5% FBS (Biowest) in PBS) for 2 h at room temperature. The cells were incubated at 4 °C for 16 h with the following primary antibodies: anti-Nestin (Millipore, MAB353, 1:200), anti-Sox2 (Santa Cruz Biotechnology, sc-17320, 1:200), anti-SSEA1 (Santa Cruz Biotechnology, sc-21702, 1:100), anti-Olig2 (Millipore, AB9610, 1:200), anti-Tuj1 (Covance, MMS-435P, 1:500), and anti-GFAP (DAKO, Z0334, 1:500). The next day, cells were washed three times with PBS and twice with blocking solution. Appropriate secondary antibodies were applied to the cells for 2 h at room temperature. The cell nuclei were stained with Hoechst 33342 (Sigma).
Quantitative RT-PCR (qRT-PCR)
To analyze the relative abundance of mRNA transcripts of inflammation, apoptosis, neural factor, and neural regeneration marker genes, we isolated spinal cord tissues from each of three individual SCI rats at different time points, 5 days, 4 weeks, and 12 weeks after injecting vehicle, cNSCs, and iNSCs, respectively. Thus, we isolated spinal cord tissues from total 27 individual rats for gene expression analysis (9 rats for each vehicle, cNSCs, and iNSCs, respectively). RNA was isolated using an RNeasy mini kit (Qiagen), and a total of 1 μg of RNA was transcribed into cDNA with reverse transcriptase (Applied Biosystems) according to the manufacturer's instructions. The cDNAs of each samples were then amplified by qRT-PCR (7500 Real Time PCR system; Applied Biosystems) with specific primer pairs (Tables 1–3) using SYBR green PCR Master Mix (Applied Biosystems). The qRT-PCR was performed in triplicate. The housekeeping gene, m_Gapdh or r_B-actin, was used as an internal standard in experiments of each species.
TABLE 1.
Primers for RT-PCR
| Gene | Accession number | Sequences |
|---|---|---|
| Sox1 | NM_009233 | 5′-GTGCCCCTGACGCACATCTA-3′ |
| 5′-TCCACATAGTCATTGGCAGA-3′ | ||
| Pax6 | NM_001244198 | 5′-CAAGTTCCCGGGAGTGAACC-3′ |
| 5′-TCCACATAGTCATTGGCAGA-3′ | ||
| Olig2 | NM_016967 | 5′-ACCACCACGTGTCGGCTATG-3′ |
| 5′-TCCACATAGTCATTGGCAGA-3′ | ||
| Sox2 | NM_011443 | 5′-ACGGCCATTAACGGCACACT-3′ |
| 5′-TCCACATAGTCATTGGCAGA-3′ | ||
| Mash1 | NM_008553 | 5′-CAGAGGAACAAGAGCTGCTG-3′ |
| 5′-GATCTGCTGCCATCCTGCTT-3′ | ||
| Nestin | NM_016701 | 5′-TCCTGGTCCTCAGGGGAAGA-3′ |
| 5′-TCCACATAGTCATTGGCAGA-3′ | ||
| BLBP | NM_021272 | 5′-GGATGGCAAGATGGTCGTGA-3′ |
| 5′-TGGGACTCCAGGAAACCAAG-3′ | ||
| Glast | NM_148938 | 5′-GCTGAACACATGGCAACCAAA-3′ |
| 5′-GGCACCGGAGAGCGTTTATC-3′ | ||
| Colla1 | NM_007742 | 5′-CCCTGCCTGCTTCGTGTAAA-3′ |
| 5′-TCGTCTGTTTCCAGGGTTGG-3′ | ||
| Pdgfr | NM_001146268 | 5′-CAGGACCTCTGGCTGAAGCA-3′ |
| 5′-TCTGGGAGGCAGAAGGGAGAT-3′ | ||
| Csf1 | NM_007778 | 5′-CTGACCCAGGATGAGGACAGAC-3′ |
| 5′-AGTCCTGTGTGCCCAGCATAGA-3′ | ||
| Acta2 | NM_007392 | 5′-ATCGTCCACCGCAAATGCTT-3′ |
| 5′-AACTGGAGGCGCTGATCCAC-3′ | ||
| Thy1 | NM_009382 | 5′-CTTTCCCTCTCCCTCCTCCAAG-3′ |
| 5′-CGAGGGCTCCTGTTTCTCCTT-3′ | ||
| Actin | M12866.1 | 5′-ACTGCCGCATCCTCTTCCTC-3′ |
| 5′-CCGCTCGTTGCCAATAGTGA-3′ |
TABLE 2.
Quantitative PCR primers for mouse transcripts
| Gene | Accession number | Sequences |
|---|---|---|
| m_Gapdh | NM_008084 | 5′-CCAATGTGTCCGTCGTGGAT-3′ |
| 5′-TGCCTGCTTCACCACCTTCT-3′ | ||
| m_IL10 | NM_010548 | 5′-GCCGCTTCATCCCTGAAAAC-3′ |
| 5′-CCATTCCCAGAGGAATTGCAT-3′ | ||
| m_CD40 | M83312 | 5′-TGATCCCTGGGACTTCATGGT-3′ |
| 5′-ACATGTGCACACACGGGGTA-3′ | ||
| m_Gadd45a | NM_007836 | 5′-TGATGGCATCCGAATGGAAA-3′ |
| 5′-CAGTTTCTGTAATCCTTGCATCAGC-3′ | ||
| m_Casp3 | NM_009810 | 5′-GCAGTACAGCTTTTCCACTGA-3′ |
| 5′-AGGTCCAAAACAAGGGTTCC-3′ | ||
| m_Casp4 | NM_007609 | 5′-TCTACCTAGGAATATGGAAGCTGATGC-3′ |
| 5′-GGGTTGTAGAGTAGAAGGCAATGAAG-3′ | ||
| m_Bax | NM_007527 | 5′-GCGTCCACCAAGAAGCTGAG-3′ |
| 5′-GGAGTCCGTGTCCACGTCAG-3′ | ||
| m_Bcl2 | NM_009741 | 5′-CTCCAGCTGGCTCCCTTCAT-3′ |
| 5′-GAGAACCCCTGTCTCCAAAGGAT-3′ | ||
| m_Pycard | AF310104 | 5′-ACTTGGTGATGGACCTGGAG-3′ |
| 5′-ATGGCCTTTGGAAATGTAGC-3′ | ||
| m_Map2 | NM_001039934 | 5′-GACACTTGGGACCTGGACG-3′ |
| 5′-CAACCAACGCTCAAATGCCTT-3′ | ||
| m_Nestin | NM_016701 | 5′-GCAGGCGTTTAGACCTCTGGA-3′ |
| 5′-TCAGGGACTCCTGCCTTTCC-3′ | ||
| m_Pax6 | NM_001244198 | 5′-CAAGTTCCCGGGAGTGAACC-3′ |
| 5′-TTGGCAGAGTGAACACAATTTCC-3′ | ||
| m_HGF | NM_010427 | 5′-TGTCATTGTTCCTGGTCGTG-3′ |
| 5′-TCCATGGATGCTTCAAACAC-3′ | ||
| m_Chat | NM_009891 | 5′-CAAGCCACCAACAGCAAAGG-3′ |
| 5′-GATGGGAGCAGGGTTAGTAGGG-3′ | ||
| m_GDNF | NM_010275 | 5′-GTTTCGCATGGTTCCACACC-3′ |
| 5′-AAATGTGCTGGGCAGCTGAG-3′ | ||
| m_NT3 | NM_001164034 | 5′-ATCGACACCGCTTGCGTCT-3′ |
| 5′-TCCTCAACCTGAGCGGTTCC-3′ | ||
| m_NT4 | NM_198190 | 5′-GGCACTGGCTCTCAGAATGC-3′ |
| 5′-GACGCAAGCTGTGTCGATCC-3′ | ||
| m_NGF | NM_001112698 | 5′-AGGCTGCCTGGAGGTTCATC-3′ |
| 5′-TGCAGGCAAGTCAGCCTCTT-3′ | ||
| m_CD11b | NM_001082960 | 5′-AGCATGCGACAGTGAAAACCA-3′ |
| 5′-CCCTAAGGAGAGACCCCCAAC-3′ | ||
| m_CD18 | NM_008404 | 5′-TCCCAATGGAACAATGACAACC-3′ |
| 5′-TGCTCTAGCTTTCAGCAAACTTGG-3′ | ||
| m_Crp | AF092921 | 5′-GAACTGGCGGGCACTGAACT-3′ |
| 5′-GAGGTGCTTCAGGGCTCACA-3′ | ||
| m_Fas | NM_007987 | 5′-TTTGCTGTCAACCATGCCAAC-3′ |
| 5′-GAATCACTCCAACGGGCTGA-3′ | ||
| m_TGFb1 | NM_011577 | 5′-GGGCTACCATGCCAACTTCTG-3′ |
| 5′-CCCGGGTTGTGTTGGTTGTA-3′ |
TABLE 3.
Quantitative PCR primers for rat transcripts
| Gene | Accession number | Sequences |
|---|---|---|
| r_B-actin | NM_031144 | 5′-GCTCCTCCTGAGCGCAAGTA-3′ |
| 5′-GGCCGGACTCATCGTACTCC-3′ | ||
| r_Bax | NM_017059 | 5′-CACATGGCAGACAGTGACCATC-3′ |
| 5′-GGCCTCAGCCCATCTTCTTC-3′ | ||
| r_Bcl2 | L14680 | 5′-CCAGCATGCGACCTCTGTTT-3′ |
| 5′-CACTTGTGGCCCAGGTATGC-3′ | ||
| r_Map2 | NM_013066 | 5′-GCCGAGGTTGAGATGCCATT-3′ |
| 5′-CCCAACCCAGTGCTTCTGGT-3′ | ||
| r_GFAP | NM_017009.2 | 5′-GAAACCAGCCTGGACACCAA-3′ |
| 5′-TGTGCTCCTGCTTCGACTCC-3′ | ||
| r_Olig2 | NM_001100557 | 5′-TGCGCGATGCTAAGCTCTTT-3′ |
| 5′-TGTATGGGCCACGACACAGA-3′ | ||
| r_nestin | XM_003749314 | 5′-CAGAGCTTCCCAAGGTCTCCA-3′ |
| 5′-TCTGGCCCTCTGCTTCTTCA-3′ | ||
| r_Tnfrsf1a | NM_013091 | 5′-CTGGAGAACATCCGCGAGAC-3′ |
| 5′-TCGAGTCCCGTTCCTGAGGT-3′ | ||
| r_IL6 | NM_012589 | 5′-CCAACTCATCTTGAAAGCACTTGAA-3′ |
| 5′-TGACCACAGTGAGGAATGTCCA-3′ | ||
| r_Gadd45a | NM_024127 | 5′-AGGAGGAAGCTGTGTGGAAA-3′ |
| 5′-GAGGGCATGAAGACCAAAAA-3′ | ||
| r_Casp3 | NM_012922 | 5′-GCATTTCCCATAAGCCTCCT-3′ |
| 5′-TTGTCACATGGGAACACATTTT-3′ | ||
| r_Casp4 | NM_053736 | 5′-TTCCGGATACATGCCTGCTC-3′ |
| 5′-TCAAGGTTGCCCGATCAATG-3′ | ||
| r_Pax6 | NM_013001 | 5′-TCCAAGTTTGTATCATTCCTTTGCAT-3′ |
| 5′-TCTGTTAACAACCTTTGGAAAAACCA-3′ | ||
| r_HGF | NM_017017.2 | 5′-AGCCATAGAAGAGGCCAGTG-3′ |
| 5′-TACAACAGAAAACGCCCACA-3′ | ||
| r_Pycard | NM_172322 | 5′-ACCAGGCAGTTCGTGCAGAG-3′ |
| 5′-GCCTCAAGGCCTCAAGGAAC-3′ | ||
| r_Chat | NM_001170593 | 5′-GCTGGAGGGAACACCTCCTC-3′ |
| 5′-CGTGGCTCAGTGTTGCCTTT-3′ | ||
| r_GDNF | NM_001270630 | 5′-GGAACTCGCAATGCCGAACT-3′ |
| 5′-TGAATCGCCAGCCAATTCTC-3′ | ||
| r_NT3 | BC070504 | 5′-GCTGGCGCTGGATACGAATA-3′ |
| 5′-GGGACAGATGCCAATTCATGTT-3′ | ||
| r_NT4 | NM_013184 | 5′-ATCGACACCGCTTGCGTCT-3′ |
| 5′-TCCTCAACCTGAGCGGTTCC-3′ | ||
| r_Fas | NM_139194 | 5′-TGGCTTAGTGATTGCATCTCGTT-3′ |
| 5′-CGCAGGGTCTCTGTCCTCCT-3′ | ||
| r_Tnfrsf1b | NM_130426 | 5′-GCCGCCTAGGCAACAAAAAG-3′ |
| 5′-TCCTTAGCACGCACACACACA-3′ | ||
| r_IL1b | NM_031512 | 5′-CAGTGCACAGCCAGCACTGA-3′ |
| 5′-AATGATGTGCTTGTGCTTCATTCA-3′ | ||
| r_CDK1 | NM_019296 | 5′-TGGAAAAATGGCCCTTAAGCA-3′ |
| 5′-GCCAACAGTAAACGCCACGA-3′ | ||
| r_TNF1a | NM_012675 | 5′-ATTTGGTGACCAGGCTGTCG-3′ |
| 5′-CCAAGCGAACTTTATTTCTCTCAATG-3′ |
Whole Genome Expression Analysis
Total RNA was prepared from MEFs, cNSCs, and iNSCs using the RNeasy mini kit (Qiagen). In accordance with the Affymetrix protocol (GeneAtlasTM 3′ IVT express kit), 100 ng of total RNA was initially used to generate double-stranded cDNA using a T7-oligo(dT) primer. Amplified cRNA was purified, fragmented, and then hybridized to Affymetrix GeneChip arrays (Mouse Genome 430 PM array strip) for 16 h. The chips were then washed and stained using an Affymetrix fluidics station, and fluorescence was detected using the Affymetrix imaging station. Raw data were background-corrected and subsequently normalized using Partek Express Affymetrix Edition under the robust microarray analysis algorithm. Of the 45,141 total number of mouse genes, 1,881 genes for which expression levels were at least 4-fold different between MEFs and cNSCs were selected for analysis. Heat maps were generated using the MeV software. The original data have been uploaded to the Gene Expression Omnibus database (accession number GSE51331).
DNA Methylation Analysis
To determine the DNA methylation status, genomic DNA was treated with sodium bisulfite to convert all unmethylated cytosine residues into uracil residues using the EpiTect bisulfite kit (Qiagen) according to the manufacturer's protocol. All genomic regions selected were then amplified according to the method described in our previous studies (11, 12). Briefly, PCR amplifications were performed using SuperTaq polymerase (Ambion) in a total volume of 25 μl and a protocol of a total of 40 cycles of denaturation at 94 °C for 30 s, annealing at the appropriate temperature for each target region for 30 s, extension at 72 °C for 30 s with a first denaturation at 94 °C for 5 min, and a final extension at 72 °C for 10 min. Primer sequences and annealing temperatures used were as follows: Nestin 5′ enhancer first sense, 5′-TAAAGAGGTTGTTTGGTTTGGTAGT-3′; Nestin 5′ enhancer first antisense, 5′-CTATTCCACTCAACCTTCCTAAAA-3′ (45 °C); Nestin 5′ enhancer second sense, 5′-TAGTTTTTAGGGAGGAGATTAGAGG-3′; Nestin 5′ enhancer second antisense, 5′-CTCTTACCCCAAACACAACTAAAAC-3′ (55 °C); Col1a1 promoter first sense, 5′-GTTAGGTAGTTTTGATTGGTTGG-3′; Col1a1 promoter first antisense, 5′-ACAATAACCCCTAAAAAAAACAAAAA-3′ (55 °C); Col1a1 promoter second sense, 5′-TGGTATAAAAGGGGTTTAGGTTAGT-3′; and Col1a1 promoter second antisense, 5′-ACAATAACCCCTAAAAAAAACAAAAA-3′ (60 °C). For each primer set, 3 μl of product from the first round of PCR was used in the second round of PCR. The amplified products were verified by electrophoresis on 1% agarose gel. PCR products were subcloned using the PCR 2.1-TOPO vector (Invitrogen) according to the manufacturer's protocol. Reconstructed plasmids were purified using the QIAprep spin miniprep kit (Qiagen)m and individual clones were sequenced (Macrogen). Clones were analyzed using QUMA software.
In Vitro Differentiation
For differentiation into neurons, 5 × 104 iNSCs were plated on one poly-l-lysine/laminin-coated well from a 4-well dish in NSC medium. The next day, NSC medium was replaced with neural differentiation medium (DMEM/F-12 supplemented with 2% B27 (Invitrogen), 1× l-glutamine with Pen/Strep (Invitrogen), and 10 ng/ml of bFGF (Peprotech)). On day 4 of differentiation, the medium was replaced for 8–10 days with neural differentiation medium containing 200 mm of ascorbic acid (Sigma) but without bFGF. For differentiation into astrocytes, iNSCs were cultured in DMEM/F-12 supplemented with 10% FBS and 1× l-glutamine with Pen/Strep on one gelatin-coated well from a 4-well dish for 5 days. For differentiation into oligodendrocytes, 5 × 104 iNSCs were plated on one poly-l-lysine/laminin-coated well from a 4-well dish with NSC medium. The next day, NSC medium was replaced with oligodendrocyte differentiation medium (DMEM/F-12 supplemented with 2% B27, 1× l-glutamine with Pen/Strep, 10 ng/ml of bFGF, and 10 ng/ml of PDGF (Sigma)). On day 4 of differentiation, NSC medium was replaced with oligodendrocyte differentiation medium containing 30 ng/ml of T3 (Sigma) and 200 mm of ascorbic acid for another 4 days. In all the differentiation protocols, the differentiation medium was changed every other day.
Animal Models
Adult female Sprague-Dawley rats (12 weeks old, 230–250 g) were used for this experiment, and the Institutional Animal Care and Use Committee of Dankook University approved all animal care and surgical procedures (Approval No. 12-021). The animals were housed individually in a constant temperature (23–25 °C) and humidity (45–50%) in a specific pathogen-free room, and food and water was served without restriction. Surgical procedures have been previously described in detail (13). Briefly, rats were anesthetized by isoflurane (Forane; Choongwae Pharma) inhalation, and T8–9 spinal laminectomy was performed to expose the spinal cord at the T8–9 level. Using the MASCIS impactor (Rutgers, State University of New Jersey), a 10-g rod was dropped from 25-mm height onto the exposed T9 spinal cord and allowed to rest for 5 s. Intramuscular injection of 40 mg/kg cefotiam hydrochloride (FontiamTM; Hanmi Pharma) was performed in all operated rats during 3 days, and intraperitoneal injection of normal saline (3 ml) was performed just after surgery. The animals also received oral administration of 10 mg/kg acetaminophen syrup (TylenolTM; Janssen Pharmaceutica) for 3 days, and bladder expression was performed two times per day and continued until the amount of expressed urine was less than 0.5 ml/day. Sham-operated rats (n = 6), which received T8–9 laminectomy without spinal cord contusion, were used as controls for the urodynamic and bladder volume study.
Transplantation
At 9 days following contusion, iNSCs (1 × 106 cells in 5 μl of culture medium) were transplanted into the epicenter of the injury using a Hamilton syringe (Hamilton Company) after re-exposure of the contused spinal cord (n = 28). The transplantation of the same amount of cNSCs (n = 24) and the injection of the same volume of PBS (vehicle control) (n = 23) were used as the control. Cyclosporin A (Cipol InjTM; Chongkundang Pharmaceutical) was administered at 10 mg kg−1 day−1 subcutaneously beginning at 3 days before transplantation and continuing daily for 2 weeks after transplantation to all experimental and control groups. After that, the oral formula (Cipol SolnTM; Chongkundang Pharmaceutical) with the same quantity and concentration of cyclosporine A was administered daily until the animals were sacrificed for the analysis.
Histology
For histological analysis, all animals were deeply anesthetized and transcardially perfused with 150 ml of saline, followed by 500 ml of 4% paraformaldehyde in 0.12 m PBS (pH 7.3) via peristaltic pump. The contused thoracic spinal cord and lumbar spinal cord were removed, postfixed in 4% paraformaldehyde in PBS, and immersed for 3 days in 30% sucrose solution. The tissues were embedded in M1 compound (Thermo Scientific) and sectioned sagittally or axially (16 μm) on a cryostat. Hematoxylin and eosin staining was performed to detect the cavity size at 12 weeks post-transplantation. Sections were washed in 0.1 m PBS and immersed in Harris hematoxylin solution (Sigma) for 2 min followed by a brief wash in distilled water. Slides were then immersed briefly in 1% acid alcohol (1% HCl in 70% ethanol) and stained with eosin Y solution (BBC Biochemical) for 30 s. The slides were dehydrated with ethanol series, cleared with xylene, mounted in DPX (Sigma), and observed under microscopy (Nikon).
Luxol fast blue (LFB) staining was performed to detect myelination at 12 weeks post-transplantation. Sections were washed in 0.1 m PBS, passed through 95% ethanol, and stained in LFB (Sigma) staining solution (1% LFB in 95% ethanol with 0.5% acetic acid) overnight at 60 °C. The slides were rinsed in distilled water, differentiated in 0.005% lithium carbonate (Sigma), and then left in 70% ethanol until gray matter could be distinguished. The slides were dehydrated with ethanol series, cleared with xylene, mounted in DPX (Sigma), and observed under microscopy (Nikon).
For immunohistochemistry, the sections were incubated in 0.3% H2O2 for 30 min at room temperature to inactivate the endogenous peroxidase activity, permeabilized with 0.2% Triton X-100 in 2% BSA/PBS solution, and blocked with 10% normal serum. Primary antibodies (rabbit GAP43 polyclonal antibody, 1:1000, Novus; mouse NF-H monoclonal antibody, 1:200, Millipore; goat 5-HT polyclonal antibody, 1:100, Immunostar) diluted in 2% BSA/PBS solution were incubated overnight at 4 °C and washed three times in 0.1 m PBS. Biotinylated secondary antibodies (goat anti-rabbit IgG, 1:200, Vector Laboratories; goat anti-mouse IgG, 1:200, Vector Laboratories; horse anti-goat IgG, 1:200, Vector Laboratories) diluted in 2% BSA/PBS solution were incubated for 2 h at room temperature and washed three times. The signals were then enhanced with the Vectastain ABC kit (Vector Laboratories) for 30 min. The antigen-antibody complex was then visualized by exposing sections to 0.05% 3–3′-diaminobenzidine tetrahydrochloride (Sigma), 0.06% NiCl2 (Sigma), and 0.003% H2O2 in PBS. Sections were dehydrated with ethanol series, cleared with xylene, and mounted in DPX (Sigma).
Survival and migration of transplanted stem cells, differentiation into neuronal or glial cells, axonal outgrowth, and inflammatory responses following stem cell transplantation were all detected using immunohistochemistry. Previously mounted sections were treated with 0.2% Triton X-100 in 2% BSA/PBS solution for 5 min, washed, and blocked with 10% normal serum for 1 h. Next, slides were incubated with primary antibodies diluted in 2% BSA/PBS solution for a day at 4 °C and washed three times in 0.1 m PBS. The following primary antibodies were used for immunohistochemistry: anti-M2 (Developmental Studies Hybridoma Bank, 1:500), anti-GFAP (DAKO, Z0334, 1:1000), anti-O4 (R&D System, MAB1326, 1:1000), anti-Tuj1 (Santa Cruz Biotechnology, sc-58888, 1:500), anti-Tuj1 (Covance, PRB-435P, 1:1000), anti-NeuN (Millipore, MAB377, 1:100), anti-ChAT (Millipore, AB143, 1:100), anti-VGLUT1 (Millipore, AB5905, 1:2000), anti-VGLUT2 (Millipore, AB2251, 1:2000), anti-GAD65/67 (Millipore, AB1511, 1:2000), anti-synaptophysin (Millipore, MAB368, 1:100), anti-VEGF (Santa Cruz Biotechnology, sc-152, 1:200), anti-caspase-3 (Millipore, AB3623, 1:100), and anti-monocyte/macrophage CD68 (Millipore, MAB1435, 1:400). After washing, slides were incubated with secondary antibodies diluted in 2% BSA/PBS solution for 1 h at room temperature and then washed three times. Secondary antibodies included FITC-, rhodamine-, and Alexa 647-conjugated antibodies for monoclonal and polyclonal primary antibodies (Jackson ImmunoResearch, 1:200). Coverslips were applied to sections using Vectashield® (Vector Laboratories), and sections were observed by confocal microscopy (Carl Zeiss).
To quantify the M2-positive cells, three longitudinal sections of three different subjects from each group were selected at 12 weeks post-transplantation. The slides containing the highest amount of M2-positive cells near the lesion epicenter were selected. M2-positive cells were visualized and manually counted in all areas of the selected slides at ×200 magnification on a confocal microscope (Carl Zeiss). Double-stained NeuN-, GFAP-, O4-, GAD65/67-, VGLUT1/2-, and ChAT-positive cells were also counted from each double-stained slide. The percentage of each differentiated cell type (NeuN-, GFAP-, O4-, GAD65/67-, VGLUT1/2-, and ChAT-positive cells) was determined by dividing the number of differentiated cells by the number of M2-positive cells present in the same slide in each subject. ED1-positive cells were also counted following the same procedure used for the M2-positive cells. The observer was blinded to the experimental group. Cross-sections at the lesion epicenter, and at 3, 2.5, 2, 1.5, 1, and 0.5 mm rostral and caudal to the epicenter from three individual rats of each group were selected for analysis. The remaining spinal cord tissue in hematoxylin- and eosin-stained sections and the positive areas in LFB-stained sections were outlined manually under a light microscope (Nikon) at ×100 magnification and calculated using the National Institutes of Health ImageJ software. The GAP43-positive area at the lesion epicenter and 1 mm caudal to the epicenter from three individual rats of each group, the NF-H-positive area at the lesion epicenter and 3 mm rostral and caudal to the epicenter (n = 3 each), the 5HT-positive area at the lumbar intumescence region (n = 3 each), and the VEGF-positive area at the lesion epicenter (n = 3 each) were outlined manually in the cross-sections and calculated using the ImageJ software.
Locomotor Functional Assessments
We used two methods for the evaluation of the locomotor function of paralyzed hindlimbs after spinal cord injury, namely Basso, Beattie, and Bresnahan (BBB) scale and horizontal ladder test (n = 9 in vehicle control, n = 12 in cNSC-transplanted group, and n = 12 in iNSC-transplanted group). The BBB scale of no hindlimb movement is 0, and that of normal hindlimb movement is 21 (14). The rats were analyzed by two observers who were blinded to the treatment received by each rat and who were positioned across from each other to observe both sides of the rats during a 4-min walk in the open field (cylindrical-shaped acrylic box, 90-cm diameter, 15 cm high with a smooth floor). The horizontal ladder test was performed on a runway made of acryl walls (10 cm tall, 127 cm long, 8 cm wide between walls, 1 cm between rungs) (15). All rats were trained to walk from left to right on a runway several times for adaptation before testing, and the walk was captured with a digital camcorder. The ladder score was calculated as follows: ladder score = erroneous steps of hindlimb/total steps of hindlimb × 100 (%). The locomotor function of each group was examined every 7 days until sacrifice. All locomotor tests were recorded for at least 4 min with a digital camcorder for the coupling and ladder scores, and they were interpreted by two observers who were blinded to the identity of the rats.
Bladder Functional Assessments
The bladder volume measurement and the urodynamic study were performed as previously described (n = 8 in vehicle control, n = 3 in cNSC-transplanted group, and n = 5 in iNSC-transplanted groups) (16). For indirect evaluation of bladder volume, the horizontal and vertical diameters of the bladder were measured. The bladder volume was calculated as an imaginary ellipsoid as follows: bladder volume = (horizontal diameter of bladder/2)2 × vertical diameter of bladder/2 × π × 4/3.
For urodymanic study, a 4-Fr double lumen polyethylene catheter (CS-16402; Arrow) was used; one lumen was connected to a pressure transducer, and the other was connected to a normal saline-filled syringe and loaded into an infusion pump (KDS100; KD Scientific Inc.) with an initial rate of 10 ml h−1 and then 5 ml h−1 after the first void. Pressure signal was amplified and recorded using a data acquisition system (PowerLab 8/30 with LabChart Pro; AD Instruments). Measured parameters were the maximal micturition pressure (cmH2O) and frequency (time/min), the gross voiding patterns, and the regularity of the micturition pressure and frequency. The gross voiding patterns were described as normal, flaccid, or hyperreflexia according to the micturition frequency and the irregular micturition patterns as described previously (17).
Electrophysiological Assessment
Rats were anesthetized with isoflurane and placed over a warmed heating pad. Motor evoked potentials were recorded by stainless steel needle electrodes (AD Instruments) inserted into a gastrocnemius muscle (n = 3 in vehicle control, n = 6 in cNSC-transplanted group, and n = 7 in iNSC-transplanted groups).
The active electrode (AD Instruments) was inserted into the right lateral gastrocnemius muscle, and the reference and ground electrodes were placed on the right Achilles tendon and on the base of the tail, respectively. Magnetic stimulation was applied to the skull over the hindlimb motor cortex area using a 50-mm figure 8 double coil transducer (Magstim 2002, Magstim) with a single magnetic pulse and an 80% intensity of the determined motor threshold. The signals were amplified using Bio-amplifier (Bio Amp; AD Instruments) and analyzed using Labchart7 software (AD Instruments). A notch filter was used to remove the 60 Hz noise from the signals and a band pass filter was set from 1 to 5000 Hz. Data from five responses were averaged, and peak to peak amplitude and onset latency were used for analysis.
Statistics
All numeric data are reported as means ± S.E., and SPSS 18 (SPSS Inc.) was used for analysis. The Shapiro-Wilk test was performed to check normal distribution of all quantified histological and functional data from each group, and according to the result, parametric or nonparametric tests were chosen. For histological data, one-way analysis of variance with Scheffe's post hoc test was used to detect the differences among vehicle control, cNSC- and iNSC-transplanted groups, and the Mann-Whitney U test was performed to compare bladder volume and numeric parameters of urodynamic studies between sham-operated control and the three experimental groups. The Fisher's exact test was used to compare the pattern of regularity and the gross voiding pattern between the sham-operated control and the three experimental groups. The one-way repeated measures one-way analysis of variance was used to compare locomotor functions including the BBB and the ladder score tests among the three groups, and then the Kruskal-Wallis test with Bonferroni correction method was used at each time point. The Kruskal-Wallis test with Bonferroni correction method was used to compare electrophysiologic data among the three groups. The Spearman's correlation test was used to delineate the relationship between functional results with histological findings. Finally, linear regression analysis was performed with a correlated functional parameter as an independent factor after confirmation of a linear tendency by curve estimation. A value of p < 0.05 was considered significant.
RESULTS
Direct Reprogramming of Fibroblasts into iNSCs
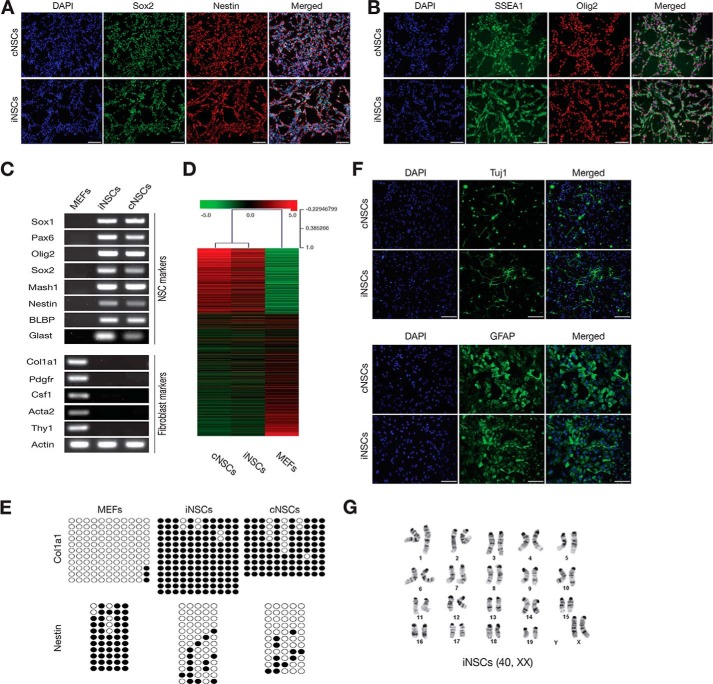
MEFs were directly converted into iNSCs using four transcription factors (Brn4, Sox2, Klf4, and c-Myc) as in our previous studies (7, 8, 18). After 4–5 weeks of transduction, we were able to observe the initial iNSC clusters and to establish stable iNSC line. Established iNSCs expressed Sox2, Nestin, Olig2, and SSEA1 at levels comparable with cNSCs, as shown by immunostaining (Fig. 1, A and B). Furthermore, iNSCs exhibited gene expression profiles similar to those of cNSCs as determined by both RT-PCR and microarray analysis (Fig. 1, C and D). We next investigated the DNA methylation status of both fibroblast marker and NSC marker to check whether iNSCs were epigenetically reprogrammed into a NSC-like state. The regulatory region of the fibroblast marker Col1a1, which was completely unmethylated in fibroblasts but highly methylated in cNSCs, became hypermethylated in iNSCs to a level similar to cNSCs. On the other hand, the second intron of Nestin (11), which was highly methylated in fibroblasts but less methylated in cNSCs, was demethylated in iNSCs as in cNSCs, indicating that iNSCs were reprogrammed into the NSC state at the epigenetic level (Fig. 1E). To examine the functionality of iNSCs prior to transplantation, we next confirmed the in vitro differentiation capacity of iNSCs. iNSCs could differentiate into both the neuronal and the glial lineages as we previously demonstrated (8) (Fig. 1F) and presented a normal karyotype (Fig. 1G).
FIGURE 1.
Direct reprogramming of fibroblasts into iNSCs. A and B, immunofluorescence microscopy images of cNSCs and iNSCs, using antibodies against Sox2/Nestin (A) and SSEA1/Olig2 (B). Scale bars, 100 μm. C, expression of NSC and fibroblast markers in established iNSCs, as shown by RT-PCR. Fibroblasts and cNSCs were used as the negative and positive controls, respectively. D, heat map representing the global gene expression profile of MEFs, iNSCs, and cNSCs. Genes that are more than 4-fold differentially expressed between MEFs and cNSCs are represented. The color bar at the top indicates gene expression in log2 scale. Red and green colors represent higher and lower gene expression levels, respectively. E, DNA methylation status on the second intron of Nestin and the promoter region of Col1a1 in fibroblasts, control NSCs, and iNSCs was assessed by bisulfite sequencing PCR. Open and filled circles represent unmethylated and methylated CpGs, respectively. F, iNSC differentiation potential into neurons and astrocytes as determined by immunostaining using antibodies against Tuj1 and GFAP, respectively. Scale bars, 100 μm. G, karyotyping of iNSCs at passage 7.
Engrafted iNSCs Could Differentiate into All Neuronal Lineages and Restore Axonal Regeneration of SCI Models
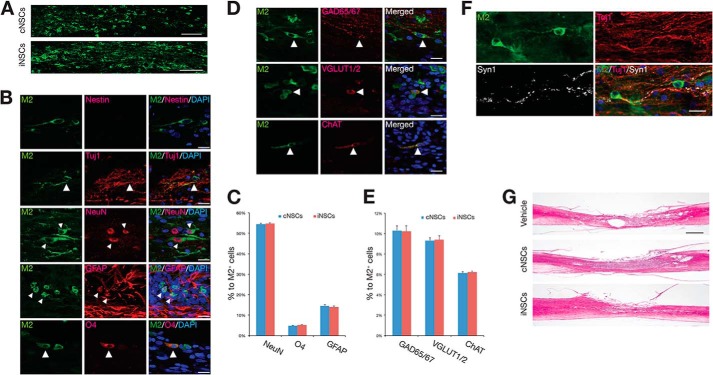
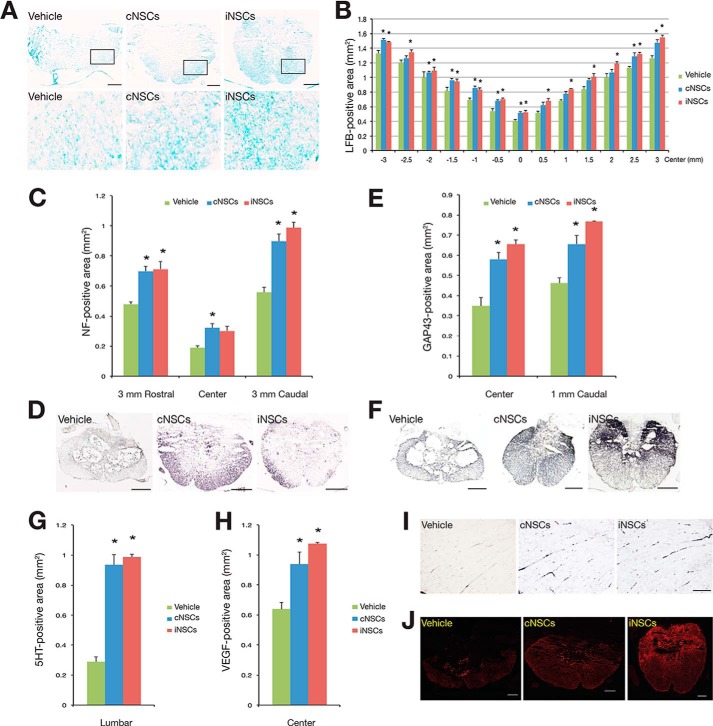
To assess the therapeutic potential of iNSCs, we transplanted 1 × 106 iNSCs into the contused thoracic spinal cord of Sprague-Dawley rats 9 days after injury. PBS and cNSCs were considered as the negative and the positive controls, respectively. The M2 antibody, which detects murine but not rat cells, was used to identify the grafted cNSCs and iNSCs and their differentiated progeny. At 12 weeks post-transplantation, grafted murine cNSCs and iNSCs were observed to have migrated into the injured cavity, with a range of 386.3 ± 14.1 and 383.0 ± 7.4 cells per slide, respectively (p > 0.05; Fig. 2A). Notably, no evidence of tumorigenesis was observed at 12 weeks after iNSC transplantation (data not shown) as in our previous study (18). In addition, the transplanted iNSCs did not express Nestin, indicating that they had lost their stem cell identity (Fig. 2B). Indeed, transplanted iNSCs had differentiated into NeuN-positive neurons (54.73 ± 0.42%), GFAP-positive astrocytes (14.06 ± 0.41%), and O4-positive oligodendrocytes (5.13 ± 0.13%) (Fig. 2, B and C). Similarly to cNSCs, most iNSCs-differentiated neurons were either GABAergic (10.22 ± 0.57%), glutamatergic (9.41 ± 0.38%), or cholinergic (6.25 ± 0.09%) (Fig. 2, D and E). Taken together, our data show that the differentiation efficiency of iNSCs into both glial cells and different subtypes of neurons is comparable with cNSCs (Fig. 2, C and E). Furthermore, we could also observe synaptic formation between host neurons and transplanted iNSC-derived neurons as shown by co-immunostaining for M2, Tuj1, and Synapsin1 (Fig. 2F). Hematoxylin and eosin staining indicated that the lesion and cavity size were significantly reduced in both cNSC- and iNSC-transplanted rats in comparison with the vehicle controls (Fig. 2G). In addition, the cNSC- and iNSC-transplanted animals exhibited an increased amount of myelin in the injured area than the vehicle-treated control, as shown by LFB staining (Fig. 3, A and B). To investigate whether engrafted iNSCs could restore axonal regrowth, we analyzed the expression of NF-H, GAP43, and 5HT. Immunohistochemical analysis revealed that the NF-H-positive area at 3 mm rostral and caudal to the epicenter (Fig. 3, C and D), the GAP43-positive area at the epicenter and 1 mm caudal to the epicenter (Fig. 3, E and F), and the 5HT-positive area at the lumbar intumescence region (Fig. 3, G and I) were significantly larger in the cNSC- and iNSC-transplanted groups than those in the vehicle control, indicating that the engrafted iNSCs promote axonal regeneration in SCI rats as control cNSCs. Finally, we investigated the effects of iNSC transplantation on angiogenesis in the injured area. As shown by immunostaining, the cNSC- and iNSC-transplanted groups exhibited an increased VEGF-positive area at the lesion epicenter than the vehicle controls (Fig. 3, H and J), demonstrating that transplantation of iNSCs enhances angiogenesis in the engrafted spinal cord.
FIGURE 2.
Histological analysis of engrafted iNSCs at 12 weeks after transplantation. A, representative images of M2-positive engrafted cNSCs or iNSCs adjacent to the transplantation site near the injured cavity at 12 weeks post-transplantation. Scale bars, 100 μm. B and C, iNSCs could differentiate into both the neuronal and the glial lineages in vivo as determined by co-staining using antibodies against M2 plus Tuj1 or NueN for neurons, GFAP for astrocytes, and O4 for oligodendrocytes, respectively. The absence of Nestin-positive cells suggests that iNSCs did not retain their stem cell identity. Error bars indicate standard error of triplicate values. Scale bars, 100 μm. D and E, iNSCs could differentiate into different subtypes of neurons as shown by immunostaining using antibodies against M2 plus GAD65/67 for GABAergic neurons, VGLUT1/2 for glutamatergic neurons, and ChAT for cholinergic neurons, respectively. Error bars indicate standard error of triplicate values. Scale bars, 100 μm. F, co-staining for M2, Tuj1, and Synapsin1 suggests synaptic formation between host neurons and iNSC-derived neurons. Scale bars, 100 μm. G, hematoxylin- and eosin-stained images of sagittal sections 3 mm around the lesion epicenter in cNSC- and iNSC-transplanted rats and vehicle control. Scale bars, 1000 μm.
FIGURE 3.
Engrafted iNSCs could restore axonal regeneration of SCI models. A, the myelinated area around the lesion epicenter was assessed by LFB staining in cNSC- and iNSC-transplanted groups, as well as in the vehicle control. Scale bars, 200 μm. B, quantitative analysis of LFB positive area in the spinal cords of cNSC- and iNSC-transplanted and vehicle rats. Error bars indicate standard error of triplicate values. *, p < 0.05. C–J, quantitative analysis of the area positive for NF (C and D), GAP43 (E and F), 5HT (G and I), and VEGF (H and J) in the spinal cords of cNSC- and iNSC-transplanted and vehicle rats. Error bars indicate standard error of triplicate values. *, p < 0.05. The NF-H-positive area at 3 mm rostral and caudal to the epicenter (D), GAP43-positive area at the epicenter and 1 mm caudal to the epicenter (F), and 5HT-positive area at lumbar intumescence (I) were assessed by immunocytochemistry in cNSC- and iNSC-transplanted groups as well as in the vehicle control. Scale bars, 100 μm. J, the VEGF-positive area was assessed by immunocytochemistry using an antibody against VEGF in cNSC- and iNSC-transplanted groups as well as in the vehicle control. Scale bars, 100 μm.
Engrafted iNSCs Promote Recovery of Motor Function and Electrophysiological Activity of SCI Models
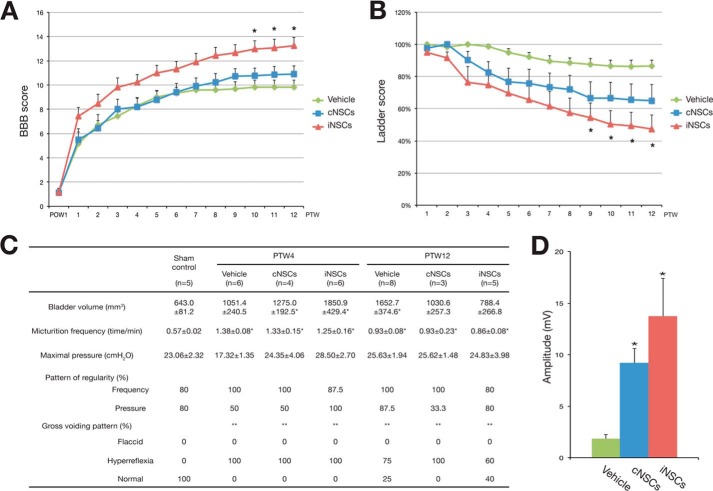
Next, we evaluated the motor functional recovery using the BBB and the ladder tests (for explanation see methods). iNSC-transplanted groups exhibited consistently higher BBB scores (Fig. 4A) and reduced ladder scores (Fig. 4B) than those of the vehicle control at 9 and 10 weeks post-transplantation, respectively. These results indicate that iNSC transplantation promotes locomotor recovery in rats with SCI. Although it appears that the therapeutic effects of iNSCs are slightly stronger than those of cNSCs, there was no statistical difference between cNSCs and iNSCs. Furthermore, we investigated the recovery of autonomic functions by measuring the bladder volume and by performing urodynamic studies at 4 and 12 weeks post-transplantation. In comparison with the sham control, the bladder volume returned to normal levels in the cNSC- and iNSC-transplanted groups at 12 weeks post-transplantation. In addition, a normal voiding pattern was restored in 40% of the iNSC-transplanted animals (Fig. 4C), although it was not statistically significant. Nevertheless these data suggest that transplantation of iNSCs could partially restore autonomic functions in rats with SCI. Finally, we measured the motor evoked potential at 12 weeks post-transplantation to determine the electrophysiological recovery. The peak to peak amplitude of the cNSC- and iNSC-transplanted groups was significantly higher (9.19 ± 1.40 and 13.74 ± 3.70 mV, respectively) compared with the vehicle control (1.83 ± 0.28 mV) (Fig. 4D). Taken together, these data indicate that transplanted iNSCs could promote recovery of motor function and electrophysiological activity.
FIGURE 4.
Transplanted iNSCs promoted motor functional and electrophysiological recovery. A and B, BBB score (A) and ladder score (B) were assessed weekly for 12 weeks. C, bladder volume and urodynamic test results at 4 and 12 weeks post-transplantation in transplanted and vehicle control rats. Error bars indicate standard error of triplicate values. *, p < 0.05. PTW, post-transplantation week. D, peak to peak amplitude of motor evoked potentials at 12 weeks post-transplantation in transplanted and vehicle control rats. Error bars indicate standard error of triplicate values. *, p < 0.05.
The Functional Recovery Is Mediated by the Combined Effects of Donor Cells and Host Environment
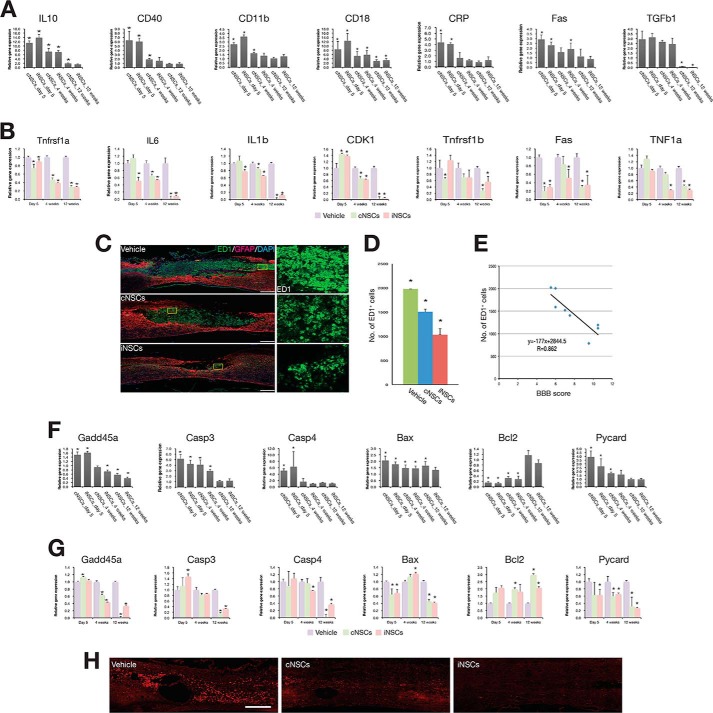
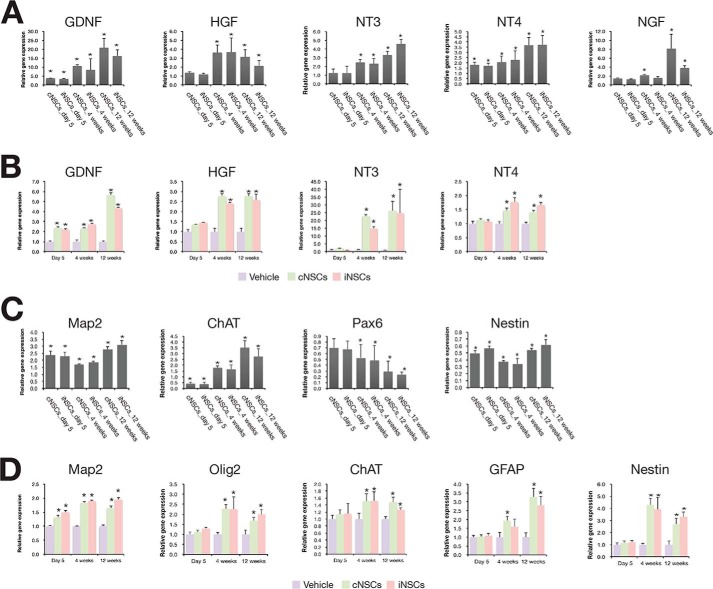
Engrafted cNSCs have been reported to promote functional recovery through the combinatorial effect of neural protection, de novo angiogenesis induction, and axonal regeneration (5, 6, 19, 20). Thus, we decided to decipher the mechanism by which engrafted iNSCs improved the locomotor function in the rat SCI model. To this end, we performed a gene expression time course analysis on the transplanted area at 5 days and at 4 and 12 weeks post-transplantation. We isolated spinal cord tissues from each of three individual SCI rats at different time points, 5 days, 4 weeks, and 12 weeks after injecting vehicle, cNSCs, and iNSCs, respectively. Thus, we isolated spinal cord tissues from total 36 individual rats for gene expression analysis (each 12 rats for vehicle, cNSCs, and iNSCs, respectively). As shown by qRT-PCR, engrafted iNSCs displayed immediate or gradual decrease of inflammation-associated genes (19) after transplantation, suggesting that transplanted iNSCs have an immunomodulative effect by which the acute inflammatory response could be ameliorated (Fig. 5A). Indeed, the number of ED1-positive monocytes and macrophages at the epicenter lesion was significantly lower in cNSC- and iNSC-transplanted groups (1506.3 ± 54.3 and 1033.7 ± 124.2 cells, respectively) compared with the vehicle control (1979.7 ± 34.1 cells) at 4 weeks post-transplantation (Fig. 5, C and D). Furthermore, the number of ED1-positive monocytes appeared to be negatively correlated with the BBB score at 4 weeks post-transplantation (r = 0.862; Fig. 5E), suggesting that inflammation impaired locomotor recovery. In addition, transplanted cNSCs and iNSCs showed a gradual decrease in the expression of apoptosis-related genes (19, 21, 22). As an exception, the expression of the anti-apoptotic gene Bcl2 increased in a time-dependent manner (Fig. 5F). Similarly, the number of caspase-3-positive cells was significantly decreased in both cNSC- and iNSC-engrafted rats compared with vehicle control (Fig. 5H). The host cells from the transplanted area also exhibited similar gene expression patterns of both apoptosis- and inflammation-related genes (Fig. 5, B and G). However, there was no dramatic change in the expression levels of those markers in the vehicle control. In addition, the expression levels of neurotropic factors (19) were increased in the engrafted iNSCs compared with the nontransplanted cultured iNSCs (Fig. 6A). Interestingly, the host cells also showed a gradual increase in the expression levels of neurotropic factors compared with that of the vehicle control (Fig. 6B), suggesting that the transplantation of iNSCs could also induce the secretion of neurohumoral factors from the host environment. Finally, we investigated the expression levels of neuronal cell markers (19, 23–25) in the injured area. cNSCs and iNSCs had differentiated into both neuronal and glial cell types in the injured area (Figs. 2, B and C, and 6C); thus, the expression levels of the NSC markers Pax6 and Nestin were found to have decreased in cNSCs and iNSCs after transplantation (Fig. 6C). Interestingly, the expression levels of the differentiation markers were found to have gradually increased in host cells until 12 weeks post-transplantation (Fig. 6D). Taken together, engrafted iNSCs not only exert their therapeutic roles through neuroprotection, immunomodulation, and cell replacement but also promote the active regeneration of the endogenous host environment as evidenced by decreased levels of both apoptosis and inflammation markers, as well as increased levels of both neurotropic factors and neuronal markers.
FIGURE 5.
Time course gene expression analysis of engrafted cNSCs, iNSCs, and host environment. A and B, expression pattern of inflammatory markers in the transplanted cells (A) and in the host environment (B). Expression levels of each marker gene were analyzed at 5 days and at 4 and 12 weeks post-transplantation by qRT-PCR. The expression levels of each marker were normalized to those of cultured cNSCs or iNSCs (A) and those of vehicle control (B). The error bars indicate standard deviation of triplicate values. *, p < 0.05. C, immunofluorescence microscopy images of the spinal cord from transplanted and vehicle control rats using antibodies against ED1 and GFAP. Scale bars, 500 μm. D, number of ED1-positive cells in transplanted and vehicle rats. Error bars indicate standard deviation of triplicate values. *, p < 0.05. E, correlation between the number of ED1-positive cells and the BBB score. F and G, expression pattern of apoptosis markers in the transplanted cells (F) and in the host environment (G). Expression levels of each marker gene were analyzed at 5 days and at 4 and 12 weeks post-transplantation by qRT-PCR. The expression levels of each marker were normalized to those of cultured cNSCs or iNSCs (F) and those of vehicle control (G). Error bars indicate standard deviation of triplicate values. *, p < 0.05. H, expression of caspase-3 in iNSC-transplanted rats with SCI. The expression of caspase-3 was assessed by immunohistochemistry using antibody against caspase-3 after 12 weeks of transplantation. Scale bars, 500 μm.
FIGURE 6.
Time course gene expression analysis of neurotropic and neuronal markers. A and B, expression pattern of neurotropic factors in the transplanted cells (A) and in the host environment (B). C and D, expression pattern of neuronal markers in the transplanted cells (C) and in the host environment (D). Expression levels of each marker gene were analyzed at 5 days and at 4 and 12 weeks post-transplantation by qRT-PCR. The expression levels of each marker were normalized to those of cultured cNSCs or iNSCs (A and C) and those of vehicle control (B and D). Error bars indicate standard deviation of triplicate values. *, p < 0.05.
DISCUSSION
In the current study, we transplanted iNSCs into the rat SCI model to investigate their therapeutic potential. Multitude immunohistochemical and histological analyses indicated that transplanted iNSCs were survived, migrated, and differentiated into neuronal and glia cells in the spinal cord. Their derivatives could restore damaged tissues and reduce the size of lesion cavity, resulting in the local circuitry reconstitution. Particularly, neurons differentiated from iNSCs extend their axons and form synapses between iNSC-derived neurons and host rat neurons that enable axonal regeneration. Thus, transplantation of iNSCs has a profound effect on axonal regeneration and local circuitry reconstitution that facilitates the locomotor function recovery. Furthermore, like cNSCs, engrafted iNSCs could also exhibit other beneficial effects such as immunomodulation, neural protection, and angiogenesis in the spinal cord, indicating that iNSCs are functionally comparable with cNSCs. Thus, iNSCs could be a promising cell source for the treatment of SCI.
Because the spinal cord injury has a limited regenerative capacity to restore lost myelin and axonal connections that cause the loss of motor, sensory, and autonomic functions (26), the cell-based therapy has been proposed to replace lost nerve cells and to remyelinate damaged spinal tracts (27). For this, several types of primary adult stem cells (or progenitors), such as mesenchymal stem cells, NSCs, and OPCs, have been transplanted into the spinal cord injury model (4). Their therapeutic potentials have been thoroughly proved using a number of criteria. However, the accessibility of these cell types is largely limited in vitro. Furthermore, their immune rejection issue upon transplantation cannot be excluded, because the primary cell type-mediated cell therapy is mostly allogenic but not autologous transplantation. In contrast, iNSCs are highly accessible and can eliminate the immune rejection issue, because they are derived from the readily accessible patient fibroblasts.
Unlike control rats, we observed a significant improvement on the forelimb-hindlimb coordination in iNSC-transplanted rats. The significant improvement appeared from 10 weeks post-transplantation. Interestingly, diminishing inflammatory events and apoptosis appeared at 4 weeks post-transplantation, implicating that the restoration of host environments occurs much earlier than the motor function recovery. If we could extensively control the host environments with a number of medications related to anti-inflammation, such as curcumin and nordihydroguaiaretic acid, at the early stage of transplantation, the significant functional recovery may bring forward to earlier than 10 weeks. Consequently, this may lead to almost complete motor function recovery.
Engrafted iNSCs were differentiated into neuronal and glia cells without forming tumors. Previous studies agreed that iNSCs were readily differentiated into neurons but rarely into oligodendrocytes in vitro (7, 8). In the line with this, we also observed a limited differentiation of iNSCs into oligodendrocytes in the spinal cords. This deficit of oligodendrocytes may associate with partial recovery of motor function (BBB score, >14). Recent studies have reported the direct reprogramming of somatic fibroblasts into induced OPCs (28, 29). Because induced OPCs could efficiently generate myelinogenic oligodendrocytes, a combinational transplantation of iNSCs and induced OPCs may compromise this issue.
This work was supported by Korea Health Technology R&D Project, Ministry of Health & Welfare Grant A120392 and Priority Research Center Program Grant 2009-0093829) of the National Research Foundation of Korea (Ministry of Education, Science and Technology).
- SCI
- spinal cord injury
- OPC
- oligodendrocytes precursor cell
- NSC
- neural stem cell
- iNSC
- induced NSC
- iPSC
- induced pluripotent stem cell
- MEF
- mouse embryonic fibroblast
- cNSC
- control NSC
- Pen/Strep
- penicillin and streptomycin
- bFGF
- basic fibroblast growth factor
- qRT-PCR
- quantitative RT-PCR
- BBB
- Basso, Beattie, and Bresnahan
- LFB
- Luxol fast blue
- GFAP
- glial fibrillary acidic protein.
REFERENCES
- 1. Willyard C. (2013) Stem cells: A time to heal. Nature 503, S4–S6 [DOI] [PubMed] [Google Scholar]
- 2. Lu P., Kadoya K., Tuszynski M. H. (2014) Axonal growth and connectivity from neural stem cell grafts in models of spinal cord injury. Curr. Opin. Neurobiol. 27, 103–109 [DOI] [PubMed] [Google Scholar]
- 3. Lee B. B., Cripps R. A., Fitzharris M., Wing P. C. (2014) The global map for traumatic spinal cord injury epidemiology: update 2011, global incidence rate. Spinal Cord 52, 110–116 [DOI] [PubMed] [Google Scholar]
- 4. Silva N. A., Sousa N., Reis R. L., Salgado A. J. (2014) From basics to clinical: a comprehensive review on spinal cord injury. Prog. Neurobiol. 114, 25–57 [DOI] [PubMed] [Google Scholar]
- 5. Nori S., Okada Y., Yasuda A., Tsuji O., Takahashi Y., Kobayashi Y., Fujiyoshi K., Koike M., Uchiyama Y., Ikeda E., Toyama Y., Yamanaka S., Nakamura M., Okano H. (2011) Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc. Natl. Acad. Sci. U.S.A. 108, 16825–16830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsuji O., Miura K., Okada Y., Fujiyoshi K., Mukaino M., Nagoshi N., Kitamura K., Kumagai G., Nishino M., Tomisato S., Higashi H., Nagai T., Katoh H., Kohda K., Matsuzaki Y., Yuzaki M., Ikeda E., Toyama Y., Nakamura M., Yamanaka S., Okano H. (2010) Therapeutic potential of appropriately evaluated safe-induced pluripotent stem cells for spinal cord injury. Proc. Natl. Acad. Sci. U.S.A. 107, 12704–12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim S. M., Flasskamp H., Hermann A., Araúzo-Bravo M. J., Lee S. C., Lee S. H., Seo E. H., Lee S. H., Storch A., Lee H. T., Schöler H. R., Tapia N., Han D. W. (2014) Direct conversion of mouse fibroblasts into induced neural stem cells. Nat. Protoc. 9, 871–881 [DOI] [PubMed] [Google Scholar]
- 8. Han D. W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M. J., Zaehres H., Wu G., Frank S., Moritz S., Greber B., Yang J. H., Lee H. T., Schwamborn J. C., Storch A., Schöler H. R. (2012) Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 [DOI] [PubMed] [Google Scholar]
- 9. Thier M., Wörsdörfer P., Lakes Y. B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M. M., Brüstle O., Edenhofer F. (2012) Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 [DOI] [PubMed] [Google Scholar]
- 10. Conti L., Pollard S. M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q. L., Cattaneo E., Smith A. (2005) Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han D. W., Do J. T., Araúzo-Bravo M. J., Lee S. H., Meissner A., Lee H. T., Jaenisch R., Schöler H. R. (2009) Epigenetic hierarchy governing Nestin expression. Stem Cells 27, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 12. Han D. W., Do J. T., Gentile L., Stehling M., Lee H. T., Schöler H. R. (2008) Pluripotential reprogramming of the somatic genome in hybrid cells occurs with the first cell cycle. Stem Cells 26, 445–454 [DOI] [PubMed] [Google Scholar]
- 13. Park W. B., Kim S. Y., Lee S. H., Kim H. W., Park J. S., Hyun J. K. (2010) The effect of mesenchymal stem cell transplantation on the recovery of bladder and hindlimb function after spinal cord contusion in rats. BMC Neurosci. 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basso D. M., Beattie M. S., Bresnahan J. C. (1996) Graded histological and locomotor outcomes after spinal cord contusion using the NYU weight-drop device versus transection. Exp. Neurol. 139, 244–256 [DOI] [PubMed] [Google Scholar]
- 15. Fouad K., Klusman I., Schwab M. E. (2004) Regenerating corticospinal fibers in the marmoset (Callitrix jacchus) after spinal cord lesion and treatment with the anti-Nogo-A antibody IN-1. Eur. J. Neurosci. 20, 2479–2482 [DOI] [PubMed] [Google Scholar]
- 16. Joo N. Y., Knowles J. C., Lee G. S., Kim J. W., Kim H. W., Son Y. J., Hyun J. K. (2012) Effects of phosphate glass fiber-collagen scaffolds on functional recovery of completely transected rat spinal cords. Acta Biomater. 8, 1802–1812 [DOI] [PubMed] [Google Scholar]
- 17. Hyun J. K., Lee Y. I., Son Y. J., Park J. S. (2009) Serial changes in bladder, locomotion, and levels of neurotrophic factors in rats with spinal cord contusion. J. Neurotrauma 26, 1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marthaler A. G., Tiemann U., Araúzo-Bravo M. J., Wu G., Zaehres H., Hyun J. K., Han D. W., Schöler H. R., Tapia N. (2013) Reprogramming to pluripotency through a somatic stem cell intermediate. PLoS One 8, e85138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumamaru H., Ohkawa Y., Saiwai H., Yamada H., Kubota K., Kobayakawa K., Akashi K., Okano H., Iwamoto Y., Okada S. (2012) Direct isolation and RNA-seq reveal environment-dependent properties of engrafted neural stem/progenitor cells. Nat. Commun. 3, 1140. [DOI] [PubMed] [Google Scholar]
- 20. Lu P., Wang Y., Graham L., McHale K., Gao M., Wu D., Brock J., Blesch A., Rosenzweig E. S., Havton L. A., Zheng B., Conner J. M., Marsala M., Tuszynski M. H. (2012) Long-distance growth and connectivity of neural stem cells after severe spinal cord injury. Cell 150, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Giovanni S., Knoblach S. M., Brandoli C., Aden S. A., Hoffman E. P., Faden A. I. (2003) Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann. Neurol. 53, 454–468 [DOI] [PubMed] [Google Scholar]
- 22. Siebert J. R., Middleton F. A., Stelzner D. J. (2010) Long descending cervical propriospinal neurons differ from thoracic propriospinal neurons in response to low thoracic spinal injury. BMC Neurosci. 11, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abematsu M., Tsujimura K., Yamano M., Saito M., Kohno K., Kohyama J., Namihira M., Komiya S., Nakashima K. (2010) Neurons derived from transplanted neural stem cells restore disrupted neuronal circuitry in a mouse model of spinal cord injury. J. Clin. Invest. 120, 3255–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ben-Hur T., Idelson M., Khaner H., Pera M., Reinhartz E., Itzik A., Reubinoff B. E. (2004) Transplantation of human embryonic stem cell-derived neural progenitors improves behavioral deficit in Parkinsonian rats. Stem Cells 22, 1246–1255 [DOI] [PubMed] [Google Scholar]
- 25. Lu P., Jones L. L., Snyder E. Y., Tuszynski M. H. (2003) Neural stem cells constitutively secrete neurotrophic factors and promote extensive host axonal growth after spinal cord injury. Exp. Neurol. 181, 115–129 [DOI] [PubMed] [Google Scholar]
- 26. Bradbury E. J., McMahon S. B. (2006) Spinal cord repair strategies: why do they work? Nat. Rev. Neurosci. 7, 644–653 [DOI] [PubMed] [Google Scholar]
- 27. Goldman S. (2005) Stem and progenitor cell-based therapy of the human central nervous system. Nat. Biotechnol. 23, 862–871 [DOI] [PubMed] [Google Scholar]
- 28. Najm F. J., Lager A. M., Zaremba A., Wyatt K., Caprariello A. V., Factor D. C., Karl R. T., Maeda T., Miller R. H., Tesar P. J. (2013) Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 31, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang N., Zuchero J. B., Ahlenius H., Marro S., Ng Y. H., Vierbuchen T., Hawkins J. S., Geissler R., Barres B. A., Wernig M. (2013) Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 31, 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]